Abstract
The envelope (Env) glycoprotein of human immunodeficiency virus type 1 (HIV-1) is the major target of neutralizing antibody responses and is likely to be a critical component of an effective vaccine against AIDS. Although monomeric HIV envelope subunit vaccines (gp120) have induced high-titer antibody responses and neutralizing antibodies against laboratory-adapted HIV-1 strains, they have failed to induce neutralizing antibodies against diverse heterologous primary HIV isolates. Most probably, the reason for this failure is that the antigenic structure(s) of these previously used immunogens does not mimic that of the functional HIV envelope, which is a trimer, and thus these immunogens do not elicit high titers of relevant functional antibodies. We recently reported that an Env glycoprotein immunogen (o-gp140SF162ΔV2) containing a partial deletion in the second variable loop (V2) derived from the R5-tropic HIV-1 isolate SF162, when used in a DNA priming-protein boosting vaccine regimen in rhesus macaques, induced neutralizing antibodies against heterologous subtype B primary isolates as well as protection to the vaccinated animals upon challenge with pathogenic SHIVSF162P4 virus. Here we describe the purification of this protein to homogeneity, its characterization as trimer, and its ability to induce primary isolate-neutralizing responses in rhesus macaques. Optimal mutations in the primary and secondary protease cleavage sites of the env gene were identified that resulted in the stable secretion of a trimeric Env glycoprotein in mammalian cell cultures. We determined the molecular mass and hydrodynamic radius (Rh) using a triple detector analysis (TDA) system. The molecular mass of the oligomer was found to be 324 kDa, close to the expected Mw of a HIV envelope trimer protein (330 kDa), and the hydrodynamic radius was 7.27 nm. Negative staining electron microscopy of o-gp140SF162ΔV2 showed that it is a trimer with considerable structural flexibility and supported the data obtained by TDA. The structural integrity of the purified trimeric protein was also confirmed by determinations of its ability to bind the HIV receptor, CD4, and its ability to bind a panel of well-characterized neutralizing monoclonal antibodies. No deleterious effect of V2 loop deletion was observed on the structure and conformation of the protein, and several critical neutralization epitopes were preserved and well exposed on the purified o-gp140SF162ΔV2 protein. In an intranasal priming and intramuscular boosting regimen, this protein induced high titers of functional antibodies, which neutralized the vaccine strain, i.e., SF162. These results highlight a potential role for the trimeric o-gp140SF162ΔV2 Env immunogen in a successful HIV vaccine.
AIDS continues to be a major health problem throughout the world, with approximately 40 million cases and 4 million deaths having been recorded so far. In certain parts of the world, nearly 30% of the total population is infected with human immunodeficiency virus (HIV). An estimated 40,000 to 80,000 primary infections occur each year in the United States alone. The situation is deteriorating as a result of rapid emergence of drug resistance against most of the effective antivirals. Therefore, there is an urgent need for an effective anti-HIV vaccine that may be used alone for protection or in conjunction with effective antivirals for treatment.
HIV Env glycoprotein is the major target of neutralizing antibody responses and thus will be an important component of a vaccine. Historically, it has been difficult to induce neutralizing antibody responses against diverse primary HIV type 1 (HIV-1) strains by utilizing monomeric HIV Env (i.e., gp120) glycoprotein as a vaccine. Therefore, it seems that there are differences between qualities of antibodies induced during the natural course of infection and those induced through vaccination. This is not due to the absence of conserved neutralization epitopes on monomeric gp120 proteins (81) but is most likely due to their level of exposure and state of presentation on monomeric Env, compared to the level and state on native trimeric Env, present on the surface of virion (37, 51, 59, 69, 77, 87). Structural studies indicated that HIV Env is present as a trimer on the surface of the virion, whereas monomers have been used for immunization (32, 84, 88). These conformational differences may affect the exposure of critical neutralizing epitopes on recombinant gp120 compared to those on native Env. In general, compared to nonneutralizing antibodies, broadly neutralizing antibodies have stronger affinities for the native envelope (trimer) (7, 8, 26, 52, 53, 60, 63, 69, 70), suggesting that the neutralizing epitopes recognized by these broadly reactive monoclonal antibodies (MAbs) are better represented on oligomeric Env. Therefore, efforts have been made to purify the HIV Env protein in its functional, trimeric conformation and evaluate its ability to induce primary isolate-neutralizing antibodies in animal models. Earlier studies have suggested that HIV Env oligomers, consisting of gp120 and the ectodomain of gp41, might be superior to monomeric gp120 for inducing strong humoral responses toward conformational epitopes (6, 18, 62, 83). In addition, antibodies induced by oligomeric Env cross-reacted with HIV envelopes of other subtypes and neutralized both T cell-adapted and some primary isolates of HIV-1 (22, 83, 92). More recent evidence has further demonstrated that oligomers may be superior at inducing primary isolate-neutralizing antibodies compared to monomers (22, 92). Therefore, trimeric HIV Env has the potential to be a better immunogen for inducing broadly cross-reactive neutralizing antibodies.
The high degree of glycosylation (approximately 50% of the molecular mass) of HIV Env is critical not only for the intracellular transport of the envelope protein, but also for providing correct functional conformation for its binding to CD4. Glycosylation also can mask T- and B-cell epitopes and thereby may decrease Env immunogenicity. Therefore, it may be that the extensive glycosylation is used by the virus not only for invading the new cells, but also as an immune shield to mask critical neutralizing epitopes. Recent evidence from simian immunodeficiency virus (SIV) and simian-human immunodeficiency virus (SHIV) rhesus infection models suggests that the emergence of viruses that are resistant to antibody-mediated neutralization is associated with modification of the glycosylation profiles and that resistance is at least partially due to the alteration in the glycosylation sites within the viral envelope (13, 41, 55-57, 66). Therefore, one strategy to increase the immunogenicity of Env would be to remove glycosylation sites without affecting the structure and conformation of the Env protein to expose more neutralizing epitopes. It was demonstrated that elimination of specific N-linked glycosylation sites from variable loops of the HIV and SIV models renders these mutant viruses more susceptible to neutralization by human or animal sera as well as by specific MAbs (1, 9, 33, 42, 44, 61, 72, 76). An important observation for HIV vaccine development is that sera collected from macaques infected with partially deglycosylated SIVmac239 viruses from which the V1 loop has been deleted have a higher neutralization potential against the parental fully glycosylated SIVmac239 viruses than the sera collected from macaques infected with the parental virus itself (61). Furthermore, a mutant of the SF162 envelope protein lacking 30 out of 40 amino acids of the V2 loop (termed ΔV2) appeared to be more effective in eliciting cross-reactive neutralizing antibodies than the parental SF162 envelope (2). On the background of the SF162 envelope, partial deletion of the V2 loop appears to increase the exposure and immunogenicity of certain neutralization epitopes. Since the oligomeric soluble ΔV2 envelope elicits cross-reactive neutralizing antibodies and such antibodies can reduce viral replication in macaques challenged with the pathogenic R5 SHIVSF162P4 isolate, we believe that this immunogen could be included in an anti-HIV vaccine (2, 14, 15).
Here we describe the purification and biochemical, biophysical, structural, and immunochemical characterization of soluble oligomeric gp140 protein containing a partial deletion in V2 loop derived from the well-characterized HIV-1 SF162 envelope (termed o-gp140SF162ΔV2). Our electron microscopic (EM) studies indicate that the purified protein is in a stable trimeric conformation, and immunochemical studies indicate that critical neutralizing epitopes are exposed and well preserved. Moreover, in an intranasal priming and intramuscular boosting regimen, o-gp140SF162ΔV2 protein induced high titers of antibodies in rhesus macaques, which neutralized the homologous primary isolate, i.e., SF162.
MATERIALS AND METHODS
Envelope plasmid construction.
The sequences encoding the open reading frame of the ectodomain of the Env protein from the HIV-1 SF162 and HIV-1 SF162ΔV2 isolates were codon modified as described elsewhere (30, 93), and constructed synthetically as a 2.1-kb EcoRI-XbaI DNA fragment. This gene cassette contained the protein-encoding region of the Env protein fused in frame to the human tissue plasminogen activator signal sequence (12). In order to stabilize the oligomeric structure of the encoded gp140 protein, we introduced a series of mutations in the primary (REKR) and secondary protease (KAKRR) cleavage sites in the Env polypeptide (21). The details of these mutations are presented in Fig. 1. The resulting Env expression cassette (gp140) was cloned into the EcoRI-XbaI sites of the pCMV3 expression vector for transient transfection of 293 cells and also for the development of stable CHO cell lines. This vector contains the cytomegalovirus enhancer/promoter elements, an ampicillin resistance gene, and sequences encoding a fusion protein composed of dihydrofolate reductase and an attenuated neomycin resistance protein.
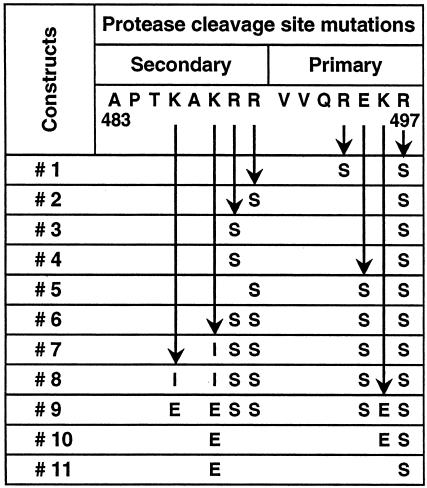
FIG. 1.
Mutations in the primary (REKR) and secondary protease (KAKRR) cleavage sites in the Env polypeptide.
Transient transfection of 293 cells.
In each well of a six-well plate, 1 × 106 293 cells were plated. At approximately 70% confluence, the cells were washed with DPBS (Mediatech, Inc., Herdon, Va.), resuspended in 3 ml of OPTI-MEM (Gibco, Grand Island, N.Y.), and incubated with DNA/LT1 complex (5 μl of LT1/μg of DNA) for 5 h. At the end of the incubation period, the cells were resuspended in regular MEM (Mediatech) containing 10% fetal bovine serum and incubated for 48 h. At the end of the incubation period, medium was collected and 1 mM EDTA and 1 mM EGTA were added before the performance of small-scale protein enrichment for expression and structural analysis.
Development of stable CHO cell lines secreting ο−gp140SF162ΔV2.
Stable CHO cell lines secreting gp140SF162ΔV2 were derived by using DG-44 cells with a double deletion mutation in the dihydrofolate reductase gene, thus making the cell line dependent on the addition of hypoxanthine, glycine, and thymidine to the growth medium, following the experimental protocol described elsewhere in detail (75).
Production of o-gp140 SF162ΔV2.
The three highest expressing gp140SF162ΔV2-CHO cell clones were used to seed a 3-liter bioreactor (one for each clone) for adaptation to medium containing a very low level of serum (0.5%) and to a cell density and perfusion rate that facilitated the maximum expression of gp140SF162ΔV2 in its oligomeric conformation. Bioreactors were monitored daily for cell density, pH, CO2, and O2 concentration, etc. The structure, conformation, and expression levels of secreted gp140SF162ΔV2 were monitored weekly. From the three bioreactor-adapted clones, the best producer clone was used to seed a 12.5-liter bioreactor for production run. At the end of the run, collected medium was concentrated 20-fold through a 100-kDa-pore-size membrane filter and stored at −80°C in presence of 1 mM EDTA and 1 mM EGTA.
Envelope antigen capture ELISA.
Envelope antigen capture enzyme-linked immunosorbent assay (ELISA) was performed by following standard protocol described elsewhere in detail (75). gp140SF162ΔV2 concentration was calculated from a standard curve derived using serial dilutions of a known concentration of recombinant gp120SF162.
Purification of o-gp140SF162ΔV2.
The concentrated CHO cell supernatant was thawed in presence of complete protease inhibitor cocktail (Boehringer Mannheim) at 4°C, and after its conductivity was adjusted, it was loaded onto a Galanthus Nivalis-agarose column (GNA) (Vector Laboratories, Burlingame, Calif.) equilibrated with 20 mM Tris-100 mM NaCl (pH 7.4). Bound gp140 was eluted with 500 mM methyl mannose pyranoside (Sigma Chemical Co., St. Louis, Mo.) in equilibration buffer. The eluate after the GNA column was loaded onto a DEAE (Pharmacia) column equilibrated with buffer (20 mM Tris, 100 mM NaCl [pH 8.0]). Under these conditions, o-gp140ΔV2 did not bind to the column, but contaminating proteins were retained on the column. The DEAE flowthrough was adjusted to 10 mM PO4 concentration, pH was adjusted to 6.8, and the flowthrough was loaded onto a ceramic hydroxyapatite (CHAP) (Bio-Rad Laboratories, Hercules, Calif.) column equilibrated with buffer (10 mM Na2HPO4, 100 mM NaCl [pH 6.8]). Under these conditions, gp140ΔV2 did not bind to CHAP column and was recovered in the flowthrough. All the column fractions were analyzed on polyacrylamide gels (native and reduced and denatured) and also in a CD4 receptor-binding assay (see below). The fractions showing strong CD4-binding activity were pooled and fractionated on a 16- by 90-mm Superdex-200 column (Pharmacia) equilibrated with 10 mM NaCitrate containing 300 mM NaCl. Fractions containing o-gp140ΔV2 were pooled, concentrated using a Stir cell (Millipore, Inc, Bedford, Mass.), and stored frozen at −80°C. During the purification process, fractions were analyzed by polyacrylamide gel electrophoresis (PAGE) both under reducing and denaturing and under native conditions following standard methods. Gels were stained with Coomassie brilliant blue or processed for immunoblotting.
Biophysical characterization of o-gp140SF162ΔV2.
The molecular mass (Mw), hydrodynamic radius (Rh), and intrinsic viscosity of o-gp140SF162ΔV2 were determined using a triple detector array (TDA) system (TDA 300, Viscotek Corp. Houston, Tex.) in conjunction with a precalibrated gel filtration high-performance liquid chromatography (HPLC) column (Bio Sil SEC-250; Bio-Rad Laboratories) using the Alliance 2690 HPLC system (Waters Corporation, Milford, Mass.). Approximately 100 μg of the purified protein (o-gp140SF162ΔV2 and gp120SF162ΔV2) was analyzed in 20 mM NaH2PO4-2 mM Na2HPO4 buffer at pH 7.4 at the flow rate of 1 ml/min. The signals from light scattering, a viscometer, and refractive index detectors were collected and analyzed using the EZ Pro software package (Viscotek Corp. Houston, Tex.).
Immunoblot detection of o-gp140SF162ΔV2.
Approximately 0.5 to 1 μg of o-gp140ΔV2 was separated on sodium dodecyl sulfate (SDS)-PAGE. After electrophoresis, the gel was treated with 50 mM Tris-HCl (pH 8.0) containing 20% glycerol for 30 min at room temperature. Proteins were transferred on to a 0.22-μM nitrocellulose membrane using a semidry transfer system (Bio-Rad Laboratories). Immunodetection of o-gp140ΔV2 was performed exactly as described previously using anti-gp120 C4 domain-specific MAb 20-2-C8.5F3 (74; Steimer et al., unpublished observations) with minor modifications.
CD4 binding assay.
To determine the capability of purified envelope protein to bind CD4, we used an HPLC-based assay with fluoresceinated CD4, as described elsewhere in detail (75).
Glycosylation profile analysis.
Enzymatic deglycosylation of the purified HIV-Env proteins was performed using reagents from Bio-Rad Research Laboratories (catalog no. 170-6500). To detect N-linked oligosaccharides, 25 to 100 μg of purified protein was treated with 2 μl of peptidyl-N glycosidase F (PNGF) (2.5 U/ml) for 1 h at 37°C. To study O-linked oligosaccharides, 25 μg of the purified protein was treated with 2 μl of NANase II (10 U/ml) and 2 μl of O-glycosidase (1 U/ml) at 37°C for 1 h. In addition, we have also performed Endo-H digestion of the purified protein following manufacturer's protocol (Boehringer Mannheim). Enzyme-treated samples were diluted in sample buffer containing 0.1% SDS and β-mercaptoethanol and analyzed by SDS-PAGE.
Immunochemical characterization of o-gp140SF162ΔV2.
The binding of well-characterized HIV-Env-specific MAbs to o-gp140SF162ΔV2 and gp120SF162 proteins was performed by a capture ELISA. Purified proteins were adsorbed on ELISA wells by an overnight incubation at room temperature (100 ng/well). The adsorbed proteins were incubated with a serial dilution of various MAbs (0.01 to 10 μg/ml in SuperBlock-0.05% Tween-20) for 4 h 37°C. For each MAb concentration, duplicate wells were used. The wells were washed with Tris-buffered saline and incubated with goat anti-human immunoglobulin G (IgG) coupled to alkaline phosphatase (Zymed) (1:30,000 dilution in SuperBlock-0.05% Tween-20) for an overnight incubation at 4°C. Substrate for alkaline phosphatase (DAKO Corp.) was added (50 μl/well) for 1 h at room temperature. Finally, 50 μl of amplifier (DAKO Corp.) was added per well, and absorbance was recorded at 490 nm using a Vmax microplate reader (Molecular Devices). A plot of the absorbance at 490 nm versus the MAb concentration was generated.
Surface plasmon resonance assay.
Surface plasmon resonance assays were performed using a BIACORE 3000 optical biosensor system (Biacore AB, Uppsala, Sweden) with simultaneous monitoring of relevant flow cells. To perform the kinetic study of the binding of o-gp140SF162ΔV2 to sCD4, sCD4 was immobilized using carbodiimide coupling onto a low-charge B1 sensor chip to attain a response of 2,000 response units. Using PBS buffer (pH 7.4) with 0.05% Tween 20, association was assessed by passing gp120SF162 or gp140SF162ΔV2 over the chip surface at a flow rate of 30 μl/min. The concentration ranges for gp120SF162 and gp140SF162ΔV2 analytes were 59 to 300 nM (59, 89, 133, 200, and 300 nM) and 4.4 to 33 nM (4.4, 6.3, 9.6, 14, 22, and 33 nM), respectively. As a negative control, we used a control surface without sCD4. For kinetic analysis, Biosensor data were prepared by subtracting binding responses obtained from a blank reference surface (without sCD4) from the specific responses. The association and dissociation phase data were fitted simultaneously to a single-site binding model by using the nonlinear data analysis program Biaevaluation 3.2.
Immunoelectron microscopy.
EM analysis of the oligomeric and monomeric gp140SF162ΔV2 was performed by negative staining as previously described (64, 65). The oligomeric (see Fig. 5A, peak A) and monomeric (see Fig. 5A, peak B) proteins obtained after the final preparative sizing column were diluted in borate buffered saline, affixed to carbon membranes, stained with 1% uranyl formate, and mounted on copper grids for analysis. EMs were recorded at a nominal magnification of ×100,000 at 100 kV on a JEOL JEM 1200EX electron microscope. Measurements were made using the image analysis software Image-Pro Plus. Fifty or more particles from fractions corresponding to the putative monomer/dimer and trimer of gp140SF162ΔV2 were measured and analyzed statistically.
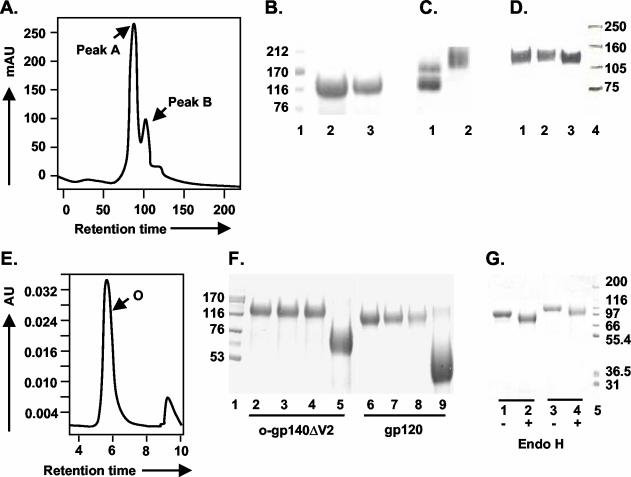
FIG. 5.
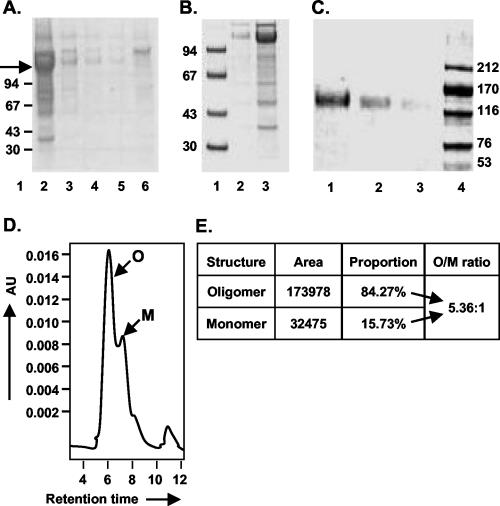
Purification and glycosylation linkage analysis of o-gp140SF162ΔV2. (A) gp140SF162ΔV2 obtained after the CHAP column was further fractionated on a precalibrated Superdex-200 sizing column to separate oligomers from the dimers and/or monomers of gp140. Peaks corresponding to oligomer (peak A) and dimer or monomer (peak B) are indicated. (B and C) Polyacrylamide gel analysis of the sizing fractions in reducing and denaturing conditions (B) (lanes: 1, molecular weight standards; 2, o-gp140SF162ΔV2; 3, gp140SF162ΔV2 dimer/monomer) and native conditions (C) (lanes: 1, dimer and monomer; 2, oligomer). (D) Immunodetection of o-gp140 using a MAb (20-2-C8.5F3) directed against the C4 domain of gp120 SF2 (lanes: 1, o-gp140SF162ΔV2;2, gp140SF162ΔV2 monomer; 3, gp120SF2). (E) Size exclusion-HPLC profile of the purified o-gp140SF162ΔV2. (F) Carbohydrate linkage analysis of purified o-gp140SF162ΔV2. Lanes: 3 and 7, o-gp140SF162ΔV2 and gp120SF162 digested with NANase; 4 and 8, O-glycosidase; 5 and 9, PNGF; 2 and 6, o-gp140SF162ΔV2 and gp120SF162 without any enzyme, used as controls; 1, molecular weight standards. (G) Endo-H digestion of gp120SF162 (lanes 1 and 2) and o-gp140SF162ΔV2 (lanes 3 and 4). + and −, presence or absence of Endo-H.
Stability studies.
Purified o-gp140 SF162ΔV2 protein collected after the final gel filtration column was used for performing the stability studies. Approximately 2 to 5 μg of the purified protein was incubated at different temperatures (25 to 100°C) for 1 h. Following this treatment the protein was analyzed by CD4 binding, size-exclusion chromatography (SEC)-HPLC, SDS-PAGE, and immunoprobing.
Antibody ELISA and virus neutralization assays.
Antibody ELISA was performed on serum samples collected 2 weeks every immunization, essentially as described previously (75).
Neutralization of homologous (SF162) and heterologous (Bal, JR-FL, and Bx08) viruses was measured in 5.25.EGFP.Luc.M7 (M7-luc) cells obtained from Nathaniel Landau. The format of this assay was identical to that of the MT-2 assay described elsewhere (50) except that virus infection was quantified by luciferase reporter gene expression using a commercial luciferase kit (Promega). In this assay, the reduction in luciferase activity is proportional to the reduction in virus input. All serum samples were heat inactivated for 1 h at 56°C prior to assay. The virus stocks of all the HIV-1 isolates (SF162, Bal, JR-FL, and Bx08) were generated in peripheral blood mononuclear cells. Neutralizing antibody titers are reported as the reciprocal serum dilution at which 80% reduction in luciferase activity was observed.
RESULTS
Expression and stabilization of o-gp140SF162ΔV2.
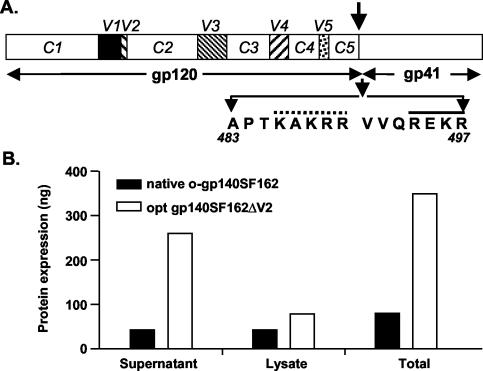
The sequence encoding a codon-optimized gp140SF162ΔV2 with a single mutation in the protease cleavage site (R at position 497 was changed to S) was cloned into an expression vector. The linear map of gp120 and gp41 is shown in Fig. 2A. To facilitate the efficient secretion of recombinant o-gp140ΔV2 protein, the native HIV signal sequence was replaced by the human tissue type plasminogen activator signal sequence. The effect of codon optimization on the extent of gp140SF162ΔV2 expression was evaluated by transiently transfecting 293 cells with native gp140 (non-codon optimized) and codon-optimized gp140SF162ΔV2 constructs and comparing the expression levels in the cell supernatants and cell lysates by capture ELISA (Fig. 2B) and immunoblotting (data not shown). It was shown previously that sequence modification of HIV gag and env significantly improves the level of expression (75, 93). Similarly, codon modifications also improved gp140SF162ΔV2 expression eightfold compared to that of the wild-type sequence construct (Fig. 2B). In addition, deletion of the V2 loop did not affect the level of expression. Using similar sequence-modified constructs, we have produced monomeric gp120 proteins derived from SF162 and SF162ΔV2.
FIG. 2.
Structure, expression, and stabilization of HIV-1 SF162ΔV2 envelope glycoprotein in oligomeric conformation. (A) Linear map of the HIV-1 gp120ΔV2 and gp41 envelope glycoproteins. The gp120 variable regions (V1 to V5) are indicated as solid squares, and arginine 483-to-serine 497 mutation in the protease cleavage site between gp120 and gp41 is indicated by an arrow. The primary (solid line) and secondary cleavage (dotted line) sites are indicated. The gp41 portion includes the N and C α helical regions known as oligomerization domain. (B) Effect of sequence modification upon expression of gp140ΔV2. Total protein expression obtained from native and sequence-modified constructs by using capture ELISA in different fractions is presented as nanograms/well.
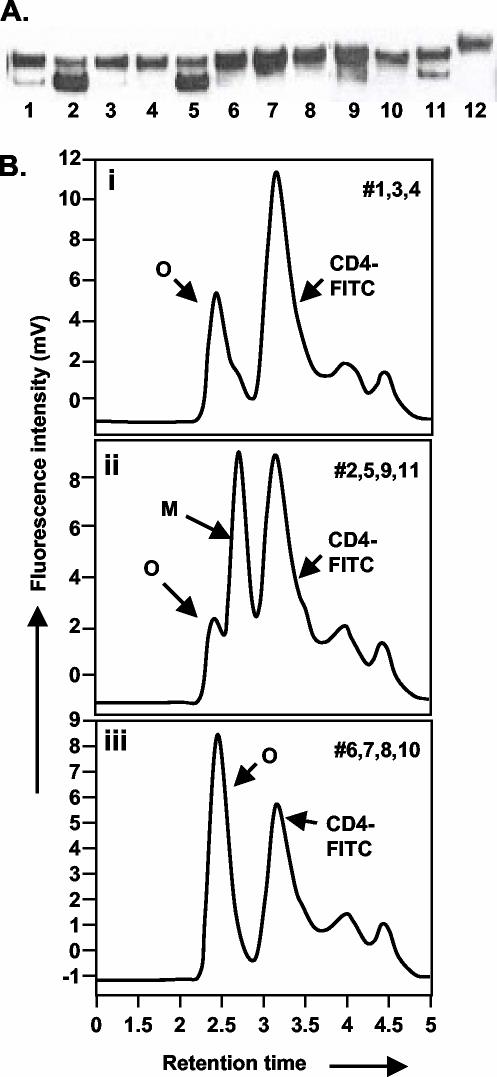
We have previously shown that introduction of a single mutation (arginine to serine) in the protease cleavage site in HIV-1 Env gene from US4, a subtype B primary isolate, was efficient at abolishing the protease cleavage site and led to the efficient secretion of stable oligomers (75). However, the same R-to-S mutation was relatively ineffective in eliminating the protease cleavage of the HIV-1 SF162 Env, and therefore monomers were the predominant form secreted (>90%) (data not shown). This indicated that the protease sensitivities at the cleavage sites differ among Env proteins derived from different primary HIV-1 isolates, and the mutations required for the secretion of stable oligomers thus differ as well. Therefore, we scanned different combinations of mutations in the primary (aa 494 to 497 [REKR]) and secondary (aa 486 to 490 [KAKRR]) protease cleavage sites in the Env protein of ΔV2 SF162 to evaluate their relative abilities to stabilize the oligomeric structure without compromising expression levels (Fig. 1). Based upon sCD4 binding in conjunction with SEC, a combination of three to seven amino acid changes in the primary and secondary protease cleavage sites (constructs 6, 7, 8, and 10) was very effective at stabilizing the oligomer (Fig. 3A and B). The highest expression, as determined by immunoblotting, was observed for construct 7, which had a total of five mutations, of which two were in primary (E495—S, R497—S) cleavage sites and three in the secondary cleavages site (K488—I, R489—S, and R490—S), as shown in Fig. 1. Hence, construct 7 was used to derive stable CHO cell lines secreting 3 to 5 μg of o-gp140SF162ΔV2/ml.
FIG. 3.
Protease cleavage site scanning for optimum mutations required for stabilization of HIV-1 SF162ΔV2 oligomers. (A) Immunoprobing data. Lanes 1 to 11 correspond to constructs 1 to 11 in Fig. 1; lane 12 is o-gp140 US4. (B) A representative CD4 binding profile obtained for protease cleavage site mutations constructs. Profile i was obtained for constructs 1, 3, and 4; profile ii was obtained for constructs 2, 5, 9, and 11; profile iii was obtained for constructs 6, 7, 8, and 10. Peaks representing oligomer (O), monomer (M), and CD4 are indicated. Based on the structure and expression data, we selected construct no. 7 for developing stable CHO cell lines.
Purification of o-gp140SF162ΔV2.
A three-step strategy was designed for efficient purification of o-gp140SF162ΔV2. First, concentrated cell supernatants were passed over a GNA lectin column. The gp140SF162ΔV2 protein bound to the column, and most contaminating proteins flowed through. The bound gp140SF162ΔV2 was eluted with 500 mM methyl mannose pyranoside. Next, the captured gp140SF162ΔV2 protein was passed over DEAE and CHAP columns. Contaminating proteins bound on one column or the other, but gp140SF162ΔV2 flowed through over both the columns. The fractions collected from each column were analyzed by SDS-PAGE, immunoblot, SEC, and a CD4-binding assay (Fig. 4). The GNA lectin was quite efficient at capturing the gp140SF162ΔV2, but there was substantial contamination with other nonrelated glycoproteins (Fig. 4A). However, a significant reduction in the number and concentration of contaminating proteins was achieved by passing the GNA lectin-captured gp140SF162ΔV2 over the DEAE (Fig. 4B) and CHAP columns (approximately 95% pure) (Fig. 4C). The gp140SF162ΔV2 obtained after the CHAP column was concentrated using 100-kDa-pore-size membrane and analyzed over BioSil 250 SEC (Fig. 4D). Based on the SEC profile, the purified gp140SF162ΔV2 preparation had approximately 80% oligomer and 20% monomer (Fig. 4E). Therefore, at the final stage of purification, o-gp140SF162ΔV2 was separated from gp140SF162ΔV2 monomer on a higher resolution, precalibrated Superdex-200 column. The SEC profile is presented in Fig. 5A. Each of the fractions constituting peak A and B migrated with an apparent molecular mass of 120 kDa on reducing and denaturing SDS-PAGE (Fig. 5B). No change in the electrophoretic mobility of these fractions was observed under denaturing conditions versus reducing and denaturing conditions (data not shown), suggesting that gp140SF162ΔV2 monomers were noncovalently associated with each other to give rise to oligomers. To ensure that the oligomers were not disulfide-linked aggregates, the purified oligomers were analyzed at increasing concentrations of β-mercaptoethanol, and no changes in the migration profile were observed, indicating that they were not disulfide-linked aggregates (data not shown). Based on the relative mobilities of fractions in peaks A and B, it appeared that peak A predominantly contained Env protein oligomers that potentially could be trimers and peak B contained Env protein predominantly in monomeric conformation (Fig. 5C). Purified o-gp140SF162ΔV2 was recognized as a single species on Western blots using MAb 20-2-C8.5F3 directed against the conserved (C4) domain of gp120 (Fig. 5D), suggesting that there is no major degradation or proteolysis of o-gp140SF162ΔV2 (Fig. 5D), and the purified preparation was homogeneous. Furthermore, we analyzed the final preparation of o-gp140SF162ΔV2 by SEC (Fig. 5E). The data suggested that the purified protein was predominantly homogeneous and potentially in trimeric conformation.
FIG. 4.
Analysis of gp140SF162ΔV2 containing fractions obtained at every step during purification. SDS-PAGE analysis of fractions obtained from GNA column (A) (lanes 2 to 6, elutions 1 to 5 from GNA column), from DEAE column (B) (lanes: 1, molecular weight standards; 2, unconcentrated flowthrough; 3, 10× concentrated DEAE flowthrough), and from CHAP column (C) (lanes: 1, 2, and 3, 10× concentrated flowthrough at 2, 1, and 0.5 μg, respectively; 4, molecular weight standards). o-gp140ΔV2 SF162 protein is indicated by an arrow. Also shown are size exclusion HPLC profile of gp140SF162ΔV2 after the CHAP column (D) and summary of the data (E).
Biochemical characterization of o-gp140SF162ΔV2. (i) Differential deglycosylation.
The nature of the oligosaccharides linkages in the purified o-gp140SF162ΔV2 was also analyzed and compared to that of monomeric gp120SF162. Purified protein was digested with PNGF, which releases asparagine-linked oligosaccharides, and with NANase II and O-glycosidase DS, which release unsubstituted Gal (β1, 3) GalNAc (α1) disaccharides attached to serine or threonine, either alone or in combination. We observed that o-gp140SF162ΔV2, like gp120SF162, had predominantly N-linked oligosaccharides, since PNGF treatment reduced o-gp140SF162ΔV2 to its protein backbone (Fig. 5F, lanes 5 and 9 for o-gp140SF162ΔV2 and gp120SF162, respectively). Furthermore, we observed that NANase and O-glycosidase DS had no effect on the electrophoretic mobility of o-gp140SF162ΔV2 (Fig. 5F, lanes 3 and 4, respectively). Similar observations were made for gp120SF162 (Fig. 5F, lanes 7 and 8, respectively) and also for o-gp140US4 (75). Furthermore, digestion of o-gp140SF162ΔV2 with a mixture of all three enzymes also had no discernible effect compared to digestion with PNGF alone (data not shown). This is similar to what was previously reported by us and others for HIV Env proteins in monomeric and oligomeric conformation (35, 75). Therefore, recombinant o-gp140SF162ΔV2 contained few or no O-linked oligosaccharides, as was observed for both native (released from virions) and recombinant gp120 (28, 49) and also for recombinant o-gp140 from US4 (75). To ensure that o-gp140SF162ΔV2 was secreted by cells after passage through the endoplasmic reticulum and Golgi, we treated o-gp140SF162ΔV2 with Endo-H, which releases terminal mannose carbohydrates. After Endo-H digestion of gp120SF162 and o-gp140SF162ΔV2, only a 15 to 20% reduction in the apparent molecular mass of either gp120SF162 or o-gp140SF162ΔV2 was observed (Fig. 5G), suggesting that not all high-mannose sugars are exposed on the surface of the molecule. This is in agreement with earlier data that demonstrates that HIV Env protein has heterogeneous glycosylation. This also suggest that carbohydrate modification of o-gp140SF162ΔV2 has taken place in the Golgi network; otherwise, EndoH digestion would have reduced o-gp140SF162ΔV2 to its protein backbone.
Finally, we performed stability studies to determine the effect of temperature upon structure and conformation of trimeric gp140SF162ΔV2. The data obtained in CD4 binding assay were analyzed as the ratio of the amounts of CD4 bound to the Env glycoprotein (CD4-B) to free CD4 (CD4-F). Based on this analysis, o-gp140SF162ΔV2 was found to be stable between 25 and 45°C (described in Materials and Methods), and only a small proportion (25%) of denaturation (not binding to CD4) was observed between 50 and 75°C. However, by heating the purified protein at temperatures of 75°C and above, the protein was completely denatured and did not bind to CD4 (data not shown).
(ii) Binding of o-gp140SF162ΔV2 to CD4.
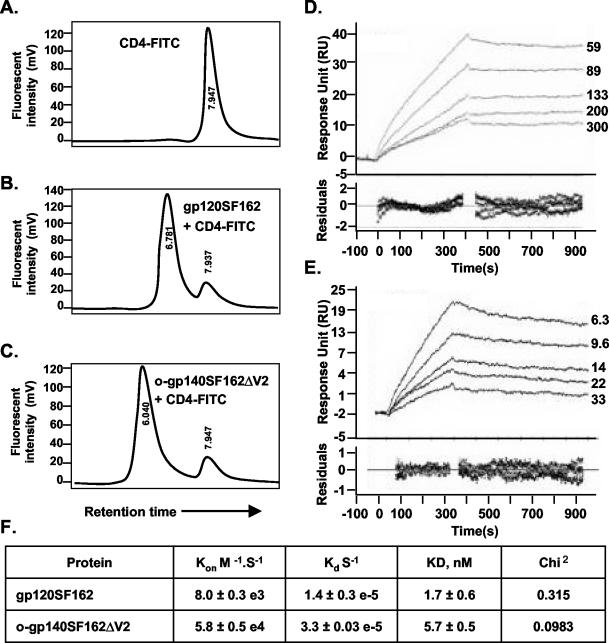
The ability of native HIV Env to bind to CD4 is essential for its biological function and can therefore serve as an indicator of native structure and functionality of recombinant Env (71). We used an HPLC-based receptor-binding assay using fluorescently labeled sCD4 (CD4-fluorescein isothiocyanate [FITC]) (75). As shown in Fig. 6, unlabeled o-gp140SF162ΔV2, when incubated with CD4-FITC, had a shorter retention time of 6.040 min (Fig. 6A) compared to 6.761 min for gp120 incubated with CD4-FITC (Fig. 6B) and 7.96 for CD4-FITC (Fig. 6C) as a consequence of the interaction between o-gp140SF162ΔV2 and CD4. Furthermore, reduced and denatured gp120 and deglycosylated gp120 (env2-3) did not bind efficiently to CD4 (data not shown).
FIG. 6.
(A to C) Shown is binding of purified gp120SF162 (B) and o-gp140SF162ΔV2 (C) to CD4, as determined by an HPLC-based assay, and an unbound CD4 profile (A). (D and E) Shown is the kinetics analysis of gp120SF162 (D) and o-gp140SF162ΔV2 (E) binding to immobilized sCD4. Different sensograms in panels D and E were generated by using a range of concentrations of gp120SF162 (59 to 300 nM) (D) and o-gp140SF162ΔV2 (6.3 to 33 nM) (E). Sensor data were prepared for kinetic analysis by subtracting binding responses collected from a blank reference surface. The association and dissociation phase data were fitted simultaneously to a single-site binding model by using Biaevaluation 3.2. (F) Summary of the kinetic data.
We performed the quantitative binding of o-gp140SF162ΔV2 to sCD4 by determining the association rates (Kon [moles per second]) and dissociation rates (Kd [per second]) and the dissociation constant (KD [nanomoles]) by BIACORE 3000. The kinetic data obtained for gp120SF162 and o-gp140SF162ΔV2 is presented in Fig. 6D and E. The summary of the binding constants is presented in Fig. 6F. The calculated on rates for gp120SF162 and o-gp140SF162ΔV2 are 8.0 × 10−3 and 5.8 × 10−4, respectively, suggesting that o-gp140SF162ΔV2 has a slower off rate compared to that of gp120SF162. The data also suggest that both these proteins have similar dissociation constants for sCD4, indicating that the deletion of the V2 loop did not affect the structure or the folding of the CD4-binding regions of the o-gp140SF162ΔV2.
Immunochemical characterization of o-gp140SF162ΔV2.
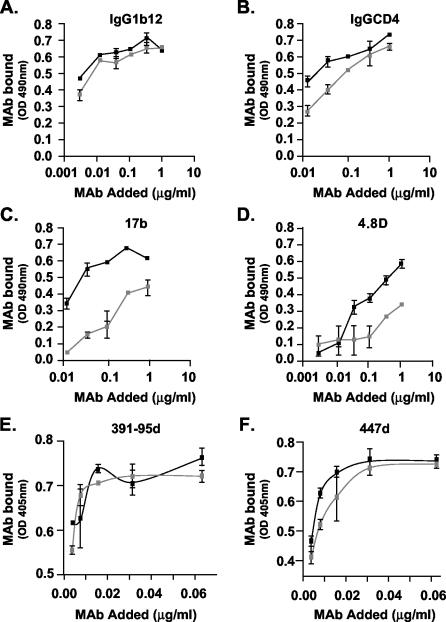
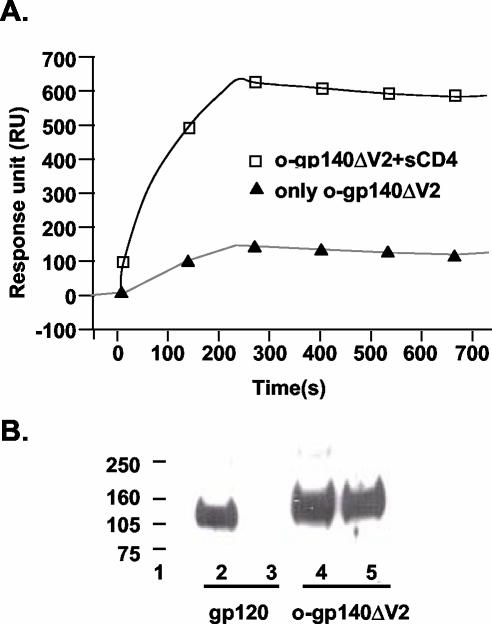
The structural integrity of o-gp140SF162ΔV2 was further evaluated by surface immunoprobing of o-gp140SF162ΔV2 and gp120SF162 using a capture ELISA with a panel of MAbs with known epitope specificities (IgG1b12, IgGCD4, 447D, 391-95d, G3136, G34, 17b, and 4.8d). The broadly reactive MAb IgG1b12, whose epitope overlaps the CD4-binding site, recognized the o-gp140SF162ΔV2 as efficiently as it did the monomeric gp120SF162 protein (Fig. 7A), suggesting that this critical neutralizing epitope is well presented in o-gp140SF162ΔV2. MAb IgGCD4, which binds to the CD4-binding site, recognized o-gp140SF162ΔV2 as efficiently as the monomeric gp120SF162 (Fig. 7B), suggesting that the oligomerization and the deletion of V2 loop did not substantially affect the CD4-binding site. In the absence of sCD4, o-gp140SF162ΔV2 was recognized more efficiently by 17b and 4.8d antibodies than was gp120SF162, suggesting that so-called CD4-inducible epitopes are better presented in the oligomer from which the V2 loop has been deleted than in the undeleted monomer. Furthermore, we observed that upon binding to sCD4, gp140SF162ΔV2 undergoes the expected conformational changes, as reflected by a significant increase in binding to the CD4-inducible epitope recognized by MAb 4.8d in oligomeric protein lacking the V2 loop in the presence of sCD4 (Fig. 8A). The structural integrity of purified o-gp140SF162ΔV2 protein was further demonstrated by its recognition by both 2G12, which recognizes a conformational carbohydrate-dependent gp120 epitope, and by 2F5, which recognizes a conserved neutralization epitope located in the ectodomain of the gp41 subunit (Fig. 8B, lanes 4 and 5, respectively, for 2G12 and 2F5). In contrast, the gp120SF162 monomer was recognized by 2G12 (Fig. 8B, lane 2) but not by 2F5, as expected (Fig. 8B, lane 3). To determine the relative exposure of the V3 loop, MAbs 447D and 391-95d were used. These antibodies recognized both gp120SF162 and the o-gp140SF162ΔV2 proteins with similar efficiency, suggesting that the V3 loop is well exposed in o-gp140SF162ΔV2 (Fig. 7E and F). As expected, compared to gp120, o-gp140SF162ΔV2 had little or no reactivity towards the V2 loop-specific antibodies (G3136 and G34) (data not shown). In addition, sera from HIV-1-infected individuals recognized both o-gp140SF162ΔV2 and gp120SF162, suggesting that the deletion of the V2 loop did not affect the exposure of the immunodominant epitopes (data not shown). Based on these observations, most of the critical neutralizing epitopes were intact and accessible in recombinant o-gp140SF162ΔV2.
FIG. 7.
Immunochemical characterization of purified gp120SF162 (gray squares) and o-gp140SF162ΔV2 (black squares) using a panel of MAbs, namely, IgG1b12 (A), IgGCD4 (B), 17b (C), 4.8d (D), 391-95d (E), and 447d (F).
FIG. 8.
Increased exposure of CD4i epitope recognized by the MAb 4.8d in o-gp140SF162ΔV2 upon binding to CD4, as determined by Biacore. (A) For this experiment, a reactive surface was prepared by capturing the MAb 4.8d on CM5 Bia-chip, and o-gp140SF162ΔV2 with (□) and without (▴) incubation with CD4 was flowed over this surface. Data were analyzed by using Biaevaluation 3.2. (B) Recognition of gp120SF162 and o-gp140SF162ΔV2 by MAbs 2F5 and 2G12 in Farr-Western assay. Lanes: 2 and 3, reactivity of gp120SF162 with 2G12 and 2F5, respectively; 4 and 5, recognition of o-gp140SF162ΔV2 by 2G12 and 2F5, respectively.
Biophysical characterization of o-gp140SF162ΔV2.
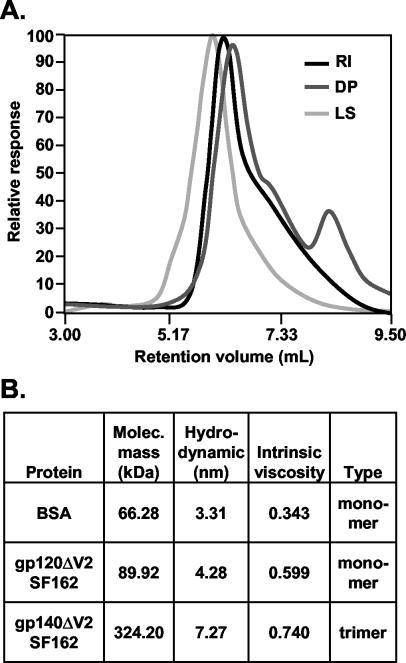
The relative responses obtained for various detectors for o-gp140SF162ΔV2 are shown in Fig. 9A. The biophysical properties of o-gp140SF162ΔV2 and gp120SF162ΔV2 (data not shown) are summarized in Fig. 9B. The calculated molecular masses of gp120SF162ΔV2 and o-gp140SF162ΔV2 were 89.92 and 324 kDa, respectively, with the molecular weight of purified o-gp140SF162ΔV2 being very close to the expected molecular weight for a trimer having a V2 loop deletion (three 110-kDa monomers have the weight of one 330-kDa trimer). The light-scattering profile for o-gp140SF162ΔV2 suggests that the purified protein was homogeneous, and the relative responses obtained from all three detectors (light scattering, refractive index, and viscosity) matched well. However, there was a slight hump in the ascending part of the light-scattering signal, suggesting that a small proportion (<2%) of the purified o-gp140SF162ΔV2 protein may have been in an aggregated state. The hydrodynamic radius (Rh) of the purified o-gp140SF162ΔV2 was determined to be larger than that of gp120SF162ΔV2 (7.27 versus 4.28 nm), suggesting a significant difference between the folding of monomeric and oligomeric proteins. In contrast, the intrinsic viscosity values for both proteins were in a similar range (Fig. 9B), indicating that the proteins had not become unfolded during the purification process. Therefore, the differences observed in the biophysical properties of these molecules likely were inherent to native proteins and were not caused by denaturation or unfolding.
FIG. 9.
Biophysical characterization of purified o-gp140SF162ΔV2 using TDA system. (A) Relative responses obtained for light scattering (LS), refractive index detector (RI), and viscometer (DP) analyses for o-gp140SF162ΔV2. The majority of the purified protein is homogeneous. (B) Summary of biophysical properties, including molecular weight, intrinsic viscosity and hydrodynamic radius, of o-gp140SF162ΔV2 compared to gp120SF162ΔV2.
Negative-stain EM.
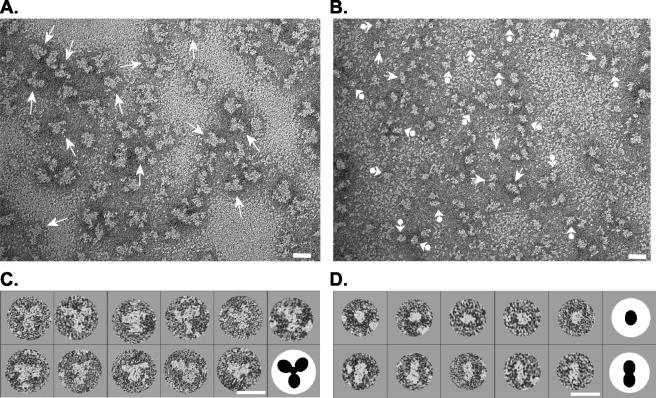
The above data indicate that most of the purified o-gp140SF162ΔV2 is in a trimeric conformation, with a smaller fraction in the monomer-to-dimer range (Fig. 5). To confirm these interpretations, we performed negative-stain EM analysis on the o-gp140SF162ΔV2. The fractions constituting peaks A (putative trimers) and B (monomers and/or dimers) were pooled and concentrated separately and analyzed by EM (panels A and B of Fig. 10 show representative fields of molecules from peaks A and B of Fig. 5, respectively). Analysis of peak B, which contains the faster-migrating molecules, revealed images consistent with both monomeric and dimeric forms (Fig. 10B), whereas molecules present in Peak A appear to be primarily composed of three-lobed, (trimeric) configurations (Fig. 10A). Examples of selected molecules from peaks A and B and diagrammatic representations are shown in Fig. 10C and D, respectively. To aid in interpretation, the images in Fig. 10C and D were enlarged, masked, and rotated such that one lobe of the dimers or trimers points downward. For the trimers, the three radial lobes are assumed to be the three gp120 moieties. Many of the trimers appear to have a centrally located mass of protein density (as described in the legend for Fig. 10C) which could represent the gp41 ectodomain of the trimer. The conformational variations seen in each fraction may represent either a degree of intramolecular flexibility or a reflection of the random orientation assumed by molecules as they adhere to the carbon membrane substrate. Some of the heterogeneity (i.e., extra lobes) could, for example, represent molecules in which the gp41 trimer is viewed in profile in addition to the three gp120 moieties. However, some degree of molecular heterogeneity cannot be ruled out.
FIG. 10.
EMs of o-gp140SF162ΔV2 obtained from preparative SEC (Fig. 5A). (A) EM of peak A (Fig. 5A) containing trilobed molecules (trimers) indicated with arrows. (B) EM of peak B (Fig. 5A) preparation with presumptive monomers and dimers indicated with arrowhead-balls and arrows, respectively. (C) Selected trimers from peak A. (D) Selected monomers (top) and dimers (bottom) from peak B. To aid interpretation, the molecules in panels C and D were enlarged, masked, and rotated such that one of the lobes of the dimers and trimers points downward. Graphic representations of a trimer and monomer and dimer (not drawn to scale) are shown in panels C and D, respectively. Bars, 20 nm.
The dimensions of the monomeric gp140SF162ΔV2molecules ranged between 60 and 100 Å (mean ± standard error of the mean [SEM], 74 ± 13 Å) from peak B and are consistent with previously reported measurements of the SOS gp140JR-FL construct (73). The presumptive dimers measured, on average, 69 ± 9 by 154 ± 18 Å (mean ± SEM) ,and the trimers were between 150 and 250 Å (mean ± SEM, 191 ± 23 Å) in their longest axis. Additional EM studies of these molecules complexed with sCD4 and epitope-specific MAbs are in progress to further define the composition and geometry of these trimers.
Immunogenicity of o-gp140SF162ΔV2 in rhesus macaques.
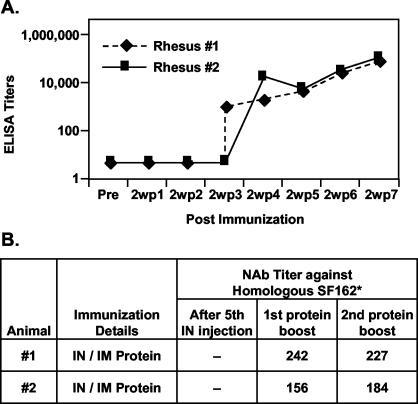
Two rhesus macaques were immunized with trimeric gp140SF162ΔV2 using an intranasal priming and intramuscular boosting regimen. Intranasal immunization induced low levels of systemic IgG responses. However, after two intramuscular boosts with o-gp140SF162ΔV2, there was a 20-fold increase in the antibody titers (Fig. 11A). Sera collected after the second intramuscular protein boosts were tested for their abilities to neutralize HIV-1 isolates (Fig. 11B). Sera collected from both the animals after the intramuscular boost with o-gp140SF162ΔV2 neutralized homologous HIV-1 isolate SF162 at significant dilutions (Fig. 11B). In addition, intranasal priming did not induce any neutralizing antibodies in these rhesus macaques. The preliminary data suggest that at 1:20 dilution, sera from animal #1 neutralized all the three heterologous HIV-1 primary isolates tested to various extents (50% for Bal, 59% for JR-FL, and 83% for Bx08). However, at the same dilution, sera from animal #2 neutralized only Bx08 (79%). Efforts are in progress to further evaluate the breadth of neutralizing antibody responses induced by o-gp140SF162ΔV2.
FIG. 11.
Binding (A) (determined by ELISA) and neutralizing (B) antibodies induced in rhesus macaques following intranasal priming and intramuscular boosting regimen using o-gp140SF162ΔV2 in combination with LTK63 or LTR72. Briefly, two rhesus macaques were immunized five times at 3-week intervals with o-gp140SF162ΔV2 (300 μg) and LTK63 or LTR72 (100 μg) in a total volume of 800 μl (400 μl per nostril). After a resting period of 4 months, both the animals were immunized twice with 100 μg of o-gp140SF162ΔV2 in MF59 intramuscularly at 4-week intervals. For determining the neutralizing activity, samples were assayed at threefold dilutions in triplicate in M7-luc cells. Neutralization antibody (NAb) titers are the serum dilutions at which luciferase activity was reduced to 80% relative to the virus control wells (cells and virus).
DISCUSSION
The HIV envelope glycoprotein is the obvious target for inducing broadly cross-reactive primary isolates neutralizing antibody responses. It is well documented that gp120 monomer elicits high-titer anti-envelope antibodies (3, 31, 45, 80). Furthermore, immunization with recombinant monomeric gp120 proteins generally induced antibodies that reacted more strongly to monomer than to oligomer (6, 82). However, these antibodies are limited in their ability to neutralize HIV-1 primary isolates, especially those of the R5 phenotype (46, 51, 82, 86). The virus has evolved ways to minimize the immunogenicity of the Env protein by extensive glycosylation of the molecule with nascent carbohydrate (high terminal mannose), thus minimizing the efficacy of antibodies by shielding the conserved region. Moreover, the receptor-binding site is buried within the molecule and is partially shielded by external peptide loops. It is not that Env glycoprotein lacks critical neutralizing epitopes, because primary isolate-neutralizing antibodies are induced in humans during the natural course of infection. The observation that most broadly reactive neutralizing antibodies isolated so far (i.e., IgG1b12, 2G12, and 2F5) have stronger affinity for the native envelope (trimer) than for monomeric gp120 or gp41 (27, 63) provides, at least conceptually, the impetus to evaluate trimeric Env glycoprotein for their ability to induce broadly cross-reactive primary isolate-neutralizing antibodies. Therefore, to achieve better immunogenicity with Env protein-based vaccines, attention has focused on oligomeric forms of Env derived from R5 isolates (5, 67, 68, 75, 89-92). Earlier studies suggested that oligomer might be more potent and effective in inducing strong antibody responses to conformational epitopes (18, 82). Yang et al. (92) demonstrated that antibodies induced by oligomeric gp140 were more effective in neutralizing heterologous primary isolates than the antibodies elicited by the corresponding monomeric gp120 protein. We have reported that a soluble oligomeric gp140 immunogen derived from the envelope of SF162 elicited cross-reactive neutralizing antibodies in rabbits (2). Furthermore, we have also demonstrated that the partial deletion of the V2 loop (o-gp140SF162ΔV2) from this immunogen not only enhanced its ability to elicit broadly neutralizing antibodies, but also partially protected vaccinated rhesus macaques upon pathogenic challenge with SHIVSF162P4 (2).
To elicit the functional antibody responses, oligomeric HIV envelope immunogens need to be stabilized so that they retain their oligomeric nature upon delivery. Several approaches are currently under evaluation, namely: (i) elimination of the protease cleavage site between gp120 and gp41 (6, 18, 20, 22), (ii) introduction of intramolecular disulfide bond formation (SOS) between gp120 and gp41 (5, 23, 67, 73), (iii) introduction, in addition to the SOS modifications, of an isoleucine-to-proline substitution at position 559 in the N-terminal heptad repeat region of gp41 (SOSIP) (68), (iv) extension of the gp41 coiled coil region by the addition of GCN4 (a transcription factor that normally forms stable homotrimers) sequences, either alone or in conjunction with added cysteines between gp120 and gp41 (23, 89, 90), and (v) addition of a trimerization domain from the C terminus of bacteriophage T4 fibritin (91). We evaluated the efficacy of both protease site mutation as well as the SOS approach to create stable trimeric HIV envelope proteins derived from different clades (B and C). Although, a single mutation in the primary protease cleavage site (R to S) is sufficient for the formation of stable oligomers from US4, an R5 using HIV-1 primary isolate (75), this modification alone is ineffective in stabilizing the oligomers with and without V2 loop derived from SF162 (data not shown). Similar observations were made by Earl et al. (18-21) and Yang et al. (90) for different isolates. In our hands, the SOS approach that has been used by Binley et al. (5) and Sanders et al. (67) for creating stable oligomers from both primary (JF-RL) and TCLA isolates has failed to generate stable oligomers with or without theV2 loop from SF162 (I. Srivastava, J. Ulmer, and S. Barnett, unpublished observations). However, these were not totally unexpected results, because follow-up studies from Schulke et al. have also demonstrated the inefficiency of this approach in creating stable oligomers from JR-FL (73). In essence, these observations suggest that strategies for the generation of stable soluble HIV envelope oligomers from a given isolate or clade will need to be determined experimentally. Sequence variation, glycosylation, size of variable loops, expression levels, and the type of protein expression system may all play a critical role in stabilizing the HIV Env glycoprotein in oligomeric conformation (89, 90).
There is no consensus regarding the structure and the mass of recombinant HIV Env glycoproteins. Differences in observed mass of the trimeric molecule may be because these molecules were derived from different isolates and characterized by different techniques in different laboratories. Although it is expected that the trimeric Env glycoprotein derived from different isolates (such as o-gp140 US4 and SF162) will differ in their molecular masses due to sequence differences, they should be at least in a similar mass range. Most of the soluble recombinant Env oligomeric proteins purified to date have been shown to be either dimers, tetramers (6, 19, 20), or monomers (73), with the possible exception of those described by Grundner et al. (29), Yang et al. (89, 91), and Chakrabarti et al. (11). Most of the studies used either SEC or sucrose gradient centrifugation to determine the mass of the oligomeric molecule. This may not be the most appropriate means to determine the molecular weight of purified proteins due to a lack of direct relationship between molecular size and molecular weight of proteins, even under strongly denaturing conditions (47). In addition, proteins with extensive posttranslational modifications, such as glycosylation (85), as is the case with HIV envelope proteins, may further complicate this type of analysis. The most appropriate and precise methods for determining the molecular mass are osmometry, analytical centrifugation, and classical light scattering. Osmometry and analytical ultracentrifugation are labor intensive and time consuming. In addition, the high shear force induced during the ultracentrifugation may cause the dissociation of noncovalently associated HIV envelope subunits. In contrast, classical light scattering can be performed quickly and relatively easily and under very mild conditions; therefore, it is a reasonable choice for determining the molecular mass of the proteins (17, 36, 43). We previously used this approach to successfully characterize o-gp140-US4 and demonstrated that the purified protein was potentially in trimeric conformation (75). The calculated molecular mass for o-gp140 US4 was found to be 474 kDa, which is in good agreement with the molecular mass of 486 kDa of the intact Env glycoprotein isolated from the surface of virion as demonstrated recently by Center et al. using transmission-scanning electron microscope images (10). To ascertain the oligomeric status of o-gp140SF162ΔV2 produced in the CHO cell lines, we performed a detailed biophysical characterization of the purified protein using a TDA system coupled to a size exclusion column to determine the Mw, Rh and intrinsic viscosity. The Mw of the purified HIV envelope oligomer was estimated to be 324 kDa, very close to the expected Mw of a trimer (330 kDa). The differences in the calculated molecular mass of o-gp140 US4 and o-gp140SF162ΔV2 may be partially explained by the deletion of V2 loop, which also includes the deletion of one N-linked glycosylation site at position 186 (42, 43). Furthermore, the Env glycoprotein (trimers and monomers) derived from SF162 is slightly smaller than a similar protein derived from US4, as observed by SDS-PAGE analysis, suggesting that sequence differences may indeed play a role in determining the molecular mass of the protein. The reported molecular mass for transiently expressed Env protein oligomers derived from JR-FL, YU2, and HxBc2 is smaller than expected (400 to 410 kDa) (5, 89), and the molecular mass of the oligomer derived from JR-FL using a SOSIP approach is considerably larger than expected (29, 68). However, further characterization will be needed to confirm the trimeric nature of these purified proteins.
In the present study, we performed an in-depth biophysical characterization of the purified o-gp140SF162ΔV2 protein by using triple detectors and negative-stain EM of the purified Env glycoproteins to determine the structure and conformation of the purified protein. Combined, these methods strongly support the conclusion that the purified o-gp140SF162ΔV2 is predominantly in a trimeric conformation. As shown earlier, we found that the monomeric proteins are loosely folded globular structures with higher Rh values (5.07 nm for gp120 US4 [75] and 4.33 nm for gp120SF162ΔV2). In contrast, Env glycoprotein trimers are tightly folded structures, as indicated by their smaller-than-expected Rh values (8.4 nm for o-gp140 US4 [75] and 7.6 nm for o-gp140SF162ΔV2) for oligomers. These data are consistent with earlier observations that there are significant differences in the structure and folding of monomer and trimer (54, 87).
It has been proposed that cleavable trimers may be better than uncleaved trimers for inducing the primary isolate-neutralizing antibody responses, mainly because native Env is believed to undergo structural rearrangement on the viral surface and thus be more likely to express native epitopes following cleavage. However, recombinant cleavable Env proteins are structurally labile and need to be stabilized in the trimeric conformation in a manner that does not compromise the exposure of critical neutralizing epitopes. While there are no clear immunogenicity nor challenge data to suggest that cleaved oligomers are more potent for inducing strong neutralizing or protective antibodies, it may suffice to use cleavable Env immunogens during the DNA-priming phase (C. Buckner et al., submitted for publication).
The binding of HIV envelope protein to CD4, an obligatory step in viral replication (16, 34), is dependent upon both conformation (38, 48) and glycosylation (4, 40, 58). Evidence so far suggests that N-linked glycosylation is important for the formation of correct disulfide bonds and for providing functional conformation to the Env protein (24, 25, 39, 40) to foster CD4 binding, even though the carbohydrate side chains are not directly involved in CD4 binding (40). The V2 loop deletion, which also involves the deletion of a glycosylation site at position 186 (42, 43), did not affect the structure and conformation of HIV Env because purified o-gp140SF162ΔV2 bound to sCD4 with an affinity and efficiency similar to that of gp120 monomer, as determined qualitatively (HPLC assay) and quantitatively (Biacore). This finding suggests that the purified protein is in its correct native conformation. This was not entirely unexpected as it was previously demonstrated that the SF162 envelope from which V2 has been deleted maintains a functional structure and allows the virus to efficiently replicate in both peripheral blood mononuclear cells and macrophages in a CD4/CCR5-dependent manner (76), and transiently expressed protein was efficiently recognized by CD4-binding site MAbs (78). We have also demonstrated that the purified o-gp140SF162ΔV2, like native and recombinant monomeric and oligomeric Env proteins, contains predominantly N-linked glycosylation (35, 75).
We have determined the structural integrity of the purified gp140ΔV2 trimers with the use of several MAbs of known epitope specificity. Important epitopes exposed on purified trimers include those near or on the CD4-binding site (recognized by MAbs IgG1b12 and CD4-IgG2), the CD4-inducible site that overlaps the coreceptor binding site (recognized by MAbs 17b and 4.8d), gp41 (2F5), a carbohydrate-dependent epitope on gp120 (recognized by 2G12), and V3 loop epitopes (recognized by MAbs 447D and 391-95D). The monoclonal binding results obtained for o-gp140SF162ΔV2 are similar to those observed with corresponding native Env present on infectious SF162ΔV2 virions (79). It seems that irrespective of the cleavage status, critical neutralizing epitopes are properly exposed on recombinantly produced Env oligomers. In fact, the oligomers from which the V2 loop was deleted were recognized more efficiently by MAbs 17b and 4.8d in the absence of sCD4 compared to the monomeric form (Fig. 7C and D). In addition, upon binding to sCD4, there was significant up regulation of CD4-inducible epitope recognized by 4.8d.
In an intranasal priming and intramuscular boosting immunogenicity study in rhesus macaques, we have clearly demonstrated that o-gp140SF162ΔV2 induced strong Env-specific antibodies, and these antibodies were able to neutralize a homologous HIV-1 clade B isolate, i.e., SF162. The preliminary data suggest that the immunization regimen used in this study has induced cross-reactive neutralizing antibody responses against Bx08. In-depth analysis of the breadth of neutralizing antibody responses is ongoing and will be presented separately in a follow-up manuscript. Furthermore, intranasal priming with o-gp140SF162ΔV2 also induced mucosal responses (M. Vajdy et al., unpublished observations).
These data suggest that the deletion of V2 loop from the SF162 envelope increases the exposure of CD4-inducible epitope on o-gp140SF162ΔV2 compared to monomer. The preservation of critical neutralizing epitopes on the recombinant trimers and the ability of this immunogen to induce neutralizing antibody activity against heterologous viruses further emphasize the potential of trimeric gp140SF162ΔV2 for vaccine application.
Acknowledgments
We thank Margaret Liu for her support and encouragement during her tenure as Vice President for Vaccines and Gene Therapy Research at Chiron. We also thank Dennis Burton, Susan Zolla-Pazner, Michael Gorny, James Robinson, and Herman Katinger for providing monoclonal antibodies for the structural characterization of the purified proteins. We also thank Michael Houghton and Gib Otten for critically reading the manuscript and Nelle Cronen and Randy Deck for excellent editorial and administrative support. We thank Mark Wininger, Karen Matsuoka, Erika Calderon, and Robert Chow for their help in carrying out large-scale cell culture experiments.
This work was supported by financial support provided by NIH CRDA (NO-AI-95367, TO#3) and by D&DT contract NO-1-AI-05396 and R21 AI50430-01 to M.V. Research in the laboratories of Leonidas Stamatatos and Kenneth Roux was supported by investigator-initiated NIH research grants RO1 AI47708 and R21 A144291, respectively.
REFERENCES
- 1.Back, N. K., L. Smit, J. J. De Jong, W. Keulen, M. Schutten, J. Goudsmit, and M. Tersmette. 1994. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199:431-438. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, S. W., S. Rajasekar, H. Legg, B. Doe, D. H. Fuller, J. R. Haynes, C. M. Walker, and K. S. Steimer. 1997. Vaccination with HIV-1 gp120 DNA induces immune responses that are boosted by a recombinant gp120 protein subunit. Vaccine 15:869-873. [DOI] [PubMed] [Google Scholar]
- 4.Barr, P. J., K. S. Steimer, E. A. Sabin, D. Parkes, C. George-Nascimento, J. C. Stephans, M. A. Powers, A. Gyenes, G. A. Van Nest, E. T. Miller, et al. 1987. Antigenicity and immunogenicity of domains of the human immunodeficiency virus (HIV) envelope polypeptide expressed in the yeast Saccharomyces cerevisiae. Vaccine 5:90-101. [DOI] [PubMed] [Google Scholar]
- 5.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broder, C. C., P. L. Earl, D. Long, S. T. Abedon, B. Moss, and R. W. Doms. 1994. Antigenic implications of human immunodeficiency virus type 1 envelope quaternary structure: oligomer-specific and -sensitive monoclonal antibodies. Proc. Natl. Acad. Sci. USA 91:11699-11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton, D. R., and D. C. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl. A):S87-S98. [PubMed] [Google Scholar]
- 8.Burton, D. R., and P. W. Parren. 2000. Vaccines and the induction of functional antibodies: time to look beyond the molecules of natural infection? Nat. Med. 6:123-125. [DOI] [PubMed] [Google Scholar]
- 9.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Center, R. J., R. D. Leapman, J. Lebowitz, L. O. Arthur, P. L. Earl, and B. Moss. 2002. Oligomeric structure of the human immunodeficiency virus type 1 envelope protein on the virion surface. J. Virol. 76:7863-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakrabarti, B. K., W. P. Kong, B. Y. Wu, Z. Y. Yang, J. Friborg, X. Ling, S. R. King, D. C. Montefiori, and G. J. Nabel. 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J. Virol. 76:5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman, B. S., R. M. Thayer, K. A. Vincent, and N. L. Haigwood. 1991. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 19:3979-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng-Mayer, C., A. Brown, J. Harouse, P. A. Luciw, and A. J. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherpelis, S., X. Jin, A. Gettie, D. D. Ho, S. W. Barnett, I. Shrivastava, and L. Stamatatos. 2001. DNA-immunization with a V2 deleted HIV-1 envelope elicits protective antibodies in macaques. Immunol. Lett. 79:47-55. [DOI] [PubMed] [Google Scholar]
- 15.Cherpelis, S., I. Srivastava, A. Gettie, X. Jin, D. D. Ho, S. W. Barnett, and L. Stamatatos. 2001. DNA vaccination with the human immunodeficiency virus type 1 SF162ΔV2 envelope elicits immune responses that offer partial protection from simian/human immunodeficiency virus infection to CD8+ T-cell-depleted rhesus macaques. J. Virol. 75:1547-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 17.Dollinger, G., B. Cunico, M. Kunitani, D. Johnson, and R. Jones. 1992. Practical on-line determination of biopolymer molecular weights by high-performance liquid chromatography with classical light-scattering detection. J. Chromatogr. 592:215-228. [Google Scholar]
- 18.Earl, P. L., C. C. Broder, D. Long, S. A. Lee, J. Peterson, S. Chakrabarti, R. W. Doms, and B. Moss. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J. Virol. 68:3015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earl, P. L., R. W. Doms, and B. Moss. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 87:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earl, P. L., S. Koenig, and B. Moss. 1991. Biological and immunological properties of human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J. Virol. 65:31-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Earl, P. L., and B. Moss. 1993. Mutational analysis of the assembly domain of the HIV-1 envelope glycoprotein. AIDS Res. Hum. Retrovir. 9:589-594. [DOI] [PubMed] [Google Scholar]
- 22.Earl, P. L., W. Sugiura, D. C. Montefiori, C. C. Broder, S. A. Lee, C. Wild, J. Lifson, and B. Moss. 2001. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J. Virol. 75:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farzan, M., H. Choe, E. Desjardins, Y. Sun, J. Kuhn, J. Cao, D. Archambault, P. Kolchinsky, M. Koch, R. Wyatt, and J. Sodroski. 1998. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J. Virol. 72:7620-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fennie, C., and L. A. Lasky. 1989. Model for intracellular folding of the human immunodeficiency virus type 1 gp120. J. Virol. 63:639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fenouillet, E., B. Clerget-Raslain, J. C. Gluckman, D. Guetard, L. Montagnier, and E. Bahraoui. 1989. Role of N-linked glycans in the interaction between the envelope glycoprotein of human immunodeficiency virus and its CD4 cellular receptor. Structural enzymatic analysis. J. Exp Med. 169:807-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Interaction of polyclonal and monoclonal anti-glycoprotein 120 antibodies with oligomeric glycoprotein 120-glycoprotein 41 complexes of a primary HIV type 1 isolate relationship to neutralization. AIDS Res. Hum. Retrovir. 14:591-597. [DOI] [PubMed] [Google Scholar]
- 27.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geyer, H., C. Holschbach, G. Hunsmann, and J. Schneider. 1988. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J. Biol. Chem. 263:11760-11767. [PubMed] [Google Scholar]
- 29.Grundner, C., T. Mirzabekov, J. Sodroski, and R. Wyatt. 2002. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J. Virol. 76:3511-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 31.Haigwood, N. L., P. L. Nara, E. Brooks, G. A. Van Nest, G. Ott, K. W. Higgins, N. Dunlop, C. J. Scandella, J. W. Eichberg, and K. S. Steimer. 1992. Native but not denatured recombinant human immunodeficiency virus type 1 gp120 generates broad-spectrum neutralizing antibodies in baboons. J. Virol. 66:172-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, W. E., J. Morgan, J. Reitter, B. A. Puffer, S. Czajak, R. W. Doms, and R. C. Desrosiers. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 76:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klatzmann, D., E. Champagne, S. Chamaret, J. Gruest, D. Guetard, T. Hercend, J. C. Gluckman, and L. Montagnier. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767-768. [DOI] [PubMed] [Google Scholar]
- 35.Kozarsky, K., M. Penman, L. Basiripour, W. Haseltine, J. Sodroski, and M. Krieger. 1989. Glycosylation and processing of the human immunodeficiency virus type 1 envelope protein. J. Acquir. Immune Defic. Syndr. 2:163-169. [PubMed] [Google Scholar]
- 36.Krull, I. S., H. H. Stuting, and S. C. Krzysko. 1988. Conformational studies of bovine alkaline phosphatase in hydrophobic interaction and size-exclusion chromatography with linear diode array and low-angle laser light scattering detection. J. Chromatogr. 442:29-52. [DOI] [PubMed] [Google Scholar]
- 37.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lasky, L. A., G. Nakamura, D. H. Smith, C. Fennie, C. Shimasaki, E. Patzer, P. Berman, T. Gregory, and D. J. Capon. 1987. Delineation of a region of the human immunodeficiency virus type 1 gp120 glycoprotein critical for interaction with the CD4 receptor. Cell 50:975-985. [DOI] [PubMed] [Google Scholar]
- 39.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 40.Li, Y., L. Luo, N. Rasool, and C. Y. Kang. 1993. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J. Virol. 67:584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lue, J., M. Hsu, D. Yang, P. Marx, Z. Chen, and C. Cheng-Mayer. 2002. Addition of a single gp120 glycan confers increased binding to dendritic cell-specific ICAM-3-grabbing nonintegrin and neutralization escape to human immunodeficiency virus type 1. J. Virol. 76:10299-10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maezawa, S., Y. Hayashi, T. Nakae, J. Ishii, K. Kameyama, and T. Takagi. 1983. Determination of molecular weight of membrane proteins by the use of low-angle laser light scattering combined with high-performance gel chromatography in the presence of a non-ionic surfactant. Biochim. Biophys. Acta 747:291-297. [DOI] [PubMed] [Google Scholar]
- 44.Malenbaum, S. E., D. Yang, L. Cavacini, M. Posner, J. Robinson, and C. Cheng-Mayer. 2000. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J. Virol. 74:11008-11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, D. S. Burke, et al. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 47.McDonnell, M. E., and A. M. Jamieson. 1976. Rapid characterization of protein molecular weights and hydrodynamic structures by quasielastic laser-light scattering. Biopolymers 15:1283-1299. [DOI] [PubMed] [Google Scholar]
- 48.McDougal, J. S., J. K. Nicholson, G. D. Cross, S. P. Cort, M. S. Kennedy, and A. C. Mawle. 1986. Binding of the human retrovirus HTLV-III/LAV/ARV/HIV to the CD4 (T4) molecule: conformation dependence, epitope mapping, antibody inhibition, and potential for idiotypic mimicry. J. Immunol. 137:2937-2944. [PubMed] [Google Scholar]
- 49.Mizuochi, T., M. W. Spellman, M. Larkin, J. Solomon, L. J. Basa, and T. Feizi. 1988. Structural characterization by chromatographic profiling of the oligosaccharides of human immunodeficiency virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese hamster ovary cells. Biomed. Chromatogr. 2:260-270. [DOI] [PubMed] [Google Scholar]
- 50.Montefiori, D. C., W. E. Robinson, Jr., S. S. Schuffman, and W. M. Mitchell. 1988. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J. Clin. Microbiol. 26:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117-S136. [PubMed] [Google Scholar]
- 53.Moore, J. P., P. W. Parren, and D. R. Burton. 2001. Genetic subtypes, humoral immunity, and human immunodeficiency virus type 1 vaccine development. J. Virol. 75:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore, J. P., Q. J. Sattentau, R. Wyatt, and J. Sodroski. 1994. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J. Virol. 68:469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Narayan, S. V., S. Mukherjee, F. Jia, Z. Li, C. Wang, L. Foresman, C. McCormick-Davis, E. B. Stephens, S. V. Joag, and O. Narayan. 1999. Characterization of a neutralization-escape variant of SHIVKU-1, a virus that causes acquired immune deficiency syndrome in pig-tailed macaques. Virology 256:54-63. [DOI] [PubMed] [Google Scholar]
- 56.Overbaugh, J., and L. M. Rudensey. 1992. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J. Virol. 66:5937-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Overbaugh, J., L. M. Rudensey, M. D. Papenhausen, R. E. Benveniste, and W. R. Morton. 1991. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J. Virol. 65:7025-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papandreou, M. J., T. Idziorek, R. Miquelis, and E. Fenouillet. 1996. Glycosylation and stability of mature HIV envelope glycoprotein conformation under various conditions. FEBS Lett. 379:171-176. [DOI] [PubMed] [Google Scholar]
- 59.Parren, P. W., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13(Suppl. A):S137-S162. [PubMed] [Google Scholar]
- 60.Poignard, P., E. O. Saphire, P. W. Parren, and D. R. Burton. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 61.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 62.Richardson, T. M., B. L. Stryjewski, C. C. Broder, J. A. Hoxie, J. R. Mascola, P. L. Earl, and R. W. Doms. 1996. Humoral response to oligomeric human immunodeficiency virus type 1 envelope protein. J. Virol. 70:753-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roux, K. H. 1989. Immunoelectron microscopy of idiotype-anti-idiotype complexes. Methods Enzymol. 178:130-144. [DOI] [PubMed] [Google Scholar]
- 65.Roux, K. H. 1996. Negative-stain immunoelectron-microscopic analysis of small macromolecules of immunologic significance. Methods 10:247-256. [DOI] [PubMed] [Google Scholar]
- 66.Rudensey, L. M., J. T. Kimata, E. M. Long, B. Chackerian, and J. Overbaugh. 1998. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 72:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanders, R. W., L. Schiffner, A. Master, F. Kajumo, Y. Guo, T. Dragic, J. P. Moore, and J. M. Binley. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J. Virol. 74:5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanders, R. W., M. Vesanen, N. Schuelke, A. Master, L. Schiffner, R. Kalyanaraman, M. Paluch, B. Berkhout, P. J. Maddon, W. C. Olson, M. Lu, and J. P. Moore. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76:8875-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sattentau, Q. J., S. Zolla-Pazner, and P. Poignard. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713-717. [DOI] [PubMed] [Google Scholar]
- 71.Scandella, C. J., J. Kilpatrick, W. Lidster, C. Parker, J. P. Moore, G. K. Moore, K. A. Mann, P. Brown, S. Coates, B. Chapman, et al. 1993. Nonaffinity purification of recombinant gp120 for use in AIDS vaccine development. AIDS Res. Hum. Retrovir. 9:1233-1244. [DOI] [PubMed] [Google Scholar]
- 72.Schonning, K., B. Jansson, S. Olofsson, J. O. Nielsen, and J. S. Hansen. 1996. Resistance to V3-directed neutralization caused by an N-linked oligosaccharide depends on the quaternary structure of the HIV-1 envelope oligomer. Virology 218:134-140. [DOI] [PubMed] [Google Scholar]
- 73.Schulke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Srivastava, I. K., M. Schmidt, U. Certa, H. Dobeli, and L. H. Perrin. 1990. Specificity and inhibitory activity of antibodies to Plasmodium falciparum aldolase. J. Immunol. 144:1497-1503. [PubMed] [Google Scholar]
- 75.Srivastava, I. K., L. Stamatatos, H. Legg, E. Kan, A. Fong, S. R. Coates, L. Leung, M. Wininger, J. J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2002. Purification and characterization of oligomeric envelope glycoprotein from a primary r5 subtype B human immunodeficiency virus. J. Virol. 76:2835-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stamatatos, L., and C. Cheng-Mayer. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stamatatos, L., and C. Cheng-Mayer. 1995. Structural modulations of the envelope gp120 glycoprotein of human immunodeficiency virus type 1 upon oligomerization and differential V3 loop epitope exposure of isolates displaying distinct tropism upon virion-soluble receptor binding. J. Virol. 69:6191-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stamatatos, L., M. Lim, and C. Cheng-Mayer. 2000. Generation and structural analysis of soluble oligomeric gp140 envelope proteins derived from neutralization-resistant and neutralization-susceptible primary HIV type 1 isolates.. AIDS Res. Hum. Retrovir. 16:981-994. [DOI] [PubMed] [Google Scholar]
- 79.Stamatatos, L., M. Wiskerchen, and C. Cheng-Mayer. 1998. Effect of major deletions in the V1 and V2 loops of a macrophage-tropic HIV type 1 isolate on viral envelope structure, cell entry, and replication. AIDS Res. Hum. Retrovir. 14:1129-1139. [DOI] [PubMed] [Google Scholar]
- 80.Steimer, K. S., and N. L. Haigwood. 1991. Importance of conformation on the neutralizing antibody response to HIV-1 gp120. Biotechnol. Ther. 2:63-89. [PubMed] [Google Scholar]
- 81.Steimer, K. S., C. J. Scandella, P. V. Skiles, and N. L. Haigwood. 1991. Neutralization of divergent HIV-1 isolates by conformation-dependent human antibodies to Gp120. Science 254:105-108. [DOI] [PubMed] [Google Scholar]
- 82.VanCott, T. C., F. R. Bethke, D. S. Burke, R. R. Redfield, and D. L. Birx. 1995. Lack of induction of antibodies specific for conserved, discontinuous epitopes of HIV-1 envelope glycoprotein by candidate AIDS vaccines. J. Immunol. 155:4100-4110. [PubMed] [Google Scholar]
- 83.VanCott, T. C., J. R. Mascola, R. W. Kaminski, V. Kalyanaraman, P. L. Hallberg, P. R. Burnett, J. T. Ulrich, D. J. Rechtman, and D. L. Birx. 1997. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J. Virol. 71:4319-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 85.Wen, J., T. Arakawa, and J. S. Philo. 1996. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal. Biochem. 240:155-166. [DOI] [PubMed] [Google Scholar]
- 86.Wrin, T., and J. H. Nunberg. 1994. HIV-1MN recombinant gp120 vaccine serum, which fails to neutralize primary isolates of HIV-1, does not antagonize neutralization by antibodies from infected individuals. AIDS 8:1622-1623. [DOI] [PubMed] [Google Scholar]
- 87.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 88.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 89.Yang, X., M. Farzan, R. Wyatt, and J. Sodroski. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 74:5716-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang, X., L. Florin, M. Farzan, P. Kolchinsky, P. D. Kwong, J. Sodroski, and R. Wyatt. 2000. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J. Virol. 74:4746-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang, X., J. Lee, E. M. Mahony, P. D. Kwong, R. Wyatt, and J. Sodroski. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76:4634-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang, X., R. Wyatt, and J. Sodroski. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 75:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.zur Megede, J., M. C. Chen, B. Doe, M. Schaefer, C. E. Greer, M. Selby, G. R. Otten, and S. W. Barnett. 2000. Increased expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 gag gene. J. Virol. 74:2628-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]