Abstract
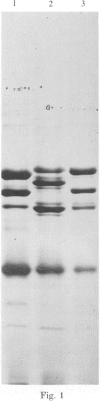
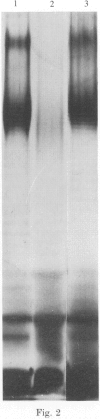
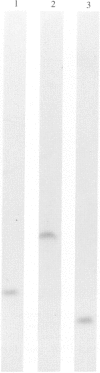
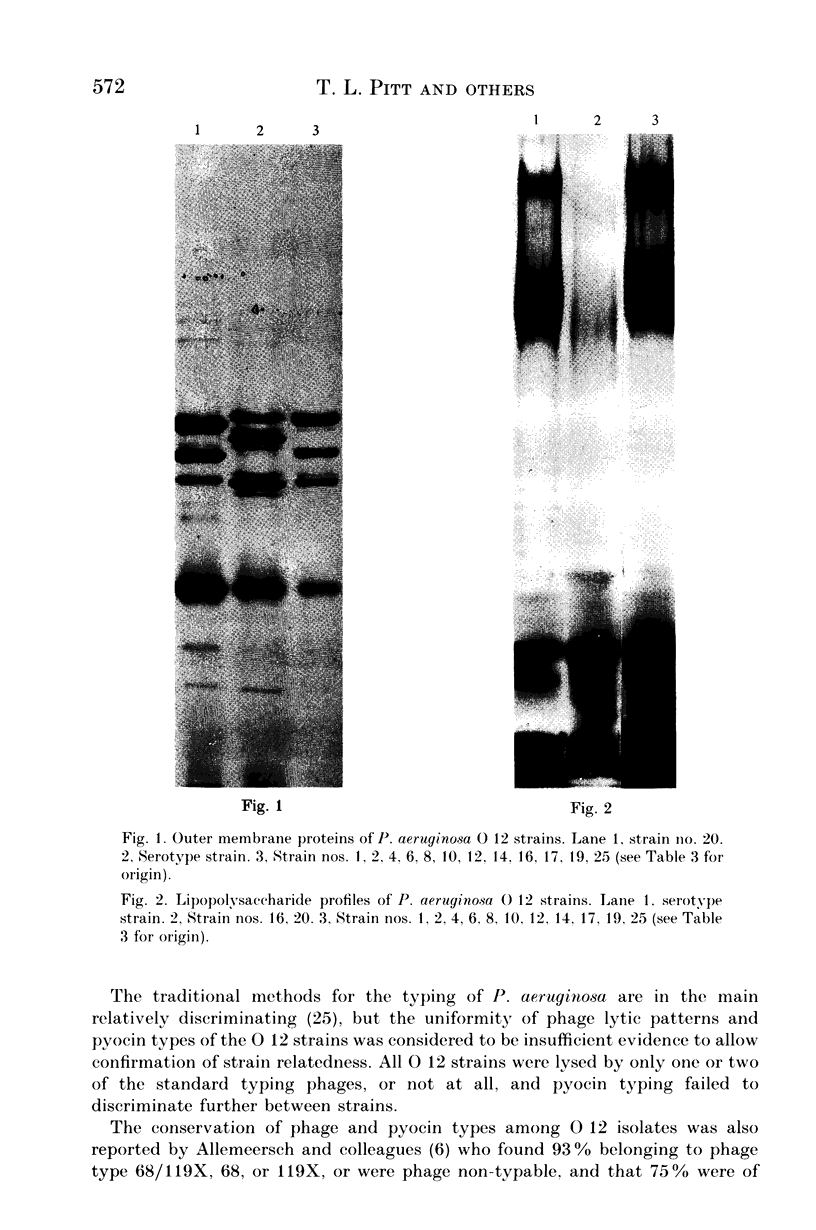
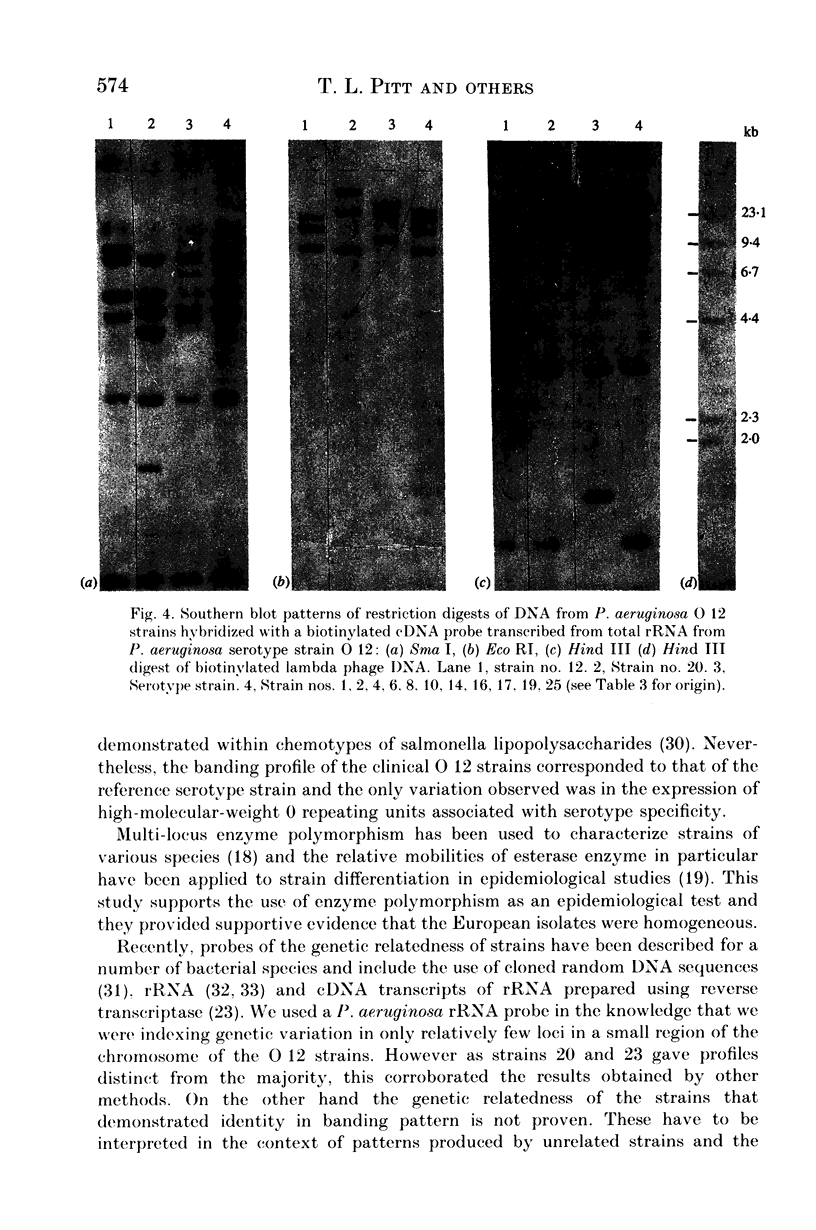
A survey was made of serotype association and multiple antibiotic resistance in strains of Pseudomonas aeruginosa in Europe. Of 208 epidemiologically distinct strains from 16 laboratories in 10 countries, 48 were resistant to carbenicillin (MIC greater than 128 micrograms/ml) and gentamicin (MIC greater than 4 micrograms/ml), and 12 of these strains were of serotype O 12. Representative O 12 strains from different countries were compared with two multiresistant O 12 strains isolated 5 years apart, from a British burns unit and the antibiotic sensitive serotype reference strain. All O 12 strains were similar by phage and pyocin typing but lysogenic phage profiles indicated that two strains (the later burns isolate and the serotype strain) were distinct from the others. Electrophoretic characterization of outer membrane proteins, lipopolysaccharides and esterase enzymes corroborated the relationship of the strains and restriction fragment length polymorphism of DNA fragments hybridized with a cDNA probe copy of rRNA from P. aeruginosa provided further proof of their relatedness. We propose that the uniformity of characters of multiresistant O 12 P. aeruginosa in Europe is suggestive of a common origin for the strains.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allemeersch D., Beumer J., Devleeschouwer M., De Maeyer S., Dony J., Godard C., Osterrieth P., Pithsy A., Van der Auwera P., Van Poppel H. Marked increase of Pseudomonas aeruginosa serotype 012 in Belgium since 1982. Eur J Clin Microbiol Infect Dis. 1988 Apr;7(2):265–269. doi: 10.1007/BF01963099. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Del Piano M., La Palombara P., Picci A., Nicosia R. Serological and pyocin typing and antibiotic sensitivity of Pseudomonas aeruginosa strains. Microbiologica. 1986 Apr;9(2):253–258. [PubMed] [Google Scholar]
- Fyfe J. A., Harris G., Govan J. R. Revised pyocin typing method for Pseudomonas aeruginosa. J Clin Microbiol. 1984 Jul;20(1):47–50. doi: 10.1128/jcm.20.1.47-50.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giammanco A., Di Stefano R., Arista S., Sinatra A., Chiarini A. Infections caused by Pseudomonas aeruginosa: relatively frequent isolation of serogroup 12 from clinical specimens. Eur J Epidemiol. 1985 Jun;1(2):104–109. doi: 10.1007/BF00141801. [DOI] [PubMed] [Google Scholar]
- Grimont F., Grimont P. A. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986 Sep-Oct;137B(2):165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- HABS I. Untersuchungen über die O-Antigene von Pseudomonas aeruginosa. Z Hyg Infektionskr. 1957;144(3):218–228. [PubMed] [Google Scholar]
- Hancock R. E., Carey A. M. Outer membrane of Pseudomonas aeruginosa: heat- 2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979 Dec;140(3):902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983 Apr;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H., Hagen L., Sutton R. A. Determination of urinary oxalate by high-performance liquid chromatography. Anal Biochem. 1982 Jan 1;119(1):1–3. doi: 10.1016/0003-2697(82)90656-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Legakis N. J., Koukoubanis N., Malliara K., Michalitsianos D., Papavassiliou J. Importance of carbenicillin and gentamicin cross-resistant serotype 0:12 Pseudomonas aeruginosa in six Athens hospitals. Eur J Clin Microbiol. 1987 Jun;6(3):300–303. doi: 10.1007/BF02017618. [DOI] [PubMed] [Google Scholar]
- Livermore D. M., Pitt T. L. Dissociation of surface properties and "intrinsic" resistance to beta lactams in Pseudomonas aeruginosa. J Med Microbiol. 1986 Nov;22(3):217–224. doi: 10.1099/00222615-22-3-217. [DOI] [PubMed] [Google Scholar]
- Orskov F., Orskov I. From the national institutes of health. Summary of a workshop on the clone concept in the epidemiology, taxonomy, and evolution of the enterobacteriaceae and other bacteria. J Infect Dis. 1983 Aug;148(2):346–357. doi: 10.1093/infdis/148.2.346. [DOI] [PubMed] [Google Scholar]
- Perinpanayagam R. M., Grundy H. C. Outbreak of gentamicin-resistant Pseudomonas aeruginosa infection in a burns unit. J Hosp Infect. 1983 Mar;4(1):71–73. doi: 10.1016/0195-6701(83)90068-3. [DOI] [PubMed] [Google Scholar]
- Picard B., Goullet P. Comparative electrophoretic profiles of esterases, and of glutamate, lactate and malate dehydrogenases, from Aeromonas hydrophila, A. caviae and A. sobria. J Gen Microbiol. 1985 Dec;131(12):3385–3391. doi: 10.1099/00221287-131-12-3385. [DOI] [PubMed] [Google Scholar]
- Pitt T. L. Epidemiological typing of Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 1988 Apr;7(2):238–247. doi: 10.1007/BF01963095. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shionoya H., Goto S., Tsukamoto M., Homma J. Y. Relationship between pyocine and temperate phage of Pseudomonas aeruginosa. I. Isolation of temperate phages from strain P 1-III and their characteristics. Jpn J Exp Med. 1967 Oct;37(5):359–371. [PubMed] [Google Scholar]
- Stull T. L., LiPuma J. J., Edlind T. D. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988 Feb;157(2):280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- Tompkins L. S., Troup N., Labigne-Roussel A., Cohen M. L. Cloned, random chromosomal sequences as probes to identify Salmonella species. J Infect Dis. 1986 Jul;154(1):156–162. doi: 10.1093/infdis/154.1.156. [DOI] [PubMed] [Google Scholar]
- Vieu J. F., Allos G., Hassan-Massoud B., Santos-Ferreira M. O., Tselentis G. Existe-t-il une épidémiologie géographique des sérogroupes O de Pseudomonas aeruginosa? Bull Soc Pathol Exot Filiales. 1984 May-Jun;77(3):288–294. [PubMed] [Google Scholar]
- Williams R. J., Lindridge M. A., Said A. A., Livermore D. M., Williams J. D. National survey of antibiotic resistance in Pseudomonas aeruginosa. J Antimicrob Chemother. 1984 Jul;14(1):9–16. doi: 10.1093/jac/14.1.9. [DOI] [PubMed] [Google Scholar]
- de Saxe M. J., Notley C. M. Experiences with the typing of coagulase-negative staphylococci and micrococci. Zentralbl Bakteriol Orig A. 1978 Jul;241(1):46–59. [PubMed] [Google Scholar]