Abstract
In mice, activation of the Notch pathway in T cells by antigen-presenting cells overexpressing Notch ligands favors differentiation of regulatory T lymphocytes responsible for antigen-specific tolerance. To determine whether this mechanism operates in human T cells, we used Epstein-Barr virus-positive lymphoblastoid cell lines (EBV-LCL) as our (viral) antigen-presenting cells and overexpressed the Notch ligand Jagged-1 (EBV-LCL J1) by adenoviral transduction. The EBV-LCL J1s were cocultured with autologous T cells, and the proliferative and cytotoxic responses to EBV antigens were measured. Transduction had no effect on EBV-LCL expression of major histocompatibility complex (MHC) antigens or of costimulatory molecules CD80, CD86, and CD40. However, we observed a 35% inhibition of proliferation and a >65% reduction in cytotoxic-T-cell activity, and interleukin 10 production was increased ninefold. These EBV-LCL J1-stimulated T lymphocytes act as antigen-specific regulatory cells, since their addition to fresh autologous T cells cultured with autologous nontransduced EBV-LCL cells significantly inhibited both proliferation and cytotoxic effector function. Within the inhibitory population, CD4+CD25+ and CD8+CD25− T cells had the greatest activity. This inhibition appears to be antigen-specific, since responses to Candida and cytomegalovirus antigens were unaffected. Hence, transgenic expression of Jagged-1 by antigen-presenting cells can induce antigen-specific regulatory T cells in humans and modify immune responses to viral antigens.
Immunoregulatory CD4+CD25+ T cells play an important role in peripheral self-tolerance in rodents (6, 18, 39, 40) and humans (5, 15, 26, 37). These well-characterized “naturally occurring” regulatory T cells (Tr) are able to transfer tolerance in vivo, which differentiates them from other mechanisms of peripheral tolerance, including T-cell anergy (35), T-cell depletion (24), and immunological ignorance (47). Their characteristics in vitro include a low proliferative capacity after allogeneic or polyclonal stimulation, a high level of CTLA4 and CD45RO expression, and an inhibition of CD4+CD25− cell proliferation in a cell-cell contact- and dose-dependent manner (5, 15, 26, 37).
Antigen-inducible Tr also play a significant role in the development of unresponsiveness. These cells have a more heterogeneous phenotype. While they may share the CD4+CD25+ phenotype of naturally occurring Tr (43), they may also be CD4+CD25− (23, 46) or CD8+CD25+/− (4, 7, 8). The molecular signals involved in the induction of these regulatory cells are incompletely identified. Transforming growth factor β (TGF-β) can induce CD4+CD25+ Tr (43), as well as CD4+CD25− (46) and CD8+ Tr (8), while the 4C8 antigen can induce CD4+CD25− Tr (23). Similarly, exposure to immature dendritic cells induces CD8+ (4) and CD4+ (14) Tr, while CD40 ligand-activated plasmocytoid dendritic cells may induce CD8+ Tr (7).
The Notch pathway is another candidate molecule involved in the development of inducible Tr. In mice, antigen presented by dendritic cells overexpressing a Notch ligand leads to the differentiation of antigen-specific CD4+ T cells into regulatory cells which can transfer tolerance to naïve animals (11). Members of the Notch family are transmembrane receptors that play a role in cell fate decisions during the development of organisms from Drosophila spp. to humans (1). The Notch gene family encodes large transmembrane proteins (9), and four Notch isoforms (Notch 1 to 4) have been isolated from mammals (29). The Notch receptor has a series of ligands, which are classified into two groups based on the prototype Serrate and Delta ligands first identified in Drosophila spp. In mammals, two Delta-like molecules (Delta 1 and Delta 3) and two Serrate-like molecules (Jagged-1 and Jagged-2) have been identified. Little is currently known about the specificity of the various Notch receptors for each ligand (1, 9). Signals generated through Notch-Jagged interactions result in proteolytic processing of Notch and translocation of the Notch intracellular domain to the nucleus, where it interacts with transcriptional regulators. Activation of the intracellular domain inhibits differentiation along a particular pathway but leaves cells competent to adopt different fates (1).
In the hemopoietic system, Notch is expressed by stem cells while Notch ligands are found in bone marrow stroma, which provides the microenvironment necessary for stem cell survival and differentiation. The Notch signal modulates responses to cell type specification cues mediated by the multiplicity of growth and differentiation factors present in this environment and renders the most primitive progenitor cells more resistant to differentiation (13, 42). The importance of these receptors in hemopoietic and lymphoid development has become increasingly evident (3, 25, 30).
Because Notch and its ligands play an important role in T-cell development and in the recruitment of inducible Tr in mice, we investigated whether or not the Notch pathway may play a similar role in humans. We looked at the effects on T-cell function of the coexpression of high levels of the Notch ligand Jagged-1 by autologous B cells infected with Epstein-Barr virus (EBV). This is a well-defined antigen-specific system in which EBV-lymphoblastoid cell line cells (EBV-LCL) function as antigen-presenting cells (33) which readily induce proliferative and cytotoxic effector functions that are viral-antigen specific when the cells are cocultured with T lymphocytes from EBV-immune donors (32).
We found that EBV-LCL overexpressing the Notch ligand Jagged-1 induce Tr (in both the CD4 and CD8 subpopulations) that specifically inhibit the proliferative and cytotoxic memory responses to EBV proteins. These Tr produce interleukin-10 (IL-10) and are also able to inhibit the proliferative and cytotoxic anti-EBV T-cell responses of autologous T lymphocytes that have not been exposed to Notch ligand.
MATERIALS AND METHODS
Cells and cell lines.
Peripheral blood mononuclear cells (PBMC) were obtained from healthy EBV-seropositive adults. EBV-LCL were obtained by EBV (B95-8) immortalization of mature B cells from the same donors. A bone marrow stromal cell line was used as the positive control for Jagged-1 protein expression in Western blotting (41).
All cells were cultured in complete medium prepared with RPMI 1640 (BioWhittaker, Walkersville, Md.) supplemented with 10% heat-inactivated fetal calf serum (HyClone), antibiotics, and l-glutamine and maintained at 37°C in an atmosphere of 5% CO2. For TGF-β cytokine assessment, cells were cultured in X-VIVO-15 serum-free medium (BioWhittaker).
Adenoviral vector.
EBV-LCL were transduced by using the chimeric adenovirus Ad5/F35. This vector, as previously described by Shayakhmetov and Lieber (36), is an adenovirus serotype 5 (Ad5) virus in which parts of the fiber gene have been replaced by the fiber from an adenovirus serotype 35 virus. This chimeric vector is coxsackie adenovirus receptor independent and has improved transduction efficiency for coxsackie adenovirus receptor-negative lymphoid cells compared with Ad5 vectors (45). The cDNA for the full-length Jagged-1 or enhanced green fluorescent protein (EGFP; Clontech, Palo Alto, Calif.) was cloned into the shuttle plasmid pShuttle-X (Clontech). The entire region containing the cytomegalovirus (CMV) promoter, Jagged-1, or EGFP, followed by a simian virus 40 polyadenylation site, was excised by I-CeuI and pI-SceI digestion and then transferred to pAd5/F35 cleaved by using the same restriction enzymes to form pAd5/F35-Jagged-1 or pAd5/F35-EGFP. Both Ad5/F35 vectors were produced by Lipofectamine (Life Technologies, Gaithersburg, Md.) transfection of the human embryonic kidney (HEK) 293 cell line. Large-scale amplification of a single plaque generated in transfected HEK293 cells was followed by three consecutive freeze-thaw cycles for viral extraction. Both viruses were purified with a double series of cesium-chloride gradient ultracentrifugations and desalted on a dialysis cassette (Pierce, Rockford, Ill.), and titers were established by plaque assay using HEK293 cells. Viral titers were also quantitated by measurement of optical density at 260 nm, and the particle-to-PFU ratios were 160 and 200 for Ad5/F35 Jagged-1 and Ad5/F35 EGFP, respectively. These ratios are higher than those obtained in Ad5-based systems but are typical for the Ad5/F35 hybrid (45).
Western blotting.
To detect Jagged-1 protein expressed in transduced EBV-LCL and bone marrow stromal cell lines, cell extracts were prepared from pelleted cells by lysing in Laemmli's sample buffer, boiled for 5 min, and spun to remove insoluble material. Under reducing conditions, the cell lysates containing equal amounts of proteins were run in sodium dodecyl sulfate-6% polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (Bio-Rad, Hercules, Calif.). The membranes were probed with antibodies to Jagged-1 (H-114; Santa Cruz Biotechnology, Santa Cruz, Calif.) for 1 h at room temperature and then visualized with horseradish peroxidase-coupled anti-immunoglobulin (705-0350147; Jackson Immuno Research Laboratories, West Grove, Pa.) for 30 min at room temperature. Detection was achieved using the ECL system (Amersham). After stripping, the membrane was reprobed with anti-vinculin or anti-actin antibodies (Sigma-Aldrich, St Louis, Mo.). The same method was used to detect HES-1, but using sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis. The anti-HES-1 antibody (clone 35) was purchased from BD Pharmingen (San Diego, Calif.).
RNA extraction and cDNA synthesis.
Total RNA was extracted by using an RNeasy Mini kit (Qiagen, Valencia, Calif.). RNA was eluted in 40 ml of RNase-free water and stored at −80°C until use. Contaminating DNA was removed by DNase I digestion (Gibco Invitrogen Corporation, Carlsbad, Calif.). RNA was then reversed transcribed to cDNA (DNA) by using random hexamers and SuperScript II reverse transcriptase (SuperScript First-Strand Synthesis System for RT-PCR; Gibco).
Real-time quantitative-PCR assay.
For detection and quantification of Jagged-1, HES-1, and Deltex transcripts, a real-time PCR assay was developed using TaqMan technology (PE Applied Biosystems, Foster City, Calif.) (20). Primers and a probe spanning the junction between exon 22 and exon 23 of the Jagged-1 gene (GenBank database sequence accession number U73936) were designed by using Primer Express software (PE Applied Biosystems). The sequences for the designed primers and probe are as follows: forward primer (exon 22), 5′-ACC TGC CAG TGC CTG AAT G-3′; reverse primer (exon 23), 5′-AGG CAA GGT CGA GGG CC-3′; probe (junction between exons 22 and 23), 5′-FAM-ACG GAT CGC CTG CTC AAA GGT CTG-TAMRA-3′, with FAM and TAMRA representing the reporter and quencher dye, respectively, as a positive control and calibrator for relative quantification (PE Applied Biosystems) according to the manufacturer's protocol. The sequences for the HES-1 primers and probe are as follows: forward primer (exon 6), 5′-TGG GTG CCA AGC ACT GC-3′; reverse primer (exon 7), 5′-TCG-TGA-CCA-CCT-TGT-TTT-TCT-G-3′; probe (junction between exons 6 and 7), 5′-FAM-AAG-GAA-GTG-GTC-GAA-GCT-CAC-GTG-GA-TAMRA-3′. The sequences for the Deltex primers and probe are as follows: forward primer (exon 5), 5′-TGG-TTC-GAA-GAT-ACA-TGC-AGA-AG-3′; reverse primer (exon 6), 5′-ACC-AGT-CGC-TCC-ATG-CAG-AT-3′; probe (junction between exons 5 and 6), 5′-FAM-TGA-AAA-ACC-CAC-CTG-ATG-AGG-ACT-GCA-C-TAMRA-3′. RNA without reverse transcription was run in parallel for each sample. PCR amplification was performed with 2× TaqMan Universal Master Mix (PE Applied Biosystems) with 300 nM concentrations of primers and 200 nM concentrations of probe, 10 μl of cDNA, and nuclease-free water (Promega, Madison, Wis.). All samples were analyzed in duplicate. Amplification was performed by using the ABI PRISM 7700 sequence detection system (PE Applied Biosystems) and consisted of 2 min at 50°C, (inactivation of possible carryover contamination by uracil N′-glycosylase [UNG]) and 40 two-step cycles of 15 s at 95°C and 60 s at 60°C. Threshold (CT) cycles of amplifications, where the fluorescence plots crossed a defined baseline, were determined for each sample. For relative quantification, the expression in samples was normalized by comparison with the rRNA amount by using the ΔΔCT method as described in detail elsewhere (21).
Cell cultures.
Primary cocultures were performed with 5 × 104 gamma-irradiated (at 4,000 cGy) stimulator EBV-LCL transduced by Ad5/F35 Jagged-1 and 2 × 106 responder autologous PBMC or enriched T lymphocytes (see below) at a final ratio of 1:40 in a 24-well plate (Nunc, Oslo, Norway) in 2 ml of complete medium. Other ratios of responder cells to EBV-LCL were also tested as described in Results. Viable cells were harvested after Ficoll density gradient centrifugation on day 10, washed, and used in secondary cocultures. Control cocultures were stimulated with gamma-irradiated EBV-LCL either nontransduced or transduced with Ad5/F35 EGFP at identical ratios. For cytokine measurements, 400 μl of culture supernatants were harvested on days 3 and 8 and stored at −20°C for future assays. Secondary cocultures were performed with PBMC and nontransduced gamma-irradiated autologous EBV-LCL at a ratio of 10:1 with or without Tr. Tr were used unseparated or as individual CD+CD25+, CD4+CD25−, CD8+CD25+, or CD8+CD25− populations purified by positive or negative selection. CD4, CD8, and CD25 MACS microbeads and Multisort kits were used for positive selection. CD4, CD8, CD56, CD19, and CD14 MACS microbeads were used for negative selection (Miltenyi Biotec, Auburn, Calif.). The purity of the populations, evaluated by fluorescence-activated cell sorter analysis (FACScalibur; Becton Dickinson, San Jose, Calif.), was always >95%. Donor T lymphocytes were purified from PBMC by positive selection, and the purity was routinely >98%. To demonstrate the functional purity of the T cells, an adenovirus (Ad5/F35 EGFP; multiplicity of infection [MOI] = 2 PFU/cell) was used as an antigen to measure the proliferative response. T cells alone (2 × 105) had no measurable specific [3H]thymidine ([3H]thy) incorporation ([3H]thy with antigen minus [3H]thy without antigen), while in the presence of adherent cells from peripheral blood, specific [3H]thy incorporation rose to 26,000 ± 3,500 cpm after 5 days of culture.
The T cells used to detect Deltex mRNA, HES-1 mRNA, and protein were prepared by stimulating 2 × 106 PBMC with 5 × 104 gamma-irradiated (at 4,000 cGy) autologous EBV-LCL transduced by Ad5/F35 Jagged-1. T cells were harvested at 12, 24, and 48 h by using a pan-T-cell isolation kit (Miltenyi Biotec). The purity was constantly >99%, and no residual EBV-LCL (CD19 staining) were detectable.
To assess antigen specificity, soluble Candida antigens were added to 2 × 105 PBMC at a concentration of 250 μg/ml in U-bottom 96-well plates (Nunc) in 200 μl of complete medium with 2 × 105 autologous Tr. For responses to CMV, we substituted soluble CMV antigens at a concentration of 1 μg/ml to 2 × 105 PBMC.
Transwell experiments.
To assess the contribution of soluble factors to cellular inhibition, experiments were performed in 24-well plates. 106 PBMC were stimulated with 105 autologous nontransduced EBV-LCL with or without 106 Tr either added directly to the culture or placed in Transwell chambers (0.4-μm pore size; Costar). After 7 days of culture, cells were transferred to 96-well plates and proliferation was measured using liquid scintillation counting.
Monitoring of proliferation.
PBMC and T cells were cultured in 200-μl U-bottom 96-well plates (Nunc) with gamma-irradiated (at 4,000 cGy) transduced or nontransduced EBV-LCL with or without Tr. Proliferation was analyzed by [3H]thy incorporation, and 1 μCi of [3H]thy was present for the last 18 h of culture. [3H]thy uptake was measured by a liquid scintillation counter (Matrix 96 Beta counter; Canberra Packard, Meriden, Conn.) and expressed as mean counts per minute (± standard deviations [SD]) of triplicate measurements. A trypan blue exclusion before each assay constantly showed >95% viability in all conditions.
Assessment of cytokine production.
Day 3 and 8 supernatants, previously frozen at −20°C, were analyzed for their IL-2, IL-4, IL-5, IL-10, gamma interferon, and tumor necrosis factor alpha contents by using a cytometric bead array kit (Pharmingen/BD Biosciences). X-VIVO-15 culture supernatants were assessed for TGF-β content by enzyme-linked immunosorbent assay (Quantikine ELISA kit; R&D Systems, Minneapolis, Minn.) according to the manufacturer's instructions.
Flow cytometry analysis.
Cultured cells were washed, stained for 20 min at 4°C with optimal dilution for each antibody, and analyzed by flow cytometry (FACScalibur and CELLQuest software; Becton Dickinson). A total of 104 events were analyzed for each determination. Cells were stained with fluorescein isothiocyanate, phycoerythrin, or peridinin chlorophyll protein (PerCP) monoclonal antibodies (MAb) to CD4 (clone L200), CD8 (RPA-T8), CD25 (M-A251), CD40 (5C3), CD80 (L307.4), CD86 (FUN-1), HLA-A, -B, and -C (G46-2.6), and HLA-DR (L243) (BD-Biosciences).
Cytotoxicity by chromium-51 release assay.
Cytotoxic activity was assessed by 4-h chromium-51 (51Cr) release from labeled autologous EBV-LCL and K562 cell lines. The assays were performed after two stimulations by EBV-LCL (nontransduced or transduced) in primary and secondary cocultures. Briefly, 1.5 × 106 target cells were labeled with 3.7 mBq (100 mCi) of 51Cr (Amersham) and used at 5,000 cells per well. Multiple effector-to-target cell (E:T) ratios were tested in triplicate, and cytotoxic activity was expressed as a percentage of specific lysis. The percentage of specific 51Cr release was calculated as follows: percent lysis = [(experimental released cpm − spontaneous cpm)/(total lysis cpm − spontaneous cpm)] × 100. Viable effector and target cells were systematically selected by Ficoll density gradient centrifugation before each assay. The viability (>95%) was confirmed by trypan blue exclusion. The anti-class I (HLA-A, -B, and -C antigen; clone W6/32) and anti-class II (HLA DP, -DQ, and -DR antigen; clone CR3/43) antibodies used for blocking experiments were purchased from Dako (Denmark) and used at concentrations previously found to be optimal for specific blocking of cytotoxic-T-lymphocyte (CTL) activity in the EBV system (32, 33, 38).
Statistical analysis.
All data are presented as means ± 1 SD. The Student t test (Statistica Software, Tulsa, Okla.) was used to compare results between different conditions of culture. The significance level was set at a P value of <0.05. The numbers of experiments analyzed are given in each figure legend.
RESULTS
Transduction of EBV-LCL by Ad5/F35 Jagged-1 results in expression of transcripts and functional protein.
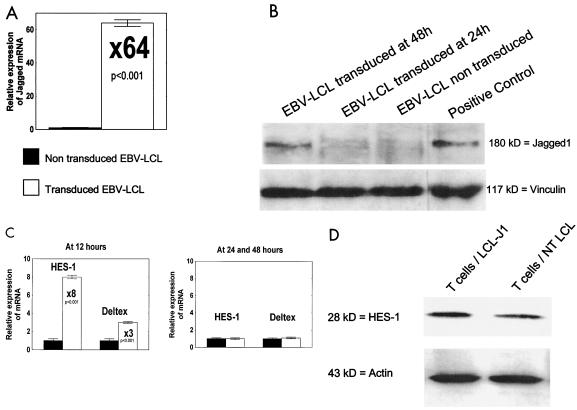
Transduction of EBV-LCL by Ad5/F35 Jagged-1 was assessed by quantitative real-time PCR and Western blotting. Forty-eight hours after transduction at an MOI of 100 PFU/cell, real-time PCR amplification showed a >60-fold increase in the Jagged-1 mRNA copy number in transduced cells (Fig. 1A). Western blotting confirmed the presence of a 180-kDa band representing the Jagged-1 protein (Fig. 1B).
FIG. 1.
Transduction of EBV-LCL by Ad5/F35 Jagged-1 with subsequent activation of Notch downstream signal in T cells. (A) Results of real-time PCR showing overexpression of Jagged-1 mRNA (×64) in transduced EBV-LCL at 48 h (MOI = 100 PFU/cell) compared with that in nontransduced EBV-LCL. Data are means ± SD from three experiments. (B) Results of Western blotting showing the Jagged-1 protein of approximately 180 kDa 48 h after transduction. Human bone marrow stromal cells were used as the positive control. Shown are the results of one experiment that is representative of three. (C) Results of real-time PCR showing HES-1 and Deltex mRNA expression levels in T cells stimulated by nontransduced autologous EBV-LCL (filled columns) compared with those for autologous EBV-LCL Jagged-1 (open columns). Overexpression after stimulation with EBV-LCL Jagged-1 was observed at 12 h but not at 24 or 48 h. Data are means ± SD from three experiments. (D) Results of Western blotting showing an overexpression of the HES-1 protein (28 kDa) in T cells 24 h after stimulation with EBV-LCL Jagged-1. Shown are the results of one experiment that is representative of three. T cells/LCL-J1, T cells stimulated by autologous EBV-LCL transduced with Ad5/F35 Jagged-1; T cells/NT LCL, T cells stimulated by nontransduced autologous EBV-LCL.
To demonstrate the functionality of the transgenic Jagged-1 protein and the ability of T cells to respond to this stimulus, we used real-time PCR to quantitate expression of the HES-1 (12) and Deltex (44) genes, since both of these encode transcriptional regulators activated by the Notch signal. As shown in Fig. 1C, both genes were overexpressed at 12 h after exposure to EBV-LCL Jagged-1 (8.5 times for HES-1 [P < 0.001] and three times for Deltex [P <0.001]) and levels normalized by 24 and 48 h. Western blotting showed overexpression of the HES-1 protein at 24 h, confirming activation of the Notch pathway (Fig. 1D).
The viability of EBV-LCL measured by trypan blue exclusion was >90% at 48 h after transduction. Transduction and expression of Jagged-1 had no effect on EBV-LCL expression of MHC molecules (HLA-A, -B, -C, -DQ, and -DR) or that of the costimulator molecules CD40, CD80, and CD86 (data not shown).
By comparison, 70 to 80% of EBV-LCL transduced with an Ad5/F35 EGFP at an MOI of 100 PFU/cell expressed GFP 48 h after transduction (data not shown).
Activated Notch inhibits proliferative and cytotoxic immune responses.
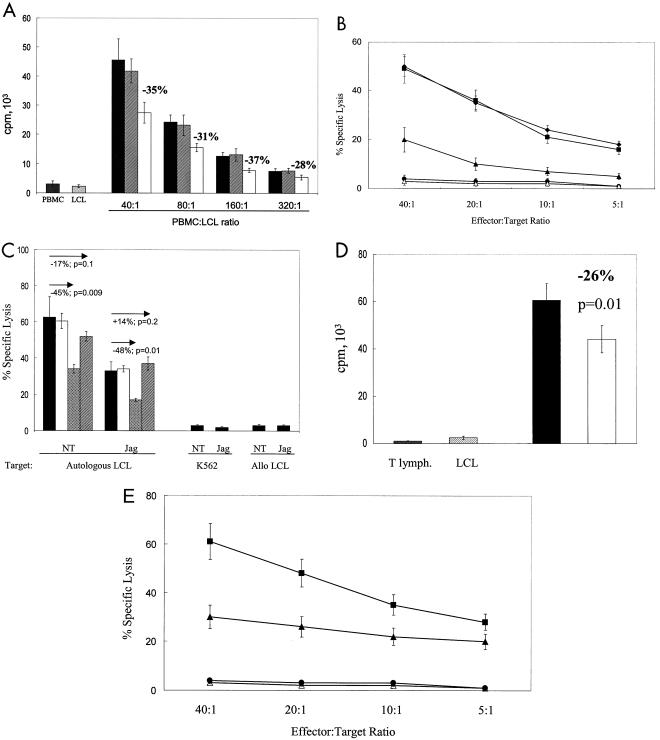
To assess the effect of activated Notch on T-cell memory immune responses to autologous EBV-LCL, PBMC from healthy EBV seropositive donors were cocultured with autologous EBV-LCL Jagged-1. In the absence of Jagged-1, this well-characterized system induces expansion of CD4+ and CD8+ EBV-specific memory T cells, manifested by a readily detectable proliferative and cytotoxic response directed toward the stimulating EBV-positive target cells (19). In five experiments, proliferation was inhibited by a mean of 35% on day 5 when the ratio of PBMC to EBV-LCL was 40:1 (P = 0.001) compared with cultures containing control vector-transduced EBV-LCL (Fig. 2A). Figure 2B shows that cytotoxic effector function against EBV-LCL was also significantly reduced (by ca. 65%) compared to controls (P < 0.02). As a confirmation that the cytotoxic activity we observed was attributable to conventional CTL, we added MAb directed against class I and class II MHC antigens. Figure 2C shows that these produced 45% (P = 0.009) and 17% (P = 0.1) blocking, respectively, of the cytotoxic response in the nontransduced cocultures. Inhibition in cocultures with EBV-LCL Jagged-1 was the same, with residual killing blocked by 48% (class I MAb, P = 0.01) and 0% (class II MAb, P = 0.2) (Fig. 2C).
FIG. 2.
Activated Notch inhibits proliferative and cytotoxic immune responses. (A) [3H]thymidine incorporation at day 5 in three different culture conditions: PBMC plus autologous LCL cells (filled columns), PBMC plus autologous LCL cells transduced by Ad5/F35 EGFP (hatched columns), and PBMC plus autologous LCL cells transduced by Ad5/F35 Jagged-1 (open columns). Four ratios of PBMC to LCL cells were tested as indicated. For each ratio, the inhibition related to Jagged-1 expression was significant (P < 0.01). Counts of PBMC alone and LCL cells alone are shown. Data shown are means ± SD from five experiments. (B) Cytotoxic activity of T cells against autologous LCL targets after CD56+-cell depletion performed just before the assay. T cells were obtained from three different culture conditions: PBMC plus autologous LCL cells (♦), PBMC plus autologous LCL cells transduced by Ad5/F35 EGFP (▪), and PBMC plus autologous LCL cells transduced by Ad5/F35 Jagged-1 (▴). No lysis of K562 cells (•) or fully HLA-mismatched allogeneic LCL cells (▵) was observed in all three culture conditions. The nontransduced condition is shown. Assays were performed between days 15 and 20 after two stimulations. The ratio of PBMC to LCL cells was 40:1 at the first stimulation and 10:1 at the second stimulation. Data shown are means ± SD from three experiments. The inhibition related to Jagged expression is significant for each E:T ratio (P < 0.02). (C) Cytotoxic activity of T lymphocytes stimulated by nontransduced LCL cells (NT) or LCL Jagged-1 (Jag). Shown are results for T lymphocytes plus autologous LCL cells (filled columns), T lymphocytes plus autologous LCL lines and isotype control (opencolumns), T lymphocytes plus autologous LCL cells plus anti-MHC class I MAb (cross-hatched columns), and T lymphocytes plus autologous LCL cells and anti-MHC class II MAb (hatched columns). 2 × 106 T lymphocytes and 5 × 104 LCL cells were used in each condition. Assays were performed between days 15 and 20 after two stimulations. The ratio of T cells to LCL cells was 40:1 at the first stimulation and 10:1 at the second stimulation. A CD56+-cell depletion was performed just before the assay. Targets were autologous LCL cells, K562 cells, or fully HLA-mismatched allogeneic LCL cells in both conditions (NT and Jag). The effector-to-target cell target ratio was 20:1. (D) [3H]thymidine uptake of T cells at day 5 in two different culture conditions: T lymphocytes (105) plus autologous LCL cells (2,500) (filled column) and T lymphocytes (105) plus autologous LCL cells transduced by Ad5/F35 Jagged-1 (2,500) (open column). Counts of T lymphocytes alone and LCL cells alone are shown. Data shown are means ± SD from five experiments. (E) Cytotoxic activity of T cells against autologous LCL targets after CD56+-cell depletion performed just before the assay. T cells were obtained from two different culture conditions: T lymphocytes plus autologous LCL cells (▪) and T lymphocytes plus autologous LCL cells transduced by Ad5/F35 Jagged1 (▴). No lysis of K562 cells (•) or fully HLA-mismatched allogeneic LCL cells (▵) was observed in both culture conditions. The nontransduced condition is shown. Assays were performed between days 15 and 20 after two stimulations. The ratio of T lymphocytes to LCL cells was 40:1 at the first stimulation and 10:1 at the second stimulation. Data shown are means ± SD from three experiments. The inhibition related to Jagged expression is significant for each E:T ratio (P < 0.05).
To discover whether inhibition could be obtained by direct activation of T cells by EBV-LCL Jagged-1 or whether more complex multilineage interactions were involved, we repeated these experiments using purified T lymphocytes instead of PBMC. In five separate experiments, identical inhibitory effects on both proliferation and cytotoxicity were obtained, indicating that direct stimulation of T cells by EBV-LCL Jagged-1 can induce inhibitory effectors (Fig. 2D and E).
Inhibition of immune response is transferable.
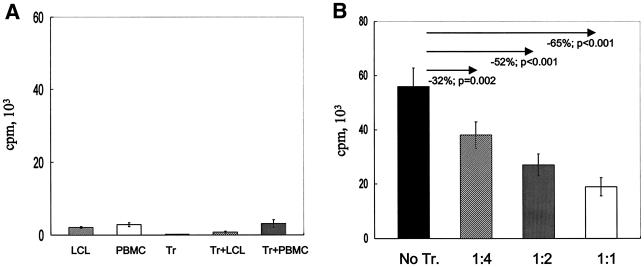
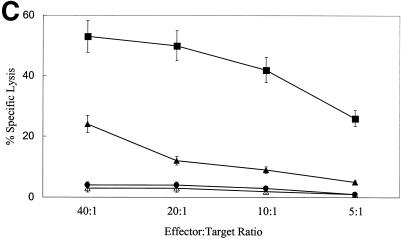
We next determined whether the EBV-LCL Jagged-1-stimulated T cells (i.e., Tr) functioned as regulators of the immune response by measuring their effects on proliferation and cytotoxicity of fresh T lymphocytes cultured with nontransduced autologous EBV-LCL. Tr alone had no measurable proliferation and minimal proliferation when cultured with fresh T cells or with nontransduced EBV-LCL (Fig. 3A). Addition of Tr to fresh autologous T cells and autologous EBV-LCL, however, inhibited proliferation proportional to the ratio of Tr to T-responder cells, and at a ratio of 1:1, the response was reduced by 65% (P < 0.001) (Fig. 3B). Moreover, addition of Tr to fresh cocultures of responder T cells and autologous EBV-LCL also significantly inhibited the generation of a cytotoxic response (Fig. 3C).
FIG. 3.
Inhibition of immune response can be transferred by Tr. (A) Proliferative counts at day 7 for LCL cells (2 × 104), PBMC (2 × 105), Tr (2 × 105), Tr (2 × 105) plus LCL cells (2 × 104), and Tr (2 × 105) plus PBMC (2 × 105). Results are means ± SD from six separate experiments. (B) Proliferative counts at day 7 for cultures of PBMC (2 × 105) plus autologous LCL cells (2 × 104) with or without autologous Tr, with ratios of Tr to PBMC of 1:4, 1:2, and 1:1. Results are means ± SD from six separate experiments. (C) Cytotoxic activity of T cells against autologous LCL targets after CD56+-cell depletion performed just before the assay. T cells were obtained from two different culture conditions: PBMC plus autologous LCL cells (▪) and PBMC plus autologous LCL cells and autologous Tr at a Tr-to-PBMC ratio of 1:1 (▴). No lysis of K562 cells (•) or fully HLA-mismatched allogeneic LCL cells (▵) was observed in both culture conditions. The condition without Tr in the culture is shown. Assays were performed between days 15 and 20 after two stimulations. The ratio of PBMC to LCL cells was 40:1 at the first stimulation and 10:1 at the second stimulation. Data shown are means ± SD from three experiments. The inhibition is significant for each E:T ratio (P < 0.01).
Phenotype of Tr induced following Jagged-1 exposure.
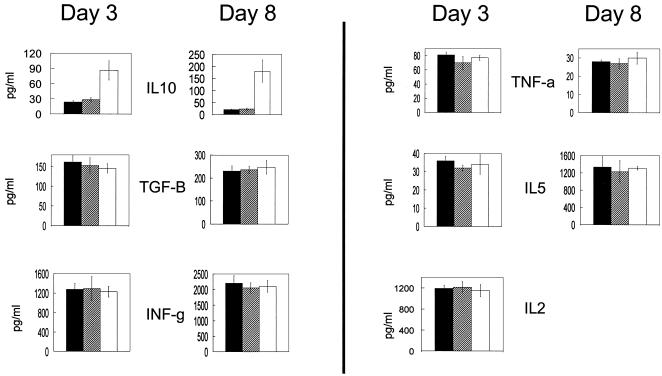
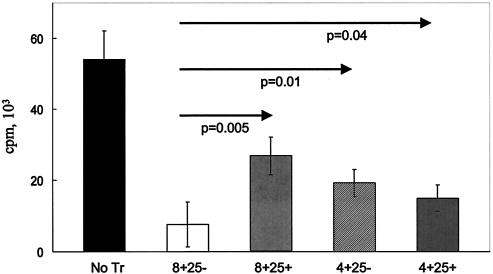
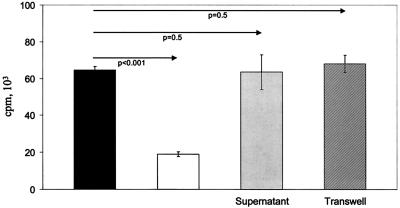
Tr populations in mice and humans may produce IL-10 (10, 22, 27, 31). Measurement of IL-10 in supernatants of primary cocultures (induction phase) showed a greater-than-ninefold increase in the level of this cytokine in the presence of Jagged-1-expressing EBV-LCL versus control EBV-LCL (P = 0.002) (Fig. 4). Productions of IL-2, IL-4, IL-5, gamma interferon, and tumor necrosis factor alpha were unchanged. Interestingly, there was no increase in the level of TGF-β, a cytokine that has been associated with some Tr subpopulations (10, 22, 31). We next compared the activities of CD4-, CD8-, and CD25-defined subsets on the proliferative response of fresh T cells to autologous EBV-LCL. As shown in Fig. 5, all populations tested had a suppressive activity, with the greatest inhibition produced by CD8+CD25− cells (88%) and the least produced by CD8+CD25+ cells (50%). Figure 6 shows the failure of fresh supernatants from Tr to inhibit proliferation and the abolition of suppression when Tr are separated from responder cells by a semipermeable membrane, implying that inhibition requires cell-cell contact and is independent of soluble factors.
FIG. 4.
Lymphocytes induced by EBV-LCL overexpressing Jagged-1 produce IL-10. Data indicate cytokine concentrations in culture supernatants at days 3 and 8 in three different culture conditions: PBMC plus autologous LCL cells (filled columns), PBMC plus autologous LCL cells transduced by Ad5/F35 EGFP (open columns), and PBMC plus autologous LCL cells transduced by Ad5/F35 Jagged-1 (hatched columns). 2 × 106 PBMC and 5 × 104 LCL cells were used in each condition. Data shown are means ± SD from four experiments. In all three conditions, no significant amount of IL-4 was detectable at either day 3 or day 8 and no significant amount of IL-2 was detectable at day 8. The increase of IL-10 was significant at days 3 and 8 (P = 0.002). No significant difference was observed for the other cytokines.
FIG. 5.
Tr do not have a characteristic phenotype definable by CD4, CD8, or CD25 antigens. Proliferative counts at day 7 in five conditions of coculture: PBMC plus autologous LCL cells at a ratio of 10:1 (No Tr), PBMC plus autologous LCL cells at a ratio 10:1 plus CD8+CD25− Tr (8+25−), PBMC plus autologous LCL cells at a ratio of 10:1 plus CD8+CD25+ Tr (8+25+), PBMC plus autologous LCL cells at a ratio of 10:1 plus CD4+CD25− Tr (4+25−), and PBMC plus autologous LCL cells at a ratio of 10:1 plus CD4+CD25+ (4+25+). The ratio of Tr to PBMC was 1:1. The inhibition was significant (P < 0.01) for each Tr subpopulation when compared to the proliferation without Tr. Data shown are means ± SD from four experiments.
FIG. 6.
The mechanism of inhibition is not mediated by soluble factors and requires cell-to-cell contact. Proliferative counts at day 7 for cultures of PBMC plus autologous LCL cells (filled column); PBMC plus autologous LCL cells and Tr, with Tr added directly to the culture (open column); PBMC plus autologous LCL cells in fresh supernatant of T lymphocytes stimulated with LCL Jagged-1 (harvested at day 8) (Supernatant); and PBMC plus autologous LCL cells and Tr placed in Transwell chambers (Transwell). The ratios of PBMC to LCL cells and of PBMC to Tr were 10:1 and 1:1, respectively. Data shown are means ± SD from four experiments.
Tr are EBV specific.
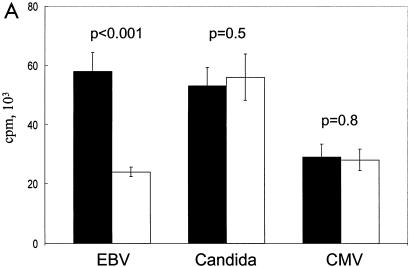
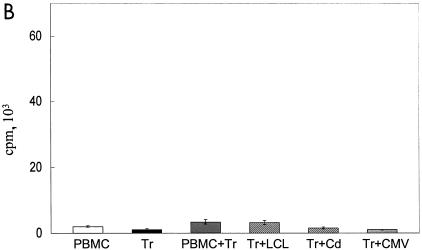
To determine the specificity of the inhibitory response, we measured the capacity of fresh T cells to respond to Candida and CMV antigens after the addition of EBV-LCL Jagged-1-generated Tr. Figure 7A shows that even when the response to autologous EBV-LCL is significantly inhibited (P < 0.001), the proliferative response of fresh T cells to Candida and CMV is fully intact. Tr alone, Tr plus Candida, Tr plus CMV, and Tr plus PBMC had no measurable proliferation (Fig. 7B). Hence, activation of Notch by Jagged-1-expressing EBV-LCL inhibits the anti-EBV response while sparing responses to viral and fungal antigens.
FIG. 7.
Tr are EBV specific. (A) Proliferative counts at day 7 of 2 × 105 PBMC stimulated with either autologous LCL cells (2 × 104) or soluble Candida antigen (250 μg/ml) or soluble CMV antigen (1 μg/ml) with (filled columns) or without (open columns) addition of 2 × 105 Tr at the beginning of the cultures. Data shown are means ± SD from six experiments. (B) [3H]thy uptake at day 7 in the following conditions of culture: PBMC (2 × 105), Tr (2 × 105), PBMC (2 × 105) plus Tr (2 × 105), Tr (2 × 105) plus LCL cells (2 × 104), Tr (2 × 105) plus Candida antigen (250 μg/ml) (Cd), and Tr (2 × 105) plus CMV antigen (1 μg/ml). Data shown are means ± SD from six experiments.
DISCUSSION
Our results indicate that Notch activation by overexpression of the Jagged-1 ligand on EBV-LCL can drive EBV-specific human T cells towards tolerance, affecting both proliferative and cytotoxic responses. Tolerized T cells are transferable, inhibiting the induction of an immune memory response to EBV. Inhibition is antigen specific and mediated by Tr which are in both the CD4 and CD8 subsets and have increased production of IL-10. Inhibition is also dose dependent and requires cell-cell contact.
Notch was originally known for its role in lateral inhibition, in which a group of cells with equivalent developmental potentials initially express both Notch and Notch ligand. Notch-mediated signaling between these cells regulates their differentiation. Feedback loops linking Notch signaling with Notch ligand expression amplify small differences in the level of Notch signaling between neighboring cells. This positive feedback eventually results in a limited number of cells becoming “signaling cells,” which express Notch ligand and differentiate along a single pathway, whereas neighboring cells become “receiving cells,” which express Notch and adopt a different fate (17). Notch can also regulate cell fate decisions by inductive signaling, the type of interaction that occurs within the hemopoietic system. In this case, Notch and its ligand are expressed separately on distinct cell types. Signals delivered to the receiving cells during cell-cell interactions induce the expression of genes that favor differentiation toward an alternative fate. Other intrinsic and extrinsic factors can influence both the activity of the Notch signaling pathway and the response of cells to Notch signals (17).
We examined the contribution of Notch activation to antigen-mediated induction of Tr in humans because there is accumulating evidence to suggest that activation of Notch receptors on T cells at the time of antigen exposure can contribute to the induction of functionally inhibitory cells in mice (5). We found that Notch activation of human T cells during antigen exposure induces both CD4+CD25+/− and CD8+CD25+/− Tr. This absence of a single phenotype defined on the basis of CD4/CD8/CD25 expression is consistent with the results of previous studies on induced Tr, which have also described populations of Tr that may be CD4+CD25+/− or CD8+ (4, 7, 8, 14, 23, 43, 46). The concept that Notch activation plays an important role in the induction of tolerance is also indirectly supported by observations that mesenchymal stem cells, which express high levels of Notch ligands (16, 41), are potent inducers of Tr in both murine and human systems and may suppress graft-versus-host disease in preclinical and clinical allogeneic stem cell transplantation models (2). We do not know what contribution Notch ligand activation makes to the induction or maintenance of naturally occurring CD4+CD25+ Tr. However, it is notable that human CD4+CD25+ Tr have high levels of Deltex and HES-1, which are transcriptional regulators of the Notch pathway, as well as increased levels of Notch 4 and Delta 1 (26).
In this study, we used EBV antigens presented on autologous B cells (EBV-LCL) to demonstrate the negative regulatory effects of overexpression of the Notch ligand Jagged-1 on the development of human antigen-specific CTL lines. We chose this system because EBV-LCL are excellent antigen-presenting cells. They express high levels of class I and class II MHC molecules as well as the costimulatory molecules CD80, CD86, and CD40. More than 90% of the adult population is EBV-seropositive, and most of these individuals have high levels of circulating memory T cells specific for EBV antigens (28). Hence, in an in vitro culture of EBV-LCL with autologous T cells from an EBV-seropositive donor, it is routinely possible to generate, reactivate, and expand EBV reactive memory CD4 and CD8 T cells (19). This system enabled us to study the consequences of forced expression of a Notch ligand by these EBV-LCL on the subsequent response to EBV. Based on our present data, our working model is that exposure of human T lymphocytes to target cells expressing antigen and appropriate costimulator signals induces reactivation of antigen-specific T cells. Exposure in the presence of Notch ligand, however, also drives the activation of antigen-specific Tr and the production of IL-10. This cytokine has a central role in the development and activity of murine Tr1 cells. Activation of murine CD4+ T cells in the presence of IL-10 produces T cells that are themselves capable of producing IL-10 and of suppressing the proliferation of fresh CD4+ T cells in an antigen-specific manner. But although IL-10 likely plays a critical role in the induction of Tr and is produced by the regulatory cells themselves, the cytokine is not by itself responsible for the subsequent inhibition of immunity, which generally requires cell-to-cell contact and is antigen specific (27). We found that fresh supernatants of Tr are not inhibitory and that separation of regulatory cells from responder cells by a semipermeable membrane abolishes the suppression, excluding a mechanism of inhibition relying on soluble factors like IL-10. These results are consistent with previous studies showing that while IL-10 may have inhibitory effects on macrophage and dendritic cell maturation and function, it has no negative effects on the ability of EBV-LCL to reactivate T-cell memory nor on the cytotoxic activity of CTL (38). In fact, perforin is upregulated on EBV-CTL by IL-10 (34).
The Tr produced following exposure to EBV-LCL Jagged-1 inhibit the response of secondary cultures of memory T cells in an antigen-specific manner even in the absence of continued activation by Notch ligand. While CD4+CD25+ T cells have high regulatory activity in this system, inhibitory effects are also seen from the CD4+CD25− and CD8+ populations. We do not yet know whether the effects of Notch activation are to “switch” a cell from a responder to a regulatory phenotype or simply to enhance the proliferation of preexisting regulatory cells. This important issue will likely best be addressed by an extensive clonal analysis of individual responding cells. Nonetheless, our data identify a molecular pathway for induction of human Tr, showing that Jagged-1 stimulation of human T cells can induce an antigen-specific population of cells capable of inhibiting both proliferative and cytotoxic effector functions. Activation of this pathway may contribute to regulation of responses to viral antigens.
Acknowledgments
This work was supported by a grant from the Association pour la Recherche sur le Cancer, France, to S.V., a grant from the Deutsche Forschungsgemeinschaft to H.-J.W., and a grant from Comitato Maria Letizia Verga per lo studio e la cura della leucemia del bambino (Monza, Italy) and Fondazione Cassa di Risparmio di Genova ed Imperia-Associazione Cristina Bassi contro le leucemie acute dell'adulto (Genova, Italy) to E.B.
REFERENCES
- 1.Artavanis-Tsakonas, S., K. Matsuno, and M. E. Fortini. 1995. Notch signaling. Science 268:225-232. [DOI] [PubMed] [Google Scholar]
- 2.Bartholomew, A., C. Sturgeon, M. Siatskas, K. Ferrer, K. McIntosh, S. Patil, W. Hardy, S. Devine, D. Ucker, R. Deans, A. Moseley, and R. Hoffman. 2002. Mesenchymal stem cells suppress lymphocyte proliferation in vitro and prolong skin graft survival in vivo. Exp. Hematol. 30:42-48. [DOI] [PubMed] [Google Scholar]
- 3.Deftos, M. L., and M. J. Bevan. 2000. Notch signaling in T cell development. Curr. Opin. Immunol. 12:166-172. [DOI] [PubMed] [Google Scholar]
- 4.Dhodapkar, M. V., and R. M. Steinman. 2002. Antigen-bearing immature dendritic cells induce peptide-specific CD8+ regulatory T cells in vivo in humans. Blood 100:174-177. [DOI] [PubMed] [Google Scholar]
- 5.Dieckmann, D., H. Plottner, S. Berchtold, T. Berger, and G. Schuler. 2001. Ex vivo isolation and characterization of CD4+CD25+ T cells with regulatory properties from human blood. J. Exp. Med. 193:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gavin, M. A., S. R. Clarke, E. Negrou, A. Gallegos, and A. Rudensky. 2002. Homeostasis and anergy of CD4+CD25+ suppressor T cells in vivo. Nat. Immunol. 3:33-41. [DOI] [PubMed] [Google Scholar]
- 7.Gilliet, M., and Y. J. Liu. 2002. Generation of human CD8 T regulatory cells by CD40 ligand-activated plasmacytoid dendritic cells. J. Exp. Med. 195:695-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray, J. D., M. Hirokawa, K. Ohtsuka, and D. A. Horwitz. 1998. Generation of an inhibitory circuit involving CD8+ T cells, IL-2, and NK cell-derived TGF-β: contrasting effects of anti-CD2 and anti-CD3. J. Immunol. 160:2248-2254. [PubMed] [Google Scholar]
- 9.Greenwald, I., and G. M. Rubin. 1992. Making a difference: the role of cell-cell interactions in establishing separate identities for equivalent cells. Cell 68:271-281. [DOI] [PubMed] [Google Scholar]
- 10.Groux, H. 2001. An overview of regulatory T cells. Microbes Infect. 3:883-889. [DOI] [PubMed] [Google Scholar]
- 11.Hoyne, G. F., I. Le Roux, M. Corsin-Jimenez, K. Tan, J. Dunne, L. M. Forsyth, M. J. Dallman, M. J. Owen, D. Ish-Horowicz, and J. R. Lamb. 2000. Serrate1-induced notch signalling regulates the decision between immunity and tolerance made by peripheral CD4+ T cells. Int. Immunol. 12:177-185. [DOI] [PubMed] [Google Scholar]
- 12.Jarriault, S., O. Le Bail, E. Hirsinger, O. Pourquie, F. Logeat, C. F. Strong, C. Brou, N. G. Seidah, and A. Israël. 1998. Delta-1 activation of Notch-1 signaling results in HES-1 transactivation. Mol. Cell. Biol. 18:7423-7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, P., G. May, L. Healy, J. Brown, G. Hoyne, S. Delassus, and T. Enver. 1998. Stromal expression of Jagged 1 promotes colony formation by fetal hematopoietic progenitor cells. Blood 92:1505-1511. [PubMed] [Google Scholar]
- 14.Jonuleit, H., E. Schmitt, G. Schuler, J. Knop, and A. H. Enk. 2000. Induction of interleukin 10-producing, nonproliferating CD4+ T cells with regulatory properties by repetitive stimulation with allogeneic immature human dendritic cells. J. Exp. Med. 192:1213-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonuleit, H., E. Schmitt, M. Stassen, A. Tuettenberg, J. Knop, and A. H. Enk. 2001. Identification and functional characterization of human CD4+CD25+ T cells with regulatory properties isolated from peripheral blood. J. Exp. Med. 193:1285-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karanu, F. N., B. Murdoch, L. Gallacher, D. M. Wu, M. Koremoto, S. Sakano, and M. Bhatia. 2000. The Notch ligand Jagged-1 represents a novel growth factor of human hematopoietic stem cells. J. Exp. Med. 192:1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimble, J., and P. Simpson. 1997. The LIN-12/Notch signaling pathway and its regulation. Annu. Rev. Cell Dev. Biol. 13:333-361. [DOI] [PubMed] [Google Scholar]
- 18.Kohm, A. P., P. A. Carpentier, H. A. Anger, and S. D. Miller. 2002. Cutting edge: CD4+CD25+ regulatory T cells suppress antigen-specific autoreactive immune responses and central nervous system inflammation during active experimental autoimmune encephalomyelitis. J. Immunol. 169:4712-4716. [DOI] [PubMed] [Google Scholar]
- 19.Lakhdar, M., A. Senik, and W. H. Fridman. 1984. Human cytotoxic T lymphocytes (CTL) against Epstein-Barr virus (EBV) infected cells: EBV specificity and involvement of major histocompatibility complex determinants in the lysis exerted by anti-EBV CTL toward HLA-compatible and allogeneic target cells. Cell Immunol. 83:414-421. [DOI] [PubMed] [Google Scholar]
- 20.Lie, Y. S., and C. J. Petropoulos. 1998. Advances in quantitative PCR technology: 5′ nuclease assays. Curr. Opin. Biotechnol. 9:43-48. [DOI] [PubMed] [Google Scholar]
- 21.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔC(T) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 22.Maloy, K. J., and F. Powrie. 2001. Regulatory T cells in the control of immune pathology. Nat. Immunol. 2:816-822. [DOI] [PubMed] [Google Scholar]
- 23.Masuyama, J., S. Kaga, S. Kano, and S. Minota. 2002. A novel costimulation pathway via the 4C8 antigen for the induction of CD4+ regulatory T cells. J. Immunol. 169:3710-3716. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. F., and A. Basten. 1996. Mechanisms of tolerance to self. Curr. Opin. Immunol. 8:815-821. [DOI] [PubMed] [Google Scholar]
- 25.Milner, L. A., and A. Bigas. 1999. Notch as a mediator of cell fate determination in hematopoiesis: evidence and speculation. Blood 93:2431-2448. [PubMed] [Google Scholar]
- 26.Ng, W. F., P. J. Duggan, F. Ponchel, G. Matarese, G. Lombardi, A. D. Edwards, J. D. Isaacs, and R. I. Lechler. 2001. Human CD4+CD25+ cells: a naturally occurring population of regulatory T cells. Blood 98:2736-2744. [DOI] [PubMed] [Google Scholar]
- 27.Read, S., and F. Powrie. 2001. CD4+ regulatory T cells. Curr. Opin. Immunol. 13:644-649. [DOI] [PubMed] [Google Scholar]
- 28.Rickinson, A. B., S. P. Lee, and N. M. Steven. 1996. Cytotoxic T lymphocyte responses to Epstein-Barr virus. Curr. Opin. Immunol. 8:492-497. [DOI] [PubMed] [Google Scholar]
- 29.Robey, E. 1997. Notch in vertebrates. Curr. Opin. Genet. Dev. 7:551-557. [DOI] [PubMed] [Google Scholar]
- 30.Robey, E. 1999. Regulation of T cell fate by Notch. Annu. Rev. Immunol. 17:283-295. [DOI] [PubMed] [Google Scholar]
- 31.Roncarolo, M. G., R. Bacchetta, C. Bordignon, S. Narula, and M. K. Levings. 2001. Type 1 T regulatory cells. Immunol. Rev. 182:68-79. [DOI] [PubMed] [Google Scholar]
- 32.Rooney, C. M., C. A. Smith, C. Y. Ng, S. Loftin, C. Li, R. A. Krance, M. K. Brenner, and H. E. Heslop. 1995. Use of gene-modified virus-specific T lymphocytes to control Epstein-Barr-virus-related lymphoproliferation. Lancet 345:9-13. [DOI] [PubMed] [Google Scholar]
- 33.Rooney, C. M., C. A. Smith, C. Y. Ng, S. K. Loftin, J. W. Sixbey, Y. Gan, D. K. Srivastava, L. C. Bowman, R. A. Krance, M. K. Brenner, and H. E. Heslop. 1998. Infusion of cytotoxic T cells for the prevention and treatment of Epstein-Barr virus-induced lymphoma in allogeneic transplant recipients. Blood 92:1549-1555. [PubMed] [Google Scholar]
- 34.Santin, A. D., P. L. Hermonat, A. Ravaggi, S. Bellone, S. Pecorelli, J. J. Roman, G. P. Parham, and M. J. Cannon. 2000. Interleukin-10 increases Th1 cytokine production and cytotoxic potential in human papillomavirus-specific CD8+ cytotoxic T lymphocytes. J. Virol. 74:4729-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz, R. H. 1996. Models of T cell anergy: is there a common molecular mechanism? J. Exp. Med. 184:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shayakhmetov, D. M., and A. Lieber. 2000. Dependence of adenovirus infectivity on length of the fiber shaft domain. J. Virol. 74:10274-10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens, L. A., C. Mottet, D. Mason, and F. Powrie. 2001. Human CD4+CD25+ thymocytes and peripheral T cells have immune suppressive activity in vitro. Eur. J. Immunol. 31:1247-1254. [DOI] [PubMed] [Google Scholar]
- 38.Stewart, J. P., and C. M. Rooney. 1992. The interleukin-10 homolog encoded by Epstein-Barr virus enhances the reactivation of virus-specific cytotoxic T cell and HLA-unrestricted killer cell responses. Virology 191:773-782. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi, T., T. Tagami, S. Yamazaki, T. Uede, J. Shimizu, N. Sakaguchi, T. W. Mak, and S. Sakaguchi. 2000. Immunologic self-tolerance maintained by CD25+CD4+ regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J. Exp. Med. 192:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thornton, A. M., and E. M. Shevach. 1998. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J. Exp. Med. 188:287-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varnum-Finney, B., L. E. Purton, M. Yu, C. Brashem-Stein, D. Flowers, S. Staats, K. A. Moore, R. Le, I., R. Mann, G. Gray, S. Artavanis-Tsakonas, and I. D. Bernstein. 1998. The Notch ligand, Jagged-1, influences the development of primitive hematopoietic precursor cells. Blood 91:4084-4091. [PubMed] [Google Scholar]
- 42.Varnum-Finney, B., L. Xu, C. Brashem-Stein, C. Nourigat, D. Flowers, S. Bakkour, W. S. Pear, and I. D. Bernstein. 2000. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat. Med. 6:1278-1281. [DOI] [PubMed] [Google Scholar]
- 43.Yamagiwa, S., J. D. Gray, S. Hashimoto, and D. A. Horwitz. 2001. A role for TGF-β in the gene ration and expansion of CD4+CD25+ regulatory T cells from human peripheral blood. J. Immunol. 166:7282-7289. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto, N., S. Yamamoto, F. Inagaki, M. Kawaichi, A. Fukamizu, N. Kishi, K. Matsuno, K. Nakamura, G. Weinmaster, H. Okano, and M. Nakafuku. 2001. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J. Biol. Chem. 276:45031-45040. [DOI] [PubMed] [Google Scholar]
- 45.Yotnda, P., H. Onishi, H. E. Heslop, D. Shayakhmetov, A. Lieber, M. Brenner, and A. Davis. 2001. Efficient infection of primitive hematopoietic stem cells by modified adenovirus. Gene Ther. 8:930-937. [DOI] [PubMed] [Google Scholar]
- 46.Zheng, S. G., J. D. Gray, K. Ohtsuka, S. Yamagiwa, and D. A. Horwitz. 2002. Generation ex vivo of TGF-β-producing regulatory T cells from CD4+. J. Immunol. 169:4183-4189. [DOI] [PubMed] [Google Scholar]
- 47.Zinkernagel, R. M., S. Ehl, P. Aichele, S. Oehen, T. Kundig, and H. Hengartner. 1997. Antigen localisation regulates immune responses in a dose- and time-dependent fashion: a geographical view of immune reactivity. Immunol. Rev. 156:199-209. [DOI] [PubMed] [Google Scholar]