Abstract
In comparisons of the pathogenicity of simian alphaherpesviruses in mice, two isolates of the baboon virus HVP2 were nearly as lethal as monkey B virus, a biological safety level 4 agent (J. W. Ritchey, K. A. Ealey, M. Payton, and R. Eberle, J. Comp. Pathol. 127:150-161, 2002). To confirm these results, mice were inoculated intramuscularly with 105 PFU of HVP2 isolates obtained from different baboon subspecies and primate centers. Some of the HVP2 isolates (6 of 13) caused paralysis and death in the mice, while 7 of 13 HVP2 isolates produced no clinical signs of disease. The apathogenic HVP2 isolates (HVP2ap) induced only low levels of serum antiviral immunoglobulin G relative to levels observed in sera from mice infected with the neurovirulent isolates of HVP2 (HVP2nv). Histological examination of tissues from mice inoculated with HVP2nv isolates showed extensive neural tissue destruction, while mice infected with HVP2ap isolates showed no lesions. Tissue samples collected at 48-h intervals postinfection suggested that HVP2ap isolates failed to replicate at the site of inoculation. There was no significant difference in the in vitro replication, plaque size, or cytopathic effect morphology of HVP2ap versus HVP2nv isolates. While HVP2 isolates replicated better in Vero monkey kidney cells than in murine L cells, plaquing efficiency of individual isolates did not correlate with the dichotomous pathogenic properties seen in mice. Phylogenetic analyses of both coding and intergenic regions (US4-6) of the HVP2 genome separated isolates into two distinct clades that correlated with the two in vivo virulence phenotypes. Taken together, these results demonstrate that two subtypes of HVP2 exist that are very closely related but differ dramatically in their ability to cause disease in a murine model.
A number of alphaherpesviruses (α-herpesviruses) related to the human herpes simplex viruses (HSV) have been isolated from various species of monkeys. These include Cercopithecine herpesvirus 1 (monkey B virus [BV]) of macaques, Cercopithecine herpesvirus 2 (SA8) of African green monkeys, Cercopithecine herpesvirus 16 (Herpesvirus papio 2 [HVP2]) of baboons, and Saimirine herpesvirus 1 (Herpesvirus saimiri 1) of squirrel monkeys. In their own natural host species, these viruses are biologically quite similar to HSV in humans. Primary infections involve mucosal surfaces, usually occurring as oral infections in sexually immature animals and as genital infections in adults (6, 13, 18, 26, 28). These viruses all establish latent infections in sensory ganglia. Latent virus can subsequently be reactivated, with infectious virus being shed in body secretions either asymptomatically or in association with recurrent lesions. With the exception of sporadic generalized infections in young animals, these viruses rarely cause serious clinical disease in their natural host species.
BV is well known for its extreme neuropathogenicity when transmitted to species other than its natural host (4, 23, 26, 28). In particular, humans bitten or scratched by macaques are prone to develop neurological infections that are approximately 80% fatal if left untreated (4, 21, 26). This has led to BV being rated as a biological safety level 4 pathogen. Herpesvirus saimiri 1 is similarly lethal when transmitted to owl monkeys or marmosets (9). Following its initial isolation, SA8 was reported to be very neurovirulent (16, 17), but subsequent studies have shown it to be increasingly less pathogenic with time (9, 11, 22). For many years the baboon virus HVP2 was assumed to be identical to SA8 (9, 11, 12, 17). However, more recent studies show that, while very closely related to SA8, baboon isolates represent a distinct virus (1, 7). Similar to the case for BV in macaques, more than 90% of adult baboons show serological evidence of infection with HVP2, and clinically evident disease due to HVP2 is uncommon (6, 27). When clinically apparent disease does occur, typical herpetic lesions are present in the mouth and/or genital region (6, 15, 18).
In contrast to BV, there have been no reports of any animal caretakers or research personnel bitten or scratched by baboons or vervets developing any signs of infection. Consequently, little attention has been paid to the simian herpesviruses, and with the exception of BV all simian α-herpesviruses are classified as biological safety level 2 agents. Because of the similarity between HVP2 and the apathogenic SA8 and the lack of any reported human infections, baboon HVP2 has largely been considered unimportant from a zoonotic standpoint. However, during the production of immune-phase sera in mice, rapid-onset death was observed in mice inoculated intraperitoneally with one isolate of HVP2 (7). In a recent study comparing SA8 and HVP2 to HSV1 in a mouse model, two strains of HVP2 were found to readily invade the central nervous system (CNS) following peripheral intramuscular inoculation (which most closely resembles a bite or scratch), producing paralysis and death (22). Here we describe further in vitro and in vivo testing of HVP2 isolates and report the existence of two distinct subtypes of HVP2 that differ markedly in their pathogenic properties in a mouse model.
MATERIALS AND METHODS
Viruses and cells.
Vero and murine L929 cells were originally obtained from the American Type Culture Collection (Rockville, Md.). DBG3 cells were used to identify infected cells by green fluorescent protein expression (2). All cells were cultured in complete Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and maintained in Dulbecco's modified Eagle's medium containing 2% fetal bovine serum.
The origins of a number of HVP2 isolates (5, 6, 7) as well as the BV strain E2490 (23) and SA8 strain B264 (8) have been previously described. HVP2 isolate A189164 was kindly provided by R. Heberling of VRL, San Antonio, Tex. Additional HVP2 isolates were obtained from oral and genital swabs taken during semiannual tuberculosis testing of baboons housed at Oklahoma University Health Sciences Center. Swab samples were processed and inoculated onto Vero or DBG3 cells as described previously (2, 6). All suspected HVP2 isolates were confirmed to be HVP2 by PCR and sequencing as described previously (1, 2). Origins and properties of HVP2 strains used in this study are summarized in Table 1.
TABLE 1.
Origins of HVP2 isolates
| HVP2 isolate | Colony of origin | Baboon species | Date of isolation |
|---|---|---|---|
| A189164 | WNPRCa | Unknown | Unknown |
| A951 | SNPRCb | Unknown | 1978 |
| OU1-76 | OUHSCc | Olive | Nov 1994 |
| OU2-5 | OUHSC | Olive | Dec 1995 |
| OU2-9 | OUHSC | Olive | Dec 1995 |
| OU2-12 | OUHSC | Olive | Dec 1995 |
| OU2-13 | OUHSC | Olive | Dec 1995 |
| OU3-1 | OUHSC | Yellow | June 1996 |
| OU3-18 | OUHSC | Yellow | June 1996 |
| OU3-40 | OUHSC | Yellow | June 1996 |
| OU4-2 | OUHSC | Chacma | Oct 1996 |
| OU4-5 | OUHSC | Olive | Oct 1996 |
| OU4-8 | OUHSC | Chacma | Oct 1996 |
| OU4-15 | OUHSC | Olive | Oct 1996 |
| OU5-35 | OUHSC | Olive | May 2002 |
| OU5-47 | OUHSC | Yellow | May 2002 |
| OU5-51 | OUHSC | Yellow | May 2002 |
| X313 | SNPRC | Olive | 1985-86 |
| 960 | SNPRC | Olive | 1985 |
| 1258 | SNPRC | Olive | 1985 |
| 1401 | SNPRC | Olive | 1985 |
WNPRC, Washington National Primate Research Center.
SNPRC, Southwest National Primate Research Center.
OUHSC, Oklahoma University Health Sciences Center.
Plaque assays were used to titrate virus. Cells were grown to near-confluency in 6- or 12-well trays. Serial 10-fold viral dilutions were made in cold maintenance medium. Medium was removed, and duplicate cell monolayers inoculated with 100 or 200 μl (12-well or 6-well, respectively) of diluted virus stock. After 1 h of incubation at 37°C, maintenance medium containing 1% methylcellulose was added. Plaques were counted at 2 (Vero and DBG3 cells) or 6 (L cells) days postinfection (p.i.).
Mouse inoculations.
Female 12- to 14-g BALB/c mice were obtained from Charles River Laboratories (Wilmington, Mass.). Mice were inoculated intramuscularly in the left hind leg as previously described (3, 22). All mice were observed twice daily for clinical signs of infection.
For HVP2ap 50% lethal dose (LD50), 50% infective dose (ID50) determinations, and 50% CNS disease values, groups of eight mice were inoculated with 104 to 107 PFU of the HVP2ap isolates OU2-5 and A951. For temporal pathology experiments, groups of nine mice were inoculated with 105 PFU of the neurovirulent isolate A189164 or the apathogenic isolate OU2-5. Two mice from each group were sacrificed at 1, 3, 5, and 7 days p.i., and one mouse was sacrificed at 9 days p.i. Four mice were inoculated with 25 μl of phosphate-buffered saline to serve as uninfected controls, one mouse being sacrificed at each time point.
Immunoassay.
Blood was collected from mice by cardiac puncture at the time of euthanasia, and the serum was stored at −80°C until tested by enzyme-linked immunosorbent assay (ELISA) to detect anti-HVP2 IgG. ELISA procedures for detection of anti-HVP2 IgG were basically as described previously (20) except that peroxidase-conjugated anti-mouse immunoglobulin G (IgG) (Vector Laboratories, Burlingame, Calif.) was used as a secondary antibody at a dilution of 1:5,000. All mouse sera were tested at a 1:100 dilution. The presence of viral antigen in tissue sections was detected by immunohistochemical staining performed on an automated stainer as previously described (3, 22)
Pathology.
Following euthanasia, mice were submerged in formalin for 18 to 24 h before being dissected for histological examination (3). Tissue samples from visceral organs, skin, skeletal muscle (site of inoculation), sciatic nerves, and brain were collected, embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin (H&E). Vertebral columns with spinal cords in situ were fixed in 10% buffered formalin and decalcified in formic acid for 1 to 3 days before being trimmed into anatomical regions (cervical, thoracic, and lumbar), sectioned at 5 μm, and stained with H&E as described previously (3).
Viral replication kinetics.
Vero cell monolayers in 12-well plates were infected with plaque-purified viral stock from three HVP2ap isolates and three HVP2nv isolates at a multiplicity of infection of 5 PFU/cell, and infectious virus was quantitated by plaque assay as described previously (8). Statistical analyses were done using an autoregressive period 1 covariance structure in the repeated measures analysis. PROC MIXED in PC SAS version 8.2 (SAS Institute, Cary, N.C.) was used for all analyses.
Plaque assays were performed on DBG3 cell monolayers with three viral isolates of each HVP2 pathogenic subtype to assess plaque size and morphology. Plaque-purified viral stock was used to infect cells to produce approximately 50 to 100 plaques in duplicate wells of a six-well plate. After 48 h at 37°C, the area of green fluorescent protein-expressing cells was calculated for 10 randomly chosen plaques for each viral isolate using MetaView software version 4.1.7 (Universal Imaging Corporation, Downington, Pa.). Statistical analyses of plaque area measurements were done using the t test function in Microsoft Excel.
DNA sequence analysis.
Restriction fragment length polymorphism (RFLP) analysis was performed as previously described using gradient-purified viral DNA (23). Gradient-purified viral DNA was also used as the PCR template for some strains of HVP2. Infected-cell DNA templates for PCR were prepared for new clinical isolates of HVP2 by inoculating Vero cells grown in a 24-well tray. Infected cells were rinsed once with phosphate-buffered saline, and total infected cell DNA was isolated with DNAzol (Molecular Research Center, Cincinnati, Ohio) following the manufacturer's protocol. PCR was used to amplify a phylogenetically informative 1.1-kbp area from the unique short region of the HVP2 genome (23, 25) using rTth polymerase (Applied Biosystems, Foster City, Calif.) with the primer set KT1 (5′TCCCGAGTTCGGTACACGCGACTG3′) and KT10 (5′CACGTCGGGGGGGTCCGTCTTCTGCTCC3′). After a 3-min denaturation at 94°C, amplification was carried out by 35 cycles of 96°C for 30 s, 68°C for 30 s, 72°C for 2 min followed by 7 min at 72°C. Several internal primers were used to sequence the region in both forward and reverse directions. PCR products were purified prior to sequencing using Wizard PCR Preps (Promega, Madison, Wis.). Vector NTI suite 7.0 (InforMax, Inc., Frederick, Md.) was used to assemble sequence files, and the MEGA version 2.1 program package (14) was used for phylogenetic analyses as described previously (19, 23).
RESULTS
Comparison of HVP2 strains in vivo.
Table 2 summarizes the clinical data for mice inoculated with 105 PFU of virus and describes the pathogenic phenotype for each of the HVP2 isolates tested in vivo. In vivo testing of the two HVP2nv isolates OU1-76 and X313 initially indicated that this virus was very neurovirulent in mice following intramuscular inoculation, with 50% CNS disease values of 103 and 102.5 PFU and LD50 values of 104.3 and 103.8 PFU, respectively (22). To confirm these results with additional HVP2 isolates, groups of five mice were inoculated with 103 to 105 PFU of six isolates of HVP2 obtained from geographically distinct breeding colonies and from different subspecies of baboons. Surprisingly, mice inoculated with four of six of these additional viruses showed no clinical signs of disease, while two of six isolates were extremely neurovirulent, similar to previous observations with HVP2 isolates OU1-76 and X313 (22). An additional seven HVP2 isolates were screened in vivo by inoculating mice in groups of five with 105 or 106 PFU. Again, several isolates (three of seven) produced no clinical signs of infection, while four of seven isolates invaded the CNS and produced severe disease. For strains that did produce clinical signs of infection, a dose of 106 PFU was uniformly lethal.
TABLE 2.
Pathogenic properties of HVP2 isolates in mice
| HVP2 isolate | No. with CNS signsa | No. of deathsb | ELISA results
|
|
|---|---|---|---|---|
| OD valuesc | No. positived | |||
| HVP2ap | ||||
| A951 | 0/5 | 0/5 | 0.254-0.459 (0.379) | 4/5 |
| OU2-5 | 0/5 | 0/5 | 0.153-0.348 (0.244) | 3/5 |
| OU2-9 | 0/5 | 0/5 | 0.122-0.404 (0.208) | 5/5 |
| OU2-12 | 0/5 | 0/5 | 0.198-0.390 (0.315) | 3/5 |
| OU4-2 | 0/5 | 0/5 | 0.196-0.334 (0.265) | 2/5 |
| OU4-8 | 0/5 | 0/5 | 0.141-0.213 (0.179) | 3/5 |
| OU4-15 | 0/5 | 0/5 | 0.105-0.500 (0.284) | 5/5 |
| HVP2nv | ||||
| A189164 | 5/5 | 1/5 | 0.999-1.407 (1.219) | 4/4 |
| OU1-76 | 5/5 | 2/5 | 1.035-1.924 (1.690) | 6/6 |
| OU3-1 | 5/5 | 5/5 | 0.731-1.122 (0.985) | 3/3 |
| OU3-18 | 5/5 | 4/5 | 0.926 | 1/1 |
| X313 | 5/5 | 5/5 | 1.135-2.078 (1.593) | 4/4 |
| 960 | 5/5 | 5/5 | NSA | |
Number of mice in groups of five inoculated with 105 PFU of virus that showed clinical signs of CNS infection, including hind limb paresis/paralysis, unsteady gait, and/or immobility.
Number of mice in each group that either died or were humanely euthanatized before 18 days p.i.
The range of OD values for positive sera for anti-HVP2 IgG in serum from mice collected at 10 to 21 days p.i. The cutoff value for positive samples was 0.100. Numbers in parentheses are the mean ODs of positive sera. Sera were not tested for mice that died at <10 days p.i. NSA, no sera available for testing.
Number of positive sera/total number of sera tested.
Groups of eight mice were inoculated with 104 to 107 PFU of apathogenic isolates OU2-5 and A951 to determine both an ID50 and an LD50. Even at the very highest dose (107 PFU), these isolates produced no clinical signs of infection. Other mice were inoculated at the same time with 105 or 106 PFU of HVP2 isolates A189164 and OU1-76, and these mice developed progressive clinical symptoms along a time course identical to that previously described (22). Briefly, mice showed edema and hyperemia of the left hind foot along with paresis of the same limb by 4 days p.i. By 6 or 7 days p.i., ulcerative lesions appeared on the left hind foot/leg, and mice showed signs of severe CNS disease, including unilateral (left) or bilateral hind limb paresis or paralysis, severe lumbar and hind limb muscle atrophy, body tremors, and depression. Most of these mice were euthanatized between days 7 and 12 p.i. due to the severity of the clinical symptoms. HVP2 isolates that did not produce any clinical signs of infection were designated HVP2ap (apathogenic), while isolates that readily invaded the CNS were designated HVP2nv (neurovirulent).
Measurement of anti-HVP2 IgG by ELISA.
Clinical observations suggested that some strains of HVP2 either are severely retarded in their ability to produce visible lesions or are unable to replicate at all following intramuscular inoculation in mice. To distinguish between these two possibilities, the induction of a humoral immune response was used as an indirect indicator of the ability of different HVP2 isolates to replicate in vivo (i.e., the ability of the virus to increase the quantity of viral antigen available to induce an immune response over the actual inoculation dose). Sera were collected from individual mice either at the time of euthanasia (7 to 12 days p.i.; mean, 10 days p.i. for HVP2nv) or at termination of the experiment (19 to 21 days p.i. for HVP2ap). Of mice infected with 105 PFU of HVP2nv that survived at least 10 days p.i., 100% were seropositive, and the optical densities (ODs) ranged from 0.731 to >2.000 (Table 2). In contrast, only 71% of mice infected with 105 PFU of HVP2ap seroconverted, and those that did had ODs ranging from <0.105 to a maximum of 0.500 (the positive/negative cutoff value was 0.100). As shown in Table 2, the group average ODs were substantially different. Mice inoculated with doses as low as 102 PFU of HVP2nv isolates seroconverted (22), while some mice inoculated with doses as high as 106 PFU of HVP2ap isolates did not seroconvert, and even mice at the highest dose of HVP2ap (107 PFU) had relatively low group ODs (0.415 and 0.440).
Histopathology of neurovirulent versus apathogenic HVP2 strains.
The clinical observations and serological results suggested that HVP2ap strains did not replicate sufficiently in the mouse to induce a strong humoral immune response. To determine how early in the pathogenic process HVP2nv and HVP2ap strains diverged, groups of mice were inoculated with 105 PFU of either HVP2nv isolate A189164 or HVP2ap isolate OU2-5, and animals were sacrificed every 2 days up to 9 days p.i. Tissues from mice were examined for microscopic signs of infection identified by either development of histological lesions or presence of viral antigen as detected by immunohistochemistry. For the temporal studies, histological examination was focused upon tissues targeted by afferent and efferent viral pathways (e.g., sciatic nerve, spinal cord, and skin). A comprehensive description of the location of virus and virus-induced lesions in all tissues (CNS, peripheral nervous system, viscera) has previously been reported for HVP2nv isolates (22).
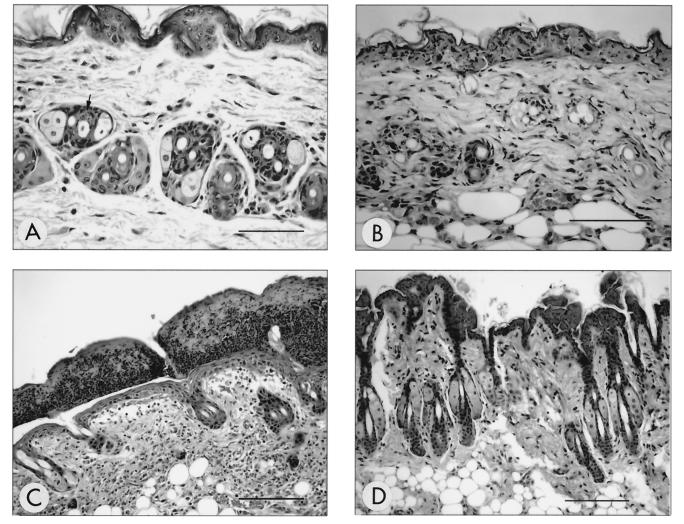
In mice inoculated with the HVP2nv strain, cutaneous lesions were evident as early as 1 day p.i. (Fig. 1A). These lesions were characterized by scattered, subtle necrosis of the epithelial cells of the hair follicles. By 5 days p.i., epithelial cell necrosis had become more conspicuous and widespread and extended into the epidermis (Fig. 1B). Erosions and ulcers were also present, with edema and inflammation within the dermis. By 9 days p.i., the cutaneous lesions were dominated by the presence of thick serocellular crusts overlying cutaneous ulcers with a mixed neutrophilic and mononuclear inflammatory infiltrate in the dermis (Fig. 1C).
FIG. 1.
Temporal development of histological lesions in haired skin. Temporal development of skin lesions in mice inoculated with HVP2nv strains was characterized by subtle necrosis of hair follicle epithelium observed as early as 1 day p.i. (A) (arrow) followed by more conspicuous necrosis involving widespread adnexal structures by 5 days p.i. (B). By 9 days p.i. (C), the epidermis was overlaid by a thick serocellular crust and the dermis contained a primarily mononuclear inflammatory infiltrate (dermatitis) with evidence of follicular destruction and effacement. Cutaneous lesions were not observed at any time point following inoculation with HVP2ap (D) (9 days p.i.). All sections were H&E stained. Bar for panels A and B = 150 μm; bar for panels C and D = 40 μm.
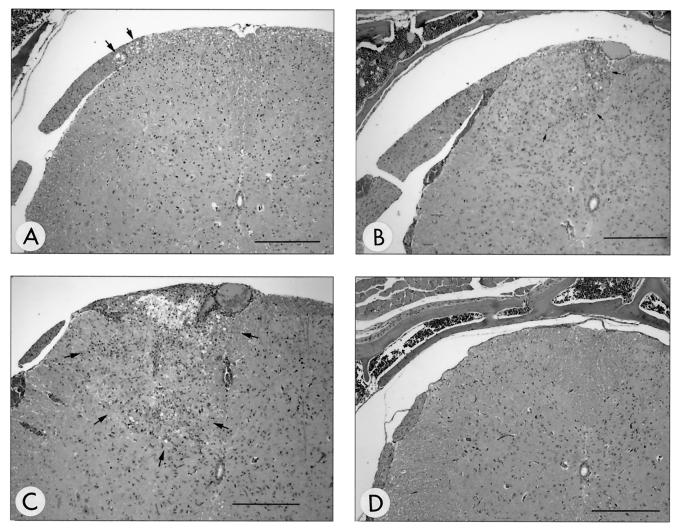
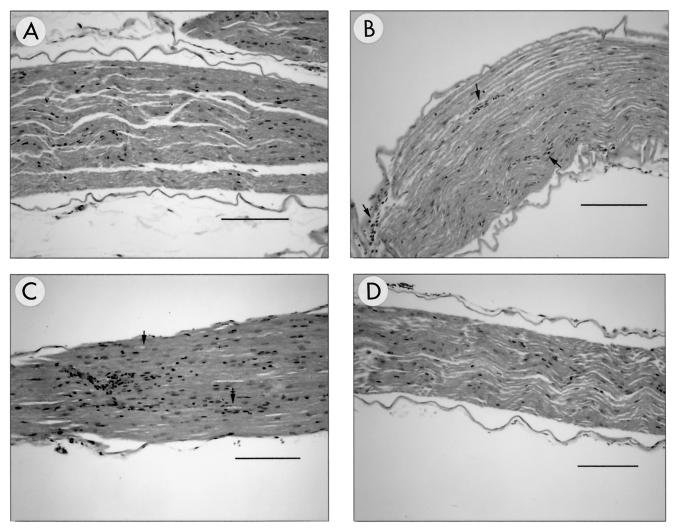
Within the nervous system, the first microscopic evidence of infection was seen at 3 days p.i. (Fig. 2A) and characterized by equivocal vacuolation of the dorsal roots present in some of the lumbar spinal cord sections. Unequivocal lesions of inflammation within the dorsal root and ipsilateral dorsal funiculus and horn of the lumbar spinal cord were present by 5 days p.i. (Fig. 2B). Mild infiltrates of primarily mononuclear inflammatory cells were also identified in left sciatic nerves at 5 days p.i. (Fig. 3B). From 5 to 9 days p.i., inflammation within the dorsal horn of the spinal cord became more severe and effacing, extending into ventral and contralateral spinal cord regions as well as progressing cranially to involve the thoracic spinal cord (Fig. 2C). Inflammation within the spinal cord was always characterized by the presence of both neutrophils and mononuclear cells. There was no evidence that one type appeared first or dominated at any point in the infection. At 7 days p.i., mild inflammation appeared at the level of the brain stem and was characterized by subtle perivascular and meningeal infiltrates. Axonal degeneration of the left sciatic nerve was observed at 7 to 9 days p.i. (Fig. 3C). Immunohistochemical staining confirmed the presence of viral antigen in lesions observed in H&E-stained tissue sections.
FIG. 2.
Temporal development of histological lesions in the lumbar spinal cord. At 3 days p.i. with HVP2nv, there was equivocal vacuolation of dorsal root structures (A) (arrows); however, inflammation was not present. By 5 days p.i., unequivocal inflammation composed of neutrophils and mononuclear inflammatory cells (lymphocytes and macrophages) appeared in the ipsilateral dorsal funiculus and horn (outlined by arrows in panel B). The inflammation increased in intensity and was accompanied by rarefaction at 9 days p.i. (outlined by arrows in panel C). No lesions were observed at any time point following inoculation with HVP2ap (D) (9 days p.i.). All sections were H&E stained. Bar for panels A and D = 235 μm; bar for panels B and C = 280 μm.
FIG. 3.
Temporal development of histological lesions in the ipsilateral sciatic nerve. Lesions were not observed in sections of sciatic nerve obtained at 1 to 3 days p.i. from HVP2nv-infected mice (A) (3 days p.i.). The first lesions appeared at 5 days p.i. and were characterized by mild mononuclear inflammatory infiltrates usually confined to perivascular locations (B) (arrows). By 9 days p.i., the inflammatory infiltrates were more conspicuous and accompanied by evidence of axonal degeneration (C) (arrows). Lesions were not seen at any time point following inoculation with HVP2ap (D) (9 days p.i.). All sections were H&E stained. Bar for panels A, B, and D = 120 μm; bar for panel C = 60 μm.
In contrast to these results, no histologic lesions indicative of viral infection were detected in mice infected with the HVP2ap strain at any time or in any tissues (Fig. 1 to 3, panels D). Consistent with these results, no evidence of viral antigen was detected by immunohistochemical staining of any tissue sections from HVP2ap-infected mice.
Replication kinetics of HVP2 subtypes.
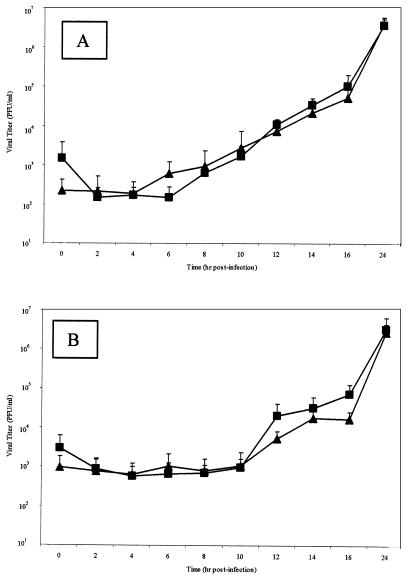
In vitro replication kinetics were examined for four HVPap isolates and four HVPnv isolates. Confluent Vero cells were infected, and both intracellular viral production and progeny virus released from the cell were measured over time. Intracellular progeny virus was first detected at significant levels between 6 to 8 h p.i. for all HVP2 isolates (Fig. 4A). As expected, release of infectious progeny virus into the extracellular fluid lagged behind the appearance of intracellular progeny virus by approximately 4 h, beginning at 10 to 12 h p.i. (Fig. 4B). Statistical analyses revealed no significant differences in the replication kinetics of HVP2ap versus HVP2nv isolates.
FIG. 4.
HVP2 in vitro growth kinetics. Vero cell monolayers were infected with 5 PFU of various HVP2 isolates representing the two HVP2 pathogenic subtypes/cell and incubated at 37°C for 48 h. Data points represent mean PFU values of four isolates from HVP2nv (▴) and HVP2ap (▪) at every time point for both intracellular virus (A) and virus present in the extracellular fluids (B). There was no statistical difference between the replication kinetics of HVP2ap and HVP2nv.
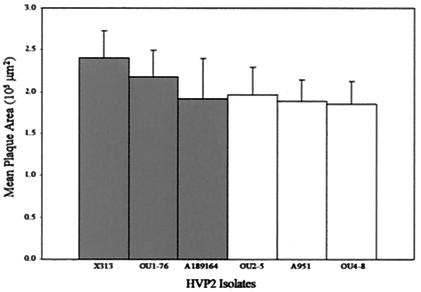
To compare plaque size and morphology of HVP2 isolates, DBG3 cells were infected with three HVP2ap and three HVP2nv isolates. At 48 h p.i., the area of 10 randomly chosen plaques was calculated for each of the six viruses (Fig. 5). There was no statistical difference between the mean plaque area of HVP2nv and HVP2ap (P value, <0.01). Similarly, no variation in cytopathic effect plaque morphology was apparent among different HVP2 isolates (data not shown).
FIG. 5.
Measurement of HVP2 plaque formation. Mean plaque area (n = 10) of three HVP2nv (▪) isolates and three HVP2ap (▪) isolates in DBG3 cells at 48 h p.i. Statistical analysis of these data showed no significant difference in plaque size between HVP2nv and HVP2ap.
The ability of HVP2 strains to replicate in murine versus primate cells in vitro was examined. Titers of six HVP2 isolates (three ap isolates and three nv isolates) were determined on both Vero and murine L cells. All six strains grew more slowly and produced smaller plaques in L cells (1- to 2-mm plaques at 6 days p.i.) than in Vero cells (3- to 4-mm plaques at 2 days p.i.). As shown in Table 3, there was considerable variation in the relative plaquing efficiency of different HVP2 isolates in the two cell lines. However, the relative plaquing efficiency did not correlate with either the in vivo phenotype of the virus or the number of times a virus had been passed in vitro.
TABLE 3.
Comparative replication efficiency of HVP2 strains on simian (Vero) and murine (L929) cells
| HVP2 isolate | Phenotype | Titer of virus stock
|
Ratio (Vero:L929) | |
|---|---|---|---|---|
| Vero | L929 | |||
| OU1-76 | nv | 2.0 × 108 | 1.9 × 107 | 10.5 |
| X313 | nv | 3.0 × 107 | 6.8 × 105 | 46 |
| OU3-1 | nv | 3.0 × 108 | 1.1 × 107 | 27 |
| A951 | ap | 1.6 × 107 | 1.2 × 105 | 33 |
| OU2-5 | ap | 2.2 × 107 | 3.5 × 105 | 63 |
| OU4-2 | ap | 5.4 × 107 | 1.9 × 106 | 2.8 |
Molecular differentiation of neurovirulent and apathogenic HVP2 strains.
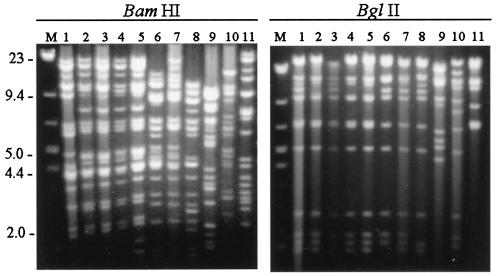
To determine if HVP2nv and HVP2ap strains could be differentiated by molecular analysis, RFLP patterns generated by restriction enzyme digestion of gradient-purified viral DNA from nine HVP-2 isolates as well as SA8 and BV were examined. As shown in Fig. 6, all HVP2 isolates showed patterns distinct from both SA8 and BV, and all HVP2 isolates had similar patterns when digested with BglII. However, different HVP2 isolates exhibited variation in their RFLPs that were apparent for a number of different restriction enzymes. While two major groups of similar RFLPs generated with BamHI were apparent among HVP2 isolates, several had RFLPs that did not fit into either of these two groups. None of the restriction enzymes tested (BamHI, SalI, MluI, and PstI) clearly separated HVP2ap from HVP2nv isolates (data not shown).
FIG. 6.
RFLP patterns generated by restriction enzyme digests with BamHI and BglII. Viral gradient DNA was digested with BamHI or BglII, and DNA fragments were separated on a 0.6% agarose gel. All HVP2 isolates were clearly distinct from both SA8 and BV. The lanes for both digests are as follows: lane M, DNA size marker; lanes 1 to 5, HVP2nv isolates OU1-76, OU3-1, A189164, OU3-18, and X313; lanes 6 to 9, HVP2ap isolates OU2-5, A951, OU4-8, and OU4-15; lane 10, SA8; lane 11, BV.
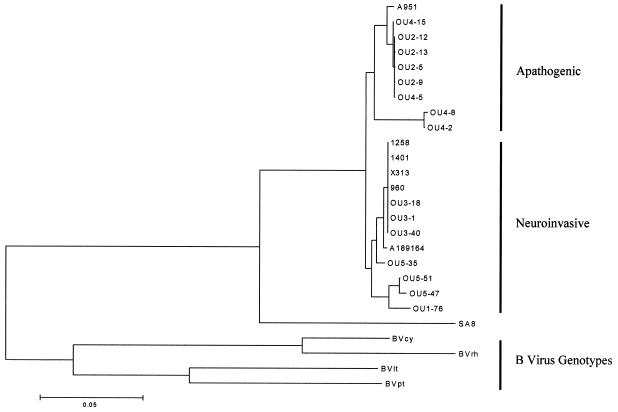
Sequence analysis of a limited area in the US region of the genome of the closely related monkey B virus previously revealed the existence of distinct genotypes of this virus (19, 23, 25). To determine if similar differences could be detected between HVP2 isolates, this same 1.1-kbp region of the HVP2 genome located between the 3′ terminus of the US4 coding sequence and the 5′ third of the US6 coding sequence was amplified from 21 HVP2 isolates by PCR and sequenced. Alignment of sequences from different BV isolates exhibited considerable sequence divergence, particularly in the US4/5 and US5/6 intergenic noncoding regions, requiring insertion of a number of gaps to optimize alignments. However, very little sequence divergence was apparent among HVP2 strains, most differences being single nucleotide substitutions or single codon deletions. Phylogenetic analysis of the aligned US sequences using several different distance algorithms resulted in trees having a consistent topology (Fig. 7). All HVP2 isolates formed a single clade distinct from both BV and SA8. However, the HVP2 clade was further subdivided into two major subgroups. Although the branch lengths separating these two subgroups are very short relative to those separating BV genotypes, the phylogenetic grouping correlated 100% with the pathogenic phenotype for the 13 HVP2 isolates tested in vivo. Isolates obtained from different baboon colonies were present in both groups, as were isolates from yellow (Papio cynocephalus) and olive (Papio anubis) baboons. Thus, HVP2ap and HVP2nv isolates were not exclusive to any single breeding colony or baboon subspecies. Although only three isolates were available from chacma (Papio ursinus) baboons, all three of these isolates grouped with the apathogenic isolates.
FIG. 7.
Phylogenetic analysis of HVP2 isolates. A 1.1-kbp region of the genome was amplified by PCR and sequenced for alignment and phylogenetic analysis (23). The tree shown was generated using Tajima-Nei distances and neighbor-joining algorithms.
DISCUSSION
The neuropathogenicity of α-herpesviruses is dependent upon a number of parameters, including the virus strain, the species used for testing, and both the route of inoculation and dose administered. For a specific virus (e.g., HSV1), different isolates or strains of the virus exhibit a range of pathogenicity, some being in a relative sense more or less pathogenic than others. Similarly, variations in virulence have been observed for several isolates of BV, with some strains being considerably more neurovirulent in mice than others (unpublished data). It was therefore surprising that with the relatively large number of distinct HVP2 isolates tested, only two distinct pathogenic phenotypes were observed: highly neurovirulent or completely apathogenic.
The typical process of α-herpesvirus infection includes local replication at the site of inoculation followed by extension into the CNS, usually via sensory afferents (24). The observed dichotomy in the pathogenic properties of HVP2ap versus HVP2nv isolates in mice may be secondary to differences in the behavior of these viruses at any number of points in this process. The complete lack of any histological lesions in the CNS of mice infected with HVP2ap strains suggests that either these viruses are unable to reach the CNS or, if they do, they are unable to replicate and produce pathological lesions. From studies on HSV, it appears that the amount of virus present at the peripheral site is directly related to the ability of the virus to enter sensory neurons and thus invade the CNS (29). The results of ELISA for detection of anti-HVP2 IgG in serum imply that HVP2ap isolates do not undergo sufficient replication at the site of inoculation to stimulate a robust immune response, suggesting that the block in HVP2ap infection occurs very early after inoculation. The lack of any visible lesions or detectable viral antigen at the site of inoculation or in any neural tissues of mice infected with HVP2ap strains supports this supposition. Thus, it is likely that insufficient progeny virus is produced at the site of inoculation to allow invasion of peripheral neurons. Moreover, given the high dose of viral inoculum tested (105 to 107 PFU), HVP2ap strains could also be defective in their ability to enter sensory or motor neurons, regardless of whether the amount of virus present locally arose from inoculation or as a result of local replication. In summary, given the excellent health of the HVP2ap-inoculated mice throughout the study, the negative to low antiviral IgG levels, and the complete lack of any detectable histological lesions or viral antigen in tissues, we suspect that HVP2ap strains most likely lack the ability to replicate at the site of inoculation, and thus, no downstream events (local inflammation, extension to CNS, generation of a strong IgG response) ever occur.
Comparison of 1.1 kbp of DNA sequence of the genome from 21 HVP2 isolates revealed that unlike BV isolates from different macaque species, genetic differences defining and separating HVP2 isolates are relatively minor. In phylogenetic analyses, branch lengths separating the two phenotypic clades of HVP2 were remarkably shorter than those separating different genotypes of BV (19, 23, 25), regardless of what distance algorithms or sequences (coding versus noncoding) were used for the analyses. While noncoding intergenic sequences of BV genotypes exhibited extensive sequence variation in aligned areas and required a number of large insertions and deletions for optimal alignment, differences in the noncoding regions between HVP2 isolates were primarily single nucleotide substitutions and small gaps (range = 1 to 10 bp; average = 2.3 bp). Similarly, there were considerable differences in the US5 (gJ glycoprotein) and US6 (gD glycoprotein) coding sequences of different BV genotypes, while differences in the gJ coding sequence of HVP2 isolates were limited to a few conservative codon changes and insertion or deletion of only three codons; no codon changes were observed in the sequenced portion of the gD open reading frame. Also unlike BV, there was no evidence for the existence of HVP2 genotypes peculiar to different baboon subspecies. Overall, the differences at the genetic level between HVP2ap and HVP2nv strains are far less than those defining BV genotypes and are much more similar to those separating individual strains of HSV1. These results indicate that although they are very closely related to each other, there appear to be two subtypes of HVP2 that differ significantly in their pathogenic phenotype in a mouse model.
The extreme polarity of the two clinical phenotypes of HVP2 isolates is somewhat surprising given the lack of major genetic or antigenic (5, 7) differences among HVP2 isolates. All HVP2 isolates tested demonstrated similar kinetic growth curves, identical cytopathic effect morphology, and plaque development rates in vitro. It is quite possible that more significant differences exist in areas of the genome of HVP2 isolates that were not examined here. The variation in RFLPs of different HVP2 isolates suggests that this may well be the case. However, it is also possible that the minor genetic differences observed between HVP2nv and HVP2ap strains (single nucleotide substitutions, small gaps) may significantly impact the clinical outcome. For instance, single nucleotide substitutions have been identified as determinant(s) of pathogenicity for HSV1 (24). Genomic sequencing and construction of recombinant viruses will be necessary to identify the specific genetic determinants of HVP2 neuroinvasiveness and virulence.
The neuroinvasiveness of HVP2nv strains is very similar to that observed for the most neurovirulent BV strains in mice (10; also unpublished observations). The ability of HVP2nv isolates to reach the CNS following peripheral intramuscular inoculation, the destruction that occurs in the CNS, and the timing of these events are all quite similar to those seen for BV (unpublished data). This raises important concerns regarding the potential for zoonotic HVP2 infections. The lack of any reports in the literature of human HVP2 infections despite the large number of animal care workers reporting baboon bites and scratches suggests that either HVP2 cannot be transmitted from baboons to humans or it is not highly pathogenic in humans, and thus, these infections go unnoticed or undifferentiated from human herpesvirus infections. Obviously, mice are a model system, and it is to be expected that there will be differences in the behavior of both the virus and host that affect the progression of a viral infection. Nonetheless, the extreme neurovirulence of HVP2nv isolates and the corresponding apathogenicity of HVP2ap isolates in mice clearly indicate that there are differences in these two subtypes of HVP2. The significance of these two subtypes relative to HVP2 infections in other species, particularly humans, remains to be determined.
Acknowledgments
We thank Denise Bream for expert technical assistance, Mark Payton for assistance with statistical analyses, and the OSU Core facility for primer synthesis and DNA sequencing.
This study was supported by PHS grants P40 RR12317 (R.E.), R01 RR07849 (R.E.), and R24 RR15289-01A2 (J.W.R.) and a Veterinary Research Scholar grant from Pfizer (K.A.E.).
REFERENCES
- 1.Black, D., and R. Eberle. 1997. Detection and differentiation of primate α-herpesviruses by PCR. J. Vet. Diagn. Investig. 9:225-231. [DOI] [PubMed] [Google Scholar]
- 2.Black, D., J. T. Saliki, and R. Eberle. 2002. Development of a green fluorescent protein reporter cell line to reduce biohazards associated with detection of infectious Cercopithecine herpesvirus 1 (monkey B virus) in clinical specimens. Comp. Med. 52:524-532. [PubMed] [Google Scholar]
- 3.Breshears, M. A., R. Eberle, and J. W. Ritchey. 2001. Characterization of gross and microscopic lesions in Balb/C mice experimentally infected with Herpesvirus saimiri 1 (HVS1). J. Comp. Pathol. 125:25-33. [DOI] [PubMed] [Google Scholar]
- 4.Davidson, W. L., and K. Hummeler. 1960. B virus infection in man. Ann. N. Y. Acad. Sci. 85:970-979. [DOI] [PubMed] [Google Scholar]
- 5.Eberle, R., D. Black, E. Blewett, and G. White. 1997. Prevalence of Herpesvirus papio 2 in baboons and identification of immunogenic viral polypeptides. Lab. Anim. Sci. 47:256-262. [PubMed] [Google Scholar]
- 6.Eberle, R., D. Black, T. Lehenbauer, and G. White. 1998. Shedding and transmission of baboon Herpesvirus papio 2 (HVP2) in a breeding colony. Lab. Anim. Sci. 48:23-28. [PubMed] [Google Scholar]
- 7.Eberle, R., D. Black, S. L. Lipper, and J. K. Hilliard. 1995. Herpesvirus papio 2, an SA8-like α-herpesvirus of baboons. Arch. Virol. 140:529-545. [DOI] [PubMed] [Google Scholar]
- 8.Eberle, R., and J. K. Hilliard. 1984. Replication of simian herpesvirus SA8 and identification of viral polypeptides synthesized in infected cells. J. Virol. 50:316-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eberle, R., and J. K. Hilliard. 1995. The simian α-herpesviruses. Infect. Agents Dis. 4:55-70. [PubMed] [Google Scholar]
- 10.Gosztonyi, G., D. Falke, and H. Ludwig. 1992. Axonal and trans-synaptic (transneuronal) spread of Herpesvirus simiae (B virus) in experimentally infected mice. Histol. Histopathol. 7:63-74. [PubMed] [Google Scholar]
- 11.Hull, R. N. 1973. The simian herpesviruses, p. 389-426. In A. S. Kaplan (ed.), The herpesviruses. Academic Press, New York, N.Y.
- 12.Kalter, S. S., S. A. Weiss, R. L. Heberling, J. E. Guajardo, and G. C. Smith III. 1978. The isolation of herpesvirus from the trigeminal ganglia of normal baboons (Papio cynocephalus). Lab. Anim. Sci. 28:705-709. [PubMed] [Google Scholar]
- 13.Keeble, S. A. 1960. B virus infection in monkeys. Ann. N. Y. Acad. Sci. 85:960-969. [DOI] [PubMed] [Google Scholar]
- 14.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Arizona State University, Tempe. [DOI] [PubMed]
- 15.Levin, J. L., J. K. Hilliard, S. L. Lipper, T. M. Butler, and W. J. Goodwin. 1988. A naturally occurring epizootic of simian agent 8 in the baboon. Lab. Anim. Sci. 38:394-397. [PubMed] [Google Scholar]
- 16.Malherbe, H., and R. Harwin. 1958. Neurotropic virus in African monkeys. Lancet ii:530.
- 17.Malherbe, H., and M. Strickland-Cholmley. 1969. Simian herpesvirus SA8 from a baboon. Lancet ii:1427. [DOI] [PubMed]
- 18.Martino, M. A., G. B. Hubbard, T. M. Butler, and J. K. Hilliard. 1988. Clinical disease associated with simian agent 8 infection in the baboon. Lab. Anim. Sci. 48:18-22. [PubMed] [Google Scholar]
- 19.Ohsawa, K., D. H. Black, R. Torii, H. Sato, and R. Eberle. 2002. Detection of a unique genotype of monkey B virus (Cercopithecine herpesvirus 1) indigenous to native Japanese macaques (Macaca fuscata). Comp. Med. 52:559-563. [PubMed] [Google Scholar]
- 20.Ohsawa, K., T. W. Lehenbauer, and R. Eberle. 1999. Herpesvirus papio 2: a safer and sensitive alternative for serodiagnosis of B virus infection in macaque monkeys. Lab. Anim. Sci. 49:605-616. [PubMed] [Google Scholar]
- 21.Palmer, A. E. 1987. B virus, Herpesvirus simiae: historical perspective. J. Med. Primatol. 16:99-130. [PubMed] [Google Scholar]
- 22.Ritchey, J. W., K. A. Ealey, M. Payton, and R. Eberle. 2002. Comparative pathology of infections with baboon and African green monkey alpha-herpesviruses in mice. J. Comp. Pathol. 127:150-161. [DOI] [PubMed] [Google Scholar]
- 23.Smith, A. L., D. Black, and R. Eberle. 1998. Molecular evidence for distinct genotypes of monkey B virus (Herpesvirus simiae) which are related to the host macaque species. J. Virol. 72:9224-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens, J. G. 1993. HSV-1 neuroinvasiveness. Intervirology 35:152-163. [DOI] [PubMed] [Google Scholar]
- 25.Thompson, S. A., J. K. Hilliard, D. Kittel, S. Lipper, W. E. Giddens, D. H. Black, and R. Eberle. 2000. Retrospective analysis of an outbreak of B virus in a colony of DeBrazza's monkeys (Cercopithecus neglectus). Comp. Med. 50:649-657. [PubMed] [Google Scholar]
- 26.Weigler, B. J. 1992. Biology of B virus in macaque and human hosts: a review. Clin. Infect. Dis. 14:555-567. [DOI] [PubMed] [Google Scholar]
- 27.Weigler, B. J., D. W. Hird, J. K. Hilliard, N. W. Lerche, J. A. Roberts, and L. M. Scott. 1993. Epidemiology of Cercopithecine herpesvirus 1 (B virus) infection and shedding in a large breeding cohort of rhesus macaques. J. Infect. Dis. 167:257-263. [DOI] [PubMed] [Google Scholar]
- 28.Whitley, R. J., and J. K. Hilliard. 2001. Cercopithecine herpesvirus (B virus), p. 2835-2848. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 29.Yamada, M., Y. Arao, F. Uro, and S. Nii. 1986. Mechanisms of difference in pathogenicity between two variants of a laboratory strain of herpes simplex virus type 1. Microbiol. Immunol. 30:1259-1270. [DOI] [PubMed] [Google Scholar]