Abstract
The MD145-12 strain (GII/4) is a member of the genus Norovirus in the Caliciviridae and was detected in a patient with acute gastroenteritis in a Maryland nursing home. The open reading frame 1 (ORF1) (encoding the nonstructural polyprotein) was cloned as a consensus sequence into various expression vectors, and a proteolytic cleavage map was determined. The virus-encoded cysteine proteinase mediated at least five cleavages (Q330/G331, Q696/G697, E875/G876, E1008/A1009, and E1189/G1190) in the ORF1 polyprotein in the following order: N-terminal protein; nucleoside triphosphatase; 20-kDa protein (p20); virus protein, genome linked (VPg); proteinase (Pro); polymerase (Pol). A time course analysis of proteolytic processing of the MD145-12 ORF1 polyprotein in an in vitro coupled transcription and translation assay allowed the identification of stable precursors and final mapped cleavage products. Stable precursors included p20VPg (analogous to the 3AB of the picornaviruses) and ProPol (analogous to the 3CD of the picornaviruses). Less stable processing intermediates were identified as p20VPgProPol, p20VPgPro, and VPgPro. The MD145-12 Pro and ProPol proteins were expressed in bacteria as active forms of the proteinase and used to further characterize their substrate specificities in trans cleavage assays. The MD145-12 Pro was able to cleave its five mapped cleavage sites in trans and, in addition, could mediate trans cleavage of the Norwalk virus (GI/I) ORF1 polyprotein into a similar proteolytic processing profile. Taken together, our data establish a model for proteolytic processing in the noroviruses that is consistent with nonstructural precursors and products identified in studies of caliciviruses that replicate in cell culture systems.
The family Caliciviridae is comprised of four genera, Vesivirus, Lagovirus, Sapovirus, and Norovirus, which are represented by prototype strains feline calicivirus (FCV), rabbit hemorrhagic disease virus (RHDV), Sapporo virus, and Norwalk virus, respectively (9). Viruses in the genus Norovirus are the major cause of nonbacterial gastroenteritis outbreaks. It has been estimated that at least 23 million cases of gastroenteritis are associated with noroviruses each year in the United States (18a). Phylogenetic analysis has shown evidence for at least three distinct norovirus genogroups designated GI, GII, and GIII (1, 17). The human pathogens described thus far belong to GI and GII and have been further divided into seven and eight genetic clusters, respectively (1, 9). Viruses in cluster GII/4 are the predominant noroviruses associated with epidemic gastroenteritis worldwide (9, 20).
The norovirus genome is a single-stranded, positive-sense RNA molecule of 7.5 kb that is polyadenylated at its 3′ end. The genome is organized into three open reading frames (ORFs): ORF1 encodes a 200-kDa nonstructural polyprotein, ORF2 encodes the 60-kDa major structural protein VP1, and ORF3 encodes the minor structural protein VP2 (7, 12, 15). The ORF1 polyprotein is processed by the “3C-like” (3CL) viral proteinase into the nonstructural proteins (10, 16, 27). From the amino to the carboxy termini, the norovirus ORF1 polyprotein can be divided into at least six functional domains that have been designated N-terminal protein (Nterm); 2C-like nucleoside triphosphatase (NTPase); p20 or p22 (depending on the genogroup); virus protein, genome linked (VPg); proteinase (Pro); and polymerase (Pol) (9, 18, 24) (Fig. 1A). The enzymatic activities or functions for only two of the norovirus nonstructural proteins have been determined. A recombinant 2C-like region from the Southampton virus has been expressed in bacteria and exhibits NTPase activity in vitro (23). The norovirus 3CL proteinase has been characterized as a chymotrypsin-like protease in which the His-30 and Cys-139 residues constitute the catalytic dyad (16, 29). Certain amino acids at positions P1 and P4 were identified as essential for efficient cis activity of the proteinase (10).
FIG. 1.
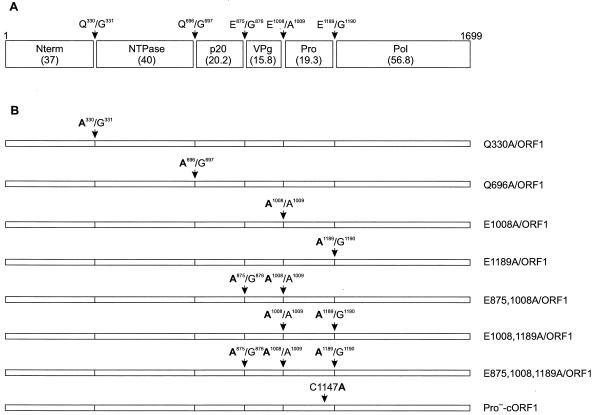
Predicted cleavage sites of the MD145-12 ORF1 polyprotein and constructs designed to study the proteolytic processing of the entire polyprotein. (A) Map of the cleavage sites for ORF1 of the MD145-12 norovirus based on the previously identified cleavage sites of the Southampton, Camberwell, and Chiba viruses (16, 18, 27, 28). Sequences of the dipeptide cleavage sites and their positions are indicated by arrows. The calculated molecular mass (shown in parentheses) of each protein is given in kilodaltons. The designation of the potential cleavage products is indicated inside boxes and is adapted from Liu et al. (18) and Green et al. (9). (B) Diagram of ORF1 constructs. The cORF1 construct corresponds to a consensus sequence of the entire ORF1. For other constructs, the mutated cleavage sites are indicated, and amino acid changes are in bold. The Pro−-cORF1 construct has a Cys to Ala substitution at position 1147, which inactivated the proteinase (16, 29).
Comparison of available norovirus ORF1 polyprotein sequences indicates the presence of at least five conserved dipeptide cleavage sites that are likely recognized by the virus-encoded 3CL proteinase (18). The first full norovirus in vitro proteolytic cleavage map was elucidated for the Southampton virus in the GI/2 genetic cluster (16, 18), and several of the individual cleavage sites have been independently confirmed in other norovirus strains (10, 27, 28). The experimental approaches for defining norovirus ORF1 cleavage sites have included a combination of site-directed mutagenesis and in vitro translation studies as well as direct N-terminal sequence analysis of cleavage products generated by bacterial overexpression of the viral Pro and surrounding polyprotein regions. Coupled transcription and translation (TNT) of the Southampton ORF1 identified three major protein products generated by cleavage of the polyprotein at the two Q/G sites bordering the NTPase (16). These three TNT products were identified as the 48-kDa N-terminal protein, the 41-kDa NTPase, and a 113-kDa precursor containing the C-terminal portion of the polyprotein. The processing of the Southampton virus 113-kDa protein was inefficient in TNT reactions, and the mapping of the cleavage sites within this precursor required overexpression in bacteria (18). The inefficient processing of this large precursor in TNT reactions has also been observed in studies of other norovirus ORF1 polyproteins (6, 10).
The MD145-12 strain (Hu/NLV/GII/MD145-12/1987/US) was described in an epidemiological survey of gastroenteritis outbreaks occurring in Maryland nursing homes during the winter of 1987-88 (8). Sequence analysis of the MD145-12 genome showed that it was closely related to Camberwell virus and belonged to cluster GII/4. In this study, we determined the proteolytic cleavage map of the MD145-12 ORF1 polyprotein and found evidence for the temporal synthesis of nonstructural precursors and products in vitro. Various forms of the viral proteinase (including mature and precursor forms) were tested for cis and trans cleavage activity, and characteristics of the dipeptide cleavage sites were examined. Comparison of our data with those of FCV and RHDV indicates striking similarities among the caliciviruses and other members of the picornavirus “superfamily.” These results indicate that early and late cleavages mediate the temporal appearance of critical precursors and products during norovirus replication.
MATERIALS AND METHODS
Virus.
Norovirus strain MD145-12 (Hu/NV/MD145-12/1987/US) was obtained from a Maryland nursing home resident with gastroenteritis, and the nucleotide sequence of the RNA genome was determined previously (8). The sequence was previously assigned GenBank accession no. AY032605.
Plasmids and primers.
The plasmids described below were selected and maintained in Escherichia coli DH5α cells, except where indicated. All plasmid constructions were sequenced to verify the presence of the viral consensus sequence, the histidine tail (pET constructs), the T7 promoter (pSPORT1 construct), the desired mutations, and any engineered start and stop codons. Oligonucleotide primers were obtained from Invitrogen (Carlsbad, Calif.).
Δlacα/T7-pSPORT1.
The plasmid pSPORT1 (Invitrogen) was modified to delete its T7 bacteriophage RNA polymerase promoter. First, a DNA fragment corresponding to nucleotides (nt) 320 through 554 of the plasmid was amplified by PCR, with the primers 5′-AACTGCAGGAGCTTGGCGTAATCATGGTCATAGC-3′ (located upstream of the T7 promoter) with an engineered PstI site (underlined), and 5′-CCACCCTGGCGCCCAATACGCAAACCG-3′, with an incorporated NarI site (underlined). The pSPORT1 plasmid and PCR amplicon were each digested with PstI and NarI, and the PCR amplicon was ligated into the compatible sites of the cut vector. The resulting plasmid was designated Δlacα/T7-pSPORT1.
Construction of cORF1 and mutagenesis.
A complete cDNA copy of the ORF1 of strain MD145-12 was cloned into Δlacα/T7-pSPORT1 under the control of a T7 promoter that was engineered to be immediately upstream of the first base of the 5′ end of the MD145-12 ORF1. This was accomplished by the construction of several intermediate plasmids. Four virus-specific overlapping cDNA fragments that contained unique internal restriction enzyme sites were amplified by reverse transcription-PCR from the virus in stool material and cloned into pCR2.1 by using the TOPO cloning system (Invitrogen) before subcloning into Δlacα/T7-pSPORT1. The first fragment (designated fragment I), corresponding to nt 1 to 932 of the virus genome, was amplified with forward primer 5′ AACTGCAGTAATACGACTCACTATAGTGAATGAAGATGGCG TCTAACG-3′, which incorporated a PstI site (underlined) and a T7 promoter (bold), and reverse primer 5′-AAGGAAAAAAGCGGCCGCAACTCCAAAGAGCTCTGCCA-3′ with an engineered NotI site (underlined). The second fragment (II), nt 876 to 2080, was amplified by using forward primer FW4 (8) and reverse primer 5′-AAGGAAAAAAGCGGCCGCCTCATCTAACCTCTCATGGAG TAGCCCTG-3′ with an engineered NotI site (underlined). The third fragment (III) corresponded to nt 1893 to 3300 and was amplified by using forward primer FW7 (8) and reverse primer 5′-AAGGAAAAAAGCGGCCGCGTAGGCCT CTTGATGAGTAGGGTGG-3′, with a NotI site (underlined). The fourth fragment (IV) corresponded to nt 3250 to 5104 (the 3′ end of ORF1) and was amplified by using the forward primer 5′-AACTGCAGAGAAGGTGCGCCCGAAGG TACCGTG-3′ and the reverse primer 5′-AAGGAAAAAAGCGGCCGCTCACTCGACGCCATCTTCATTCAC-3′, which included a NotI site (underlined) and the stop codon of ORF1 (bold). Fragment I and Δlacα/T7-pSPORT1 were digested by PstI and NotI prior to ligation. The resulting plasmid was designated T7-fragI and was used for the subcloning of fragment II into the SacI and NotI sites. The resulting construction, T7-fragI+II, was then ligated with PCR fragment III after both were treated with XmaI and NotI. The new construction, T7-fragI+II+III, and fragment IV were digested with KpnI and NotI prior to ligation and cloning. The final construction contained a complete MD145-12 ORF1 consensus sequence and was designated cORF1.
Site-directed mutagenesis of the cORF1 plasmid was performed with the QuikChange Mutagenesis kit (Stratagene, La Jolla, Calif.) to abolish potential cleavage sites. The constructs generated are diagrammed in Fig. 1B. Inactivation of the 3CL proteinase encoded in cORF1 was accomplished by replacement of the Cys-1147 of the putative active site (position 1147 in the ORF1 amino acid sequence of MD145-12) with an Ala using the mutagenesis primer (C1147A) shown in Table 1. The resulting construct was designated Pro−-cORF1 (Fig. 1B).
TABLE 1.
Oligonucleotides used for the site-directed mutagenesis
| Mutation | Oligonucleotide sequencea |
|---|---|
| C1147Ab | CACCAGGTGACGCCGGCTGCC |
| Q330Ac | GATTACGAGTTAGCAGGGCCTGAGGAC |
| Q696Ac | GTATGAGCTAGCGGGCCCAACTCTC |
| E875Ac | CGACATCAAAACTGCGGGCAAGAAAG |
| E1008Ac | GACTCAGTTTTGCGGCCCCAC |
| E1189Ac,d | GAGGCTACACTTGCAGGCGGTGACAG |
| E1189Gd | GCTACACTTGGCGGCGGTGAC |
| E1189Qd | GCTACACTTCAAGGCGGTGAC |
| G1190Qd | GCTACACTTGAACAAGGTGACAG |
| L1188Ad | GAGAGGCTACAGCTGAAGGCGGTG |
| G1191Ad | CACTTGAAGGCGCTGACAGTAAGG |
Only the positive-sense oligonucleotide is shown. Nucleotide substitutions are underlined.
The Cys at position 1147 was replaced by an Ala in the catalytic site of the proteinase.
Amino acid changes in the ORF1 polyprotein cleavage sites. The construct cORF1 was used as a template for the site-directed mutagenesis.
Amino acid changes in the cleavage site between the proteinase and the polymerase. The construct pET-ProPol was used as a template for the site-directed mutagenesis.
Cloning of MD145-12 sequences into pET vectors and mutagenesis.
The plasmid constructions pET-Pro, pET-VPg, pET-VPgPro, pET-ProPol, and pET-ΔNtermNTPasep20VPgPro, which encoded the proteinase, the VPg, a VPgPro precursor, the proteinase-polymerase precursor, and an N-terminally truncated (from amino acids 1 to 199) Nterm-NTPase-p20-VPg-proteinase complex, respectively, were engineered by using the pET-28b vector (Novagen, Madison, Wis.). The construction pET-Δp20VPgPro, encoding an N-terminally truncated (amino acids 1 to 152) p20-VPg-proteinase complex was generated by using the pET-29b vector (Novagen). Primers used for construction of the plasmids are shown in Table 2. The reverse and forward primers were designed to include the NotI and NcoI sites, respectively, and selected primers incorporated a coding sequence for an engineered His6 tag. Virus-specific DNA fragments for cloning were generated by PCR with cORF1 as a template, digested with NotI and NcoI, and cloned into the corresponding sites of the selected pET vector.
TABLE 2.
Oligonucleotides used in the construction of bacterial expression vectors
| Construct | Oligonucleotide sequencea |
|---|---|
| pET-VPg | AAGGAAAAAAGCGGCCGCTCACTCAAAACTGAGTCTCTCATTGTAGTC (reverse) |
| CATGCCATGGGCCATCATCATCATCATCATGGCAAGAAAGGGAAGAACAAGACTGGCCG (forward) | |
| pET-Pro | AAGGAAAAAAGCGGCCGCTCAGTGGTGGTGGTGGTGGTGGCCAAGTGTAGCCTCTCCCTCACTCCCCTG (reverse) |
| CATGCCATGGCCCCACCAAGCATCTGGTCGAGGATAGTC (forward) | |
| pET-Δp20VPgPro | AAGGAAAAAAGCGGCCGCTCAGTGGTGGTGGTGGTGGTGGCCAAGTGTAGCCTCTCCCTCACTCCCCTG (reverse) |
| CATGCCATGGGCCCAACTCTCACCACTTTCAACTTTGATCG (forward) | |
| pET-ΔNtermNTPasep20VPgPro | AAGGAAAAAAGCGGCCGCTCAGTGGTGGTGGTGGTGGTGGCCAAGTGTAGCCTCTCCCTCACTCCCCTG (reverse) |
| CATGCCATGGCCTTCGACAACAATTGCTATGCGTTCTG (forward) | |
| pET-VPgPro− | AAGGAAAAAAGCGGCCGCTCAGTGGTGGTGGTGGTGGTGGCCAAGTGTAGCCTCTCCCTCACTCCCCTG (reverse) |
| CATGCCATGGGCAAGAAAGGGAAGAACAAGACTGGCCG (forward) | |
| pET-ProPol | AAGGAAAAAAGCGGCCGCCTCGACGCCATCTTCATTCACAAAACTGGGAGC (reverse) |
| CATGCCATGGCCCCACCAAGCATCTGGTCGAGGATAGTC (forward) | |
| pET-Pro−Pol;E1189A/ProPol | AAGGAAAAAAGCGGCCGCTCACTCGACGCCATCTTCATTCAC (reverse) |
| CATGCCATGGGCCATCATCATCATCATCATGCCCCACCAAGCATCTGGTCGAGGATAGTC (forward) |
For the reverse and forward primers, NotI and NcoI sites (underlined) were included, respectively. His6 tag encoding sequences are indicated in bold.
The 3CL proteinase encoded in the pET-VPgPro and pET-ProPol plasmids was genetically inactivated by site-directed mutagenesis (Stratagene) by using primer C1147A as described above. The resulting plasmids were designated pET-VPgPro− and pET-Pro−Pol, respectively.
The predicted E1189/G1190 cleavage site between the proteinase and polymerase proteins was abolished by site-directed mutagenesis of the pET-ProPol plasmid. The resulting plasmid was designated pET-E1189A/ProPol. Additional plasmid constructions were generated in which amino acid residues in the P1, P1′, P2, and P2′ positions of the cleavage site, numbered according to the method of Schechter and Berger (26), between the proteinase and the polymerase were changed by site-directed mutagenesis (see diagram in Fig. 7A). The primers used for the mutagenesis of the pET-ProPol plasmid are shown in Table 1.
FIG. 7.
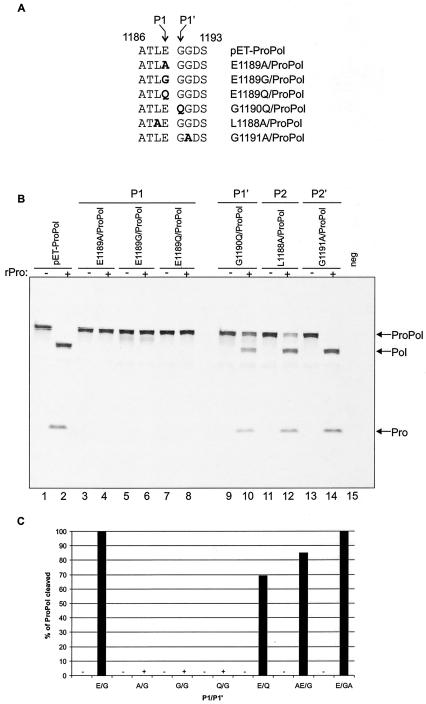
Mutational analysis of the ProPol dipeptide cleavage site in the trans cleavage assay. (A) Partial alignment of the ProPol cleavage site E1189/G1190 mutations. Changed amino acids are shown in bold type. Amino acids at the position P1 and P1′ of the cleavage site are indicated by arrows. (B) TNT reactions were conducted in a final volume of 25 μl for 1.5 h at 30°C, using the constructs indicated above the gel. For each construct, 5 μl of TNT reaction mixture was incubated with 5 μl of PBS (odd-numbered lanes, minus sign) or with 2 μg of MD145-12 rPro (even-numbered lanes, plus sign). The TNT product derived from pET-ProPol is shown in lanes 1 and 2. The negative control (neg) in lane 15 is an in vitro translation reaction mixture containing the pET-28b plasmid. Cleavage assays were resolved in 10 to 20% Tris-Gly polyacrylamide gels prior to autoradiography. ProPol, Pol, and Pro are indicated. (C) The cleavage efficiency of the ProPol precursor was determined by phosphorimaging. The intensity for the ProPol bands was determined in arbitrary units before (AUPBS) and after (AUPro) proteinase treatment. For each construct, the calculation of the cleavage efficiency was possible since the amount of [35S]Met was the same in the presence or absence of proteinase. The percentage of cleaved ProPol for the trans cleavage assay was calculated by the following formula: (1-AUPro/AUPBS) × 100.
In vitro translation analysis.
Coupled TNT reactions with T7 RNA polymerase were performed as recommended by the manufacturer (Promega Corporation, Madison, Wis.). Briefly, the TNT reaction was performed in a final volume of 25 μl with 1 to 3 μg of plasmid DNA and 15 μCi of [35S]methionine (>1,000 μCi/mmol) per reaction (Amersham Biosciences Corp, Piscataway, N.J.). The reaction mixture was incubated at 30°C for at least 1 h.
Protein analysis in SDS-PAGE and mapping of protein cleavage products.
Plasmids pET-ProPol and pET-Δp20VPgPro were used to transform E. coli BL21(DE3) cells (Novagen). Ten milliliters of bacteria was grown in Terrific Broth (TB) (Quality Biological Inc., Gaithersburg, Md.) until the optical density at 600 nm reached 0.6 to 1 and then induced for 4 h with 1 mM isopropylthio-β-d-galactoside (IPTG). The cells were pelleted and resuspended in 1 ml of lysis buffer (300 mM NaCl, 50 mM NaH2PO4 [pH 8.0], 10% glycerol) prior to one freeze-thaw cycle. The material was then sonicated to generate a total cell lysate. The soluble fraction was separated from the insoluble fraction by centrifugation of the total cell lysate at 10,000 × g for 10 min. The supernatant (soluble fraction) was collected, and the pellet (insoluble fraction) was resuspended in 1 ml of lysis buffer. Five microliters of each fraction was resolved in a 10 to 20% Tris-glycine (Tris-Gly) polyacrylamide gel by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Invitrogen) prior to staining with GelCode Blue (Pierce, Rockford, Ill.) to monitor protein expression.
Five microliters of the soluble fraction was separated in a 10 to 20% Tricine polyacrylamide gel (Invitrogen) and transferred by electroblotting to a ProBlott membrane (Applied Biosystems, Foster City, Calif.). The proteins were stained with Coomassie blue, and the bands of interest were excised for direct N-terminal amino acid sequence analysis by Edman degradation. For microsequencing of radiolabeled proteins, TNT products synthesized from the pET-ΔNtermNTPasep20VPgPro plasmid in the presence of [35S]Met were resolved in a 10 to 20% Tricine gel by SDS-PAGE and transferred by electroblotting to a Problott membrane. Autoradiography was used to locate the bands of interest on the membrane. These bands were then excised and subjected to N-terminal sequence analysis by Edman degradation, followed by analysis of each fraction in a scintillation counter.
Protein expression, purification, and production of antisera.
Plasmids pET-Pro, pET-VPg, pET-Pro−Pol, and pET-E1189A/ProPol were used to transform E. coli BL21(DE3) cells (Novagen). For each construction, 500 ml of bacteria were grown in TB medium until the optical density at 600 nm reached 0.8 to 1. Protein expression was then induced by addition of IPTG to a final concentration of 1 mM, followed by incubation of the bacteria for 4 h at 37°C. The cells were centrifuged at 4,000 × g for 20 min at 5°C and resuspended with 50 ml of lysis buffer. The cell lysate was frozen at −80°C and then thawed at room temperature before sonication. The total cell lysate was passed through a 5/8-in.-gauge needle to complete the shearing of the bacterial DNA. The mixture was centrifuged at 10,000 × g for 30 min at 5°C. The supernatant was collected and incubated with 5 ml of 50% nickel-nitrilotriacetic acid (Ni-NTA) resin slurry (Qiagen, Valencia, Calif.) for 2 h at 4°C. The resin mixture was packed to prepare a column. The column was washed with 50 and 75 ml of 10% glycerol, 10 mM β-mercaptoethanol, 300 mM NaCl, 50 mM NaH2PO4 (pH 8.0) containing 10 and 20 mM imidazole (Sigma, St. Louis, Mo.), respectively. The protein of interest was eluted with the same buffer containing either 60 mM imidazole (E1189A/ProPol and pET-Pro−Pol) or 250 mM imidazole (pET-Pro and pET-VPg). The recombinant (r) Pro and VPg proteins were dialyzed against 10% glycerol, 300 mM NaCl, 2 mM dithiothreitol, and 20 mM HEPES (pH 7.4). The rProPol complex was dialyzed against 10% glycerol, 150 mM KCl, 0.05 mM EDTA, 2 mM dithiothreitol, and 5 mM Tris-HCl (pH 7.5). The quantity of protein was estimated by the Bradford method (5). Hyperimmune sera were raised in guinea pigs against the purified rPro and rVPg proteins as previously described (32).
Immunoprecipitation assay.
A 10-μl aliquot of the TNT mixture was incubated with 5 μl of serum for 1 h at room temperature. The mixture was then rotated at 4°C overnight with protein G-Sepharose beads (Amersham Biosciences). The beads were washed, and the precipitated proteins were analyzed by SDS-PAGE as described previously (32).
trans cleavage assay.
Three to five microliters of TNT reaction product was mixed with either 5 μl of phosphate-buffered saline (PBS) (pH 7.4) containing 2 μg of purified protein or 5 μl of soluble fraction from the bacterial cell lysate. The TNT reaction product used for analysis of trans cleavage of the Norwalk virus polyprotein substrate was derived from plasmid NV FL101, which encodes a full-length “consensus” cDNA clone of the genome of Norwalk virus (cluster GI/1) in the pSPORT1 vector (S. Sosnovtsev, unpublished data). The 10-μl mixture was incubated overnight at 30°C. The entire reaction mixture was then resolved in a 12% Tris-Gly polyacrylamide gel by SDS-PAGE. Proteins were stained with Coomassie blue, after which the gel was dried and subjected to autoradiography.
Signal quantification of the [35S]Met-labeled proteins.
The intensity of radioactive bands was analyzed by using the PhosphorImager (Molecular Dynamics). The IP Lab Gel suite (Signal Analytics, Vienna, Va.) was used for the computer image analysis.
RESULTS
Establishment of the MD145-12 ORF1 polyprotein cleavage map.
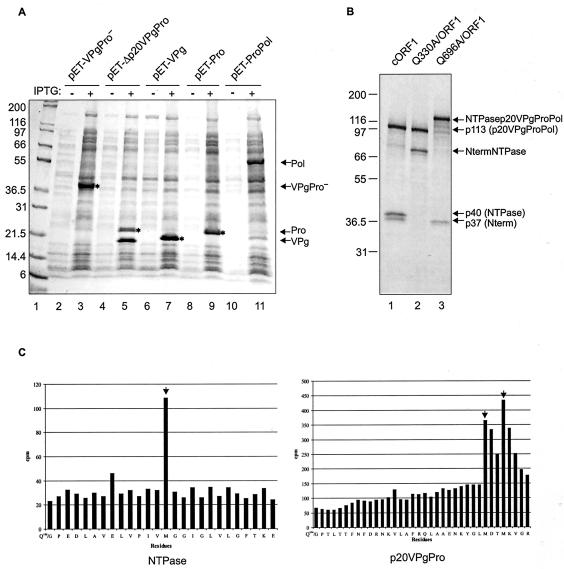
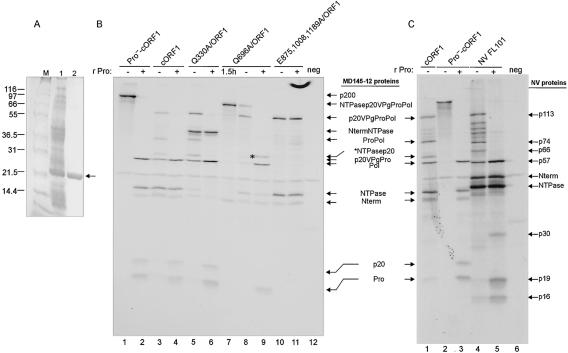
Our first goal was to determine the cleavage sites in the ORF1 polyprotein of the MD145-12 strain that are recognized by the virus-encoded 3CL proteinase. The borders of the MD145-12 VPg, Pro, and Pol proteins were identified by direct N-terminal amino acid sequence analysis (data not shown) of cleavage products generated upon expression of the proteinase and surrounding regions in bacteria (Fig. 2A). The pET-Δp20VPgPro construct encodes the C-terminal 27 amino acids of the protein p20, the complete VPg, and the predicted proteinase. The expressed proteinase was active and cleaved the protein complex to produce two major proteins of approximately 21 and 24 kDa (Fig. 2A, lane 5). The N termini of both proteins were sequenced and corresponded to cleavage at the dipeptide sites E875/G876 and E1008/A1009 to produce VPg and proteinase, respectively. The observed mass (24 kDa) of the cleaved proteinase expressed in the pET-Δp20VPgPro construct was slightly higher than the calculated mass of the mature proteinase (19.3 kDa), reflecting the presence of an engineered His tag at its C terminus. In a similar experiment, a pET-ProPol construct was engineered and expressed in bacteria to map the E1189/G1190 cleavage site between the proteinase and polymerase by direct N-terminal sequence analysis of a predominant 55-kDa protein (Fig. 2A, lane 11). Attempts to express larger regions of the ORF1 polyprotein in bacteria for mapping of upstream cleavage sites failed.
FIG. 2.
Mapping study of the MD145-12 norovirus ORF1 polyprotein. (A) Coomassie blue-stained bacterially expressed proteins used for N-terminal sequence analysis and trans cleavage assays. The constructs indicated above the gel were used to transform the strain BL21(DE3) of E. coli. Protein expression was induced as described in Materials and Methods, and the soluble proteins were separated in a 10 to 20% Tris-Gly polyacrylamide gel. The even- and odd-numbered lanes are soluble cellular fractions before (minus sign) and after (plus sign) IPTG induction, respectively. The proteins having a His6 tag are indicated by an asterisk. The proteins of interest are indicated by arrows on the right. For this figure and the following, Pro− indicates an inactivated proteinase, in which the Cys of the active site has been replaced by an Ala residue at position 1147. In a parallel experiment, proteins in the soluble fractions in lanes 5 and 11 were used to determine the N termini of the proteinase, VPg, and polymerase. The migration of VPg in lane 7 is slightly slower than in lane 5 because of an engineered His6 tag. Lane 1 contains Mark XII protein molecular weight marker (Invitrogen). Mark XII was used in all subsequent experiments. (B) Site-directed mutagenesis of the Q330/G331 and Q696/G697 MD145-12 ORF1 cleavage sites. In vitro translation assays with the plasmid constructs cORF1, Q330A/ORF1, and Q696A/ORF1 in lanes 1, 2, and 3, respectively, were performed for 1.5 h at 30°C in the presence of [35S]Met. For each construct, an aliquot of 5 μl was separated in a 10 to 20% Tris-Gly polyacrylamide gel by SDS-PAGE, and 35S-labeled products were detected by autoradiography. The deduced identity of each protein is indicated on the right. (C) Radioactive microsequencing mapping of the Q330/G331 and Q696/G697 cleavage sites. An in vitro translation reaction from the construct pET-ΔNtermNTPasep20VPgPro was performed in the presence of [35S]Met. The cleavage products NTPase and p20VPgPro were resolved by SDS-PAGE and electroblotted to a ProBlott membrane. For both proteins, an N-terminal sequencing analysis by Edman degradation was performed, and the released radioactivity (indicated in counts per minute) was determined for each cycle. Peaks corresponding to labeled Met residues are indicated by arrows. Amino acid sequences of the ORF1 polyprotein matching the radioactive profile are indicated below each graph.
The remaining cleavage sites of the MD145-12 ORF1 polyprotein were determined by using the combined methods of coupled TNT, site-directed mutagenesis, and radioactive microsequencing. Expression of the entire ORF1 polyprotein encoded in the cORF1 plasmid produced three major proteins of approximately 113, 40, and 37 kDa (Fig. 2B, lane 1). The observed cleavage products were consistent with those previously described for other noroviruses, suggesting that the conserved dipeptide cleavage sites at Q330/G331 and Q696/G697 in MD145-12 were utilized (10, 16, 27). To confirm this, the two potential cleavage sites were abolished by replacing the Gln in the P1 position by an Ala (constructs Q330A/ORF1 and Q696A/ORF1, illustrated in Fig. 1B). The TNT expression of Q330A/ORF1 produced two proteins of approximately 113 and 70 kDa, consistent with a disruption of the cleavage site that would yield the 40- and 37-kDa proteins (Fig. 2B, lane 2). For the construct Q696A/ORF1, two proteins of 140 and 37 kDa were observed, whereas the 113- and 40-kDa proteins were absent (Fig. 2B, lane 3). This protein profile was consistent with inactivation of the cleavage site between the 113- and 40-kDa proteins to yield a 140-kDa protein comprised of the 113- and 40-kDa proteins (NTPasep20VPgProPol). Taken together, these data support the identity of the 40- and 37-kDa proteins as Nterm and NTPase, respectively, and of the 113-kDa protein as the p20VPgProPol precursor. These results were confirmed by radioactive microsequencing of two major [35S]Met-labeled products derived from a TNT reaction mixture containing plasmid pET-ΔNtermNTPasep20VPgPro. The radioactive methionine profiles of the N termini corresponded to those expected of the cleaved NTPase and p20, respectively (Fig. 2C).
These data indicated that the MD145-12 cleavage map (Fig. 1A) contains five dipeptide cleavage sites that define the borders of six nonstructural proteins. These sites are consistent with those previously identified for other noroviruses (10, 16, 18, 27, 28).
Precursor and product relationships among TNT cleavage products of the MD145-12 ORF1 polyprotein.
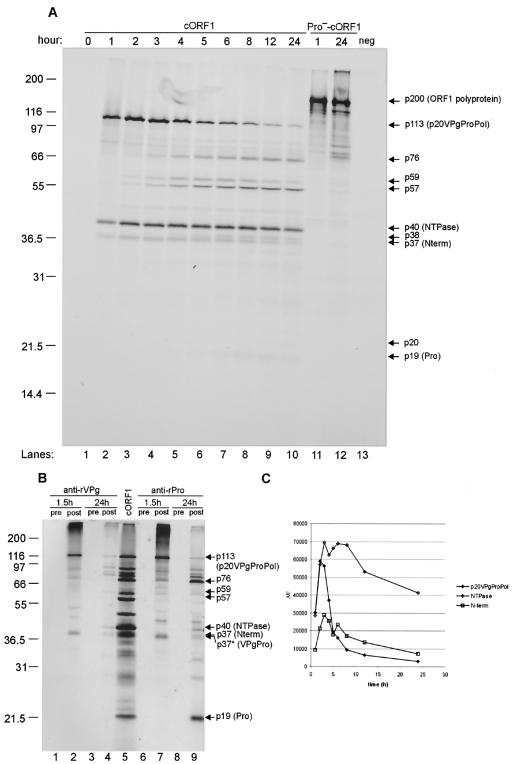
In vitro translation of cORF1 for 1 h yielded three major cleavage products (113, 40, and 37 kDa) (Fig. 2B, lane 1). We examined whether longer incubation periods of the cORF1 TNT reaction mixture would result in additional proteolytic processing of these products. As a control for this experiment, we generated a full-length ORF1 construct (Pro−-cORF1) with a genetically inactivated 3CL proteinase (Fig. 1B). A time course analysis of cORF1 was performed over a 24-h period (Fig. 3A). After 1 h, three proteins with molecular masses of 113, 40, and 37 kDa (designated p113, p40, and p37, respectively) that corresponded to the p20VPgProPol precursor, the NTPase, and the Nterm protein, respectively, as described above, were observed (Fig. 3A, lane 2). After 2 to 3 h, additional bands corresponding to proteins of 76, 59, and 57 kDa (designated p76, p59, and p57, respectively) were observed, and their intensities increased over time (up to 12 h for p59 and 24 h for p57 and p76) (Fig. 3A, lanes 2 to 10). At 4 h, a 38-kDa protein (designated p38) that migrated closely with the 37-kDa Nterm protein appeared and began to accumulate over time (Fig. 3A, lanes 5 to 10). At 5 h, a faint band corresponding to a protein of approximately 19 kDa (designated p19), which was consistent in size with the putative viral 3CL proteinase, was detected (Fig. 3A, lane 6) and increased over time. A faint band (designated p24) was observed late in the time course after prolonged exposure of the film (data not shown).
FIG. 3.
Kinetic analysis of the MD145-12 ORF1 polyprotein processing in TNT. (A) A TNT master mix using the cORF1 construct was aliquoted into 25-μl fractions that were incubated at 30°C for 1 to 24 h (lanes 1 to 10). A negative control for the processing was provided by the Pro−-cORF1 construct incubated in TNT reactions for 1 and 24 h at 30°C (lanes 11 and 12, respectively). Ten microliters of each fraction was resolved in a 12% Tris-Gly polyacrylamide gel. 35S-labeled products were detected by autoradiography. An additional control (neg) for these experiments included a TNT reaction with the pSPORT1 vector (lane 13). Protein designations are adjacent to the arrows. (B) Immunoprecipitation of the cORF1 in vitro translation products with antisera raised against the recombinant VPg or Pro proteins expressed in bacteria. The in vitro translation products were incubated either for 1.5 h or 24 h prior to immunoprecipitation analysis with preimmunization (lanes 1, 3, 6, and 8) or postimmunization sera (lanes 2, 4, 7, and 9). The precipitated products were separated in a 12% polyacrylamide gel prior to autoradiography. The positive control in lane 5 was 5 μl of a cORF1 in vitro translation mixture that was incubated 24 h at 30°C. The proteins of interest are indicated on the right with arrows. The asterisks mark the putative VPgPro complex. (C) The relative quantities of the Nterm, NTPase, and p20VPgProPol protein were calculated for each time point using a phosphorimager. Intensity values are given in arbitrary unit per Met residue (AU). This ratio is the relative light intensity divided by the number of Met residues present in the proteins Nterm (6 residues), NTPase (10 residues), or p20VPgProPol (26 residues).
The identities of these new cleavage products were examined by immunoprecipitation analysis of MD145-12 cORF1 TNT products representing the 1.5 h (early) and 24 h (late) time points. Hyperimmune sera specific for the MD145-12 recombinant VPg and Pro proteins were prepared in guinea pigs and used in the immunoprecipitation assay. The preimmunization sera from the guinea pigs did not react with the ORF1 TNT products (Fig. 3B, lanes 1, 3, 6, and 8). The VPg and Pro-specific sera precipitated p113 in the 1.5 h (Fig. 3B, lanes 2 and 7, respectively) and 24 h (Fig. 3B, lanes 4 and 9, respectively) TNT products, consistent with the identity of p113 as the p20VPgProPol precursor. The p59 protein was precipitated from the 24 h ORF1 TNT products by both sera (Fig. 3B, lanes 4 and 9). The observed mass of this protein and its recognition by both VPg- and Pro-specific antibodies suggested that it could be a precursor of p20VPgPro. An additional protein precipitated by both the VPg- and Pro-specific sera was designated p37* because it migrated to nearly the same position as the 37-kDa Nterm protein (Fig. 3B, lanes 2, 4, 7, and 9). The precipitation of this 37-kDa protein with both sera and its observed mass were consistent with its identity as a VPgPro precursor. It is interesting that this complex was present at 1.5 h (Fig. 3B, lanes 2 and 7) and showed a marked decrease in the 24 h TNT products (Fig. 3B, lanes 4 and 9) while the reverse was observed for p19 (Fig. 3B, lanes 7 and 9).
The Pro-specific serum precipitated p76 and p19 from the 24 h ORF1 products (Fig. 3B, lane 9), and these mature proteins corresponded to the expected masses of the ProPol and mature cleaved proteinase, respectively. Additional bands (including two proteins between 21.5 and 36.5 kDa) were observed in this immunoprecipitation assay (Fig. 3B, lane 9), but they were not characterized here. It should be noted that we could not detect the fully processed VPg with the VPg-specific antiserum because the MD145-12 VPg lacked methionine residues. However, radiolabeling experiments with [14C]Lys allowed the detection of a 16-kDa protein, consistent with the cleaved VPg (data not shown). Additional proteins observed in the time course (Fig. 3A) that were not characterized in the immunoprecipitation analysis were p57, p38, and p24.
The relative quantities of the p20VPgProPol, Nterm, and NTPase proteins synthesized during the TNT time course (Fig. 3A) were measured by using a phosphorimager (Fig. 3C). The values were assigned an arbitrary unit (AU = relative light unit/number of Met residues) based on the number of [35S]Met residues per protein (26, 6, and 10 for p20VPgProPol, Nterm, and NTPase, respectively). After 2 h of incubation, the intensity of the p20VPgProPol band began to decrease and declined to 5% of its maximum signal by 24 h. The intensity of the NTPase protein band remained the most stable in the reaction mixture, decreasing over time to 60% of its maximum signal. The Nterm protein signal decreased over time, but at a slower rate than the p20VPgProPol, reaching 25% of its maximum intensity at 24 h. We were not able to detect evidence for specific proteolytic processing of the Nterm or NTPase proteins in this expression system that would account for their decrease over time. However, we did observe the accumulation of products during the time course that were consistent with further processing of p20VPgProPol. These products were designated p76, p59, p57, p38, p20, and p19. The absence of these proteins in the 24 h Pro−-cORF1 reaction mixture (Fig. 3A, lane 12) supported their identities as specific cleavage products and not products of internal initiation of translation or nonspecific degradation.
The appearance of the Nterm, NTPase, and p20VPgProPol proteins within 1 h in the TNT time course suggested that cleavages at the dipeptides Q330/G331 and Q696/G697 were highly efficient and occurred cotranslationally while the proteinase was part of the larger p20VPgProPol precursor complex. Furthermore, the cleavages within the p20VPgProPol precursor occurred at a slower rate, suggesting a regulatory mechanism for the production of functional precursors and products from this region.
Analysis of the products derived by autocatalytic processing of the p20VPgProPol precursor.
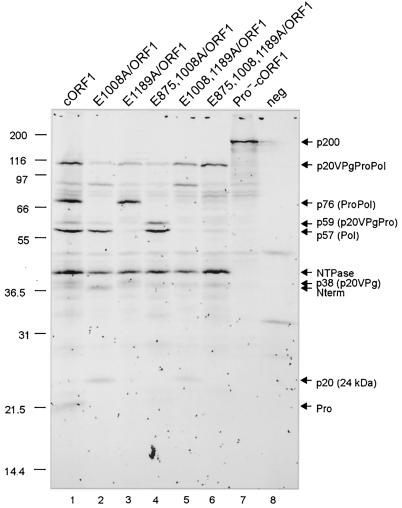
The ability to achieve additional in vitro proteolytic processing of the p20VPgProPol precursor by extension of the TNT incubation time allowed us to use a mutagenesis approach to further define the identities of the precursors and products derived from this region. As noted above, we had mapped the three cleavage sites E875/G876, E1008/A1009, and E1189/G1190 within the p20VPgProPol precursor by expression of various pET-based constructs derived from this region in bacteria (Fig. 2A). In addition, the immunoprecipitation analysis (Fig. 3B) showed evidence for the generation of VPg- and Pro-containing precursor proteins during proteolytic processing of the ORF1 in TNT. In the following experiments, we engineered various cleavage site mutations into the p20VPgProPol precursor-coding region of the cORF1 construct (illustrated in Fig. 1B) in order to examine their effects on proteolytic processing of the ORF1 polyprotein in TNT. Again, the mature form of the VPg of MD145-12 does not contain methionine and would not be detected in these TNT reactions. In addition, the VPgPro precursor (designated p37*) would be difficult to distinguish from the 37-kDa Nterm protein in these experiments. The cORF1 control TNT reaction mixture was incubated for 24 h and yielded the same products as described above: the 113-kDa p20VPgProPol precursor, the 40-kDa NTPase, the 37-kDa Nterm, and additional products designated p76, p59, p57, p38, p20, and p19 (Fig. 4, lane 1). The Pro−-cORF1 plasmid was included also as a control and yielded an approximately 200-kDa noncleaved ORF1 polyprotein and some minor unidentified proteins, including three proteins between 80 and 97 kDa that were consistently observed in this study and were likely products of internal initiation (Fig. 4, lane 7).
FIG. 4.
Site-directed mutagenesis of the E875/G876, E1008/A1009, and E1189/G1190 cleavage sites. In vitro translation assays containing the plasmid constructs indicated (lanes 1 to 7) were performed overnight at 30°C in the presence of [35S]Met. For each construct, 10 μl of a 25-μl reaction mixture was separated in a 12% Tris-Gly polyacrylamide gel. The in vitro translation product of pSPORT1 (lane 8) was included as the negative control (neg). 35S-labeled products were detected by autoradiography. Relevant proteins are indicated by arrows. It should be noted that the p20 protein showed an observed mass of 24 kDa. The constructs used in this experiment are diagrammed in Fig. 1.
The substitution of a Glu to an Ala at amino acid position 1008 (construct E1008A/ORF1) should abolish cleavage between the VPg and proteinase in the ORF1 polyprotein but allow the release of Pol and p20 from the p20VPgProPol precursor. Analysis of the TNT products from this construct showed the disappearance of the p76 and p19 bands, whereas the p57 and 24-kDa protein were detected (Fig. 4, lane 2). This pattern was consistent with the identity of p57 as the cleaved polymerase and the 24-kDa protein as p20. The slower migration of p20 and its related precursors in SDS-PAGE was consistent throughout our experiments.
The next altered cleavage site in the p20VPgProPol precursor was engineered between the proteinase and polymerase (construct E1189A/ORF1). This mutation should abolish cleavage between Pro and Pol but allow the release of the p20VPg precursor. Analysis of the TNT products from this plasmid showed an accumulation of p76, representing the intact ProPol, and the disappearance of cleaved Pol (p57) and Pro (p19) (Fig. 4, lane 3). Starting at the 5 h incubation time in the cORF1 TNT time course, we had observed a faint band of 38 kDa (p38) that accumulated over time (Fig. 3A, lanes 5 to 10). The p38 protein was consistent in size with the predicted precursor p20VPg, a protein analogous to the 3AB protein of the picornaviruses. The p38 was observed in the TNT products derived from E1189A/ORF1 (Fig. 4B, lane 3), consistent with its identity as the p20VPg precursor. Furthermore, within the TNT products from E1008A/ORF1, in which no cleavage was abolished between VPg and Pro, the 38-kDa p20VPg was not observed (Fig. 4, lane 2). The weak intensity of the p38 band was consistent with the presence of only four Met residues in the predicted p20VPg precursor protein.
To further confirm the identities of the precursors and products derived from the p20VPgProPol region, double and triple amino acid mutations were introduced into the cORF1 construct (Fig. 1B). The E875,1008A/ORF1 construct was designed to abolish cleavage between p20 and VPg as well as between VPg and the proteinase. In this construct, a p20VPgPro precursor and the mature Pol would be expected. Two proteins migrating at 59 and 57 kDa were observed in the TNT products (Fig. 4, lane 4). Given that p20VPgPro and Pol have 11 and 15 Met residues, respectively, the band intensity corresponding to Pol should be greater. The stronger band intensity and electrophoretic mobility indicated that p57 was the polymerase. The observed mass of p59, precipitated by both the VPg- and Pro-specific antisera (Fig. 3B, lanes 4 and 9, respectively), was consistent with the expected migration of p20VPgPro, assuming the altered mobility of the p20 observed above (Fig. 4, lane 2). To further confirm that p59 was p20VPgPro, the construct pET-NtermNTPasep20VPgPro (data not shown) was analyzed in a TNT reaction mixture that was incubated for 1.5 h at 30°C. Three bands corresponding to Nterm, NTPase, and p20VPgPro were observed. The latter band comigrated with p59 from an overnight TNT reaction of cORF1 (data not shown). The next double mutation was the E1008,1189A/ORF1 construct in which the cleavage sites between the VPg and proteinase as well as between the proteinase and polymerase were abolished. This construction would be expected to yield the cleaved p20 protein (which migrates at 24 kDa) and a 92-kDa VPgProPol complex. Proteins consistent with these masses were observed in the TNT reaction mixture (Fig. 4, lane 5). Mutation of all three predicted cleavage sites in the p20VPgProPol precursor resulted in the synthesis of the 113-kDa protein and none of the expected cleavage products from this region (Fig. 4, lane 6).
The proteolytic processing profiles of the TNT products observed by analysis of these ORF1 constructs verified the utilization of the cleavage sites identified in the mapping studies to release both precursors and products that were consistent with those observed in the ORF1 time course. In addition, the presence of the cleaved NTPase protein in these TNT reactions indicated that the Q330/G331 and Q696/G697upstream cleavage sites were not affected by modification of proteolytic processing events downstream in the p20VPgProPol.
trans cleavage of the ORF1 polyprotein by the MD145-12 purified proteinase.
We next developed an in vitro assay for the characterization of trans cleavage activity by the proteinase. The region of ORF1 encoding the ProPol was subcloned into pET-28b under the control of the T7 promoter. The cysteine in the active site of the proteinase was inactivated to produce Pro−Pol. Radiolabeled Pro−Pol protein was synthesized in a TNT reaction in order to provide a substrate for detection of trans cleavage at the E1189/G1190 site.
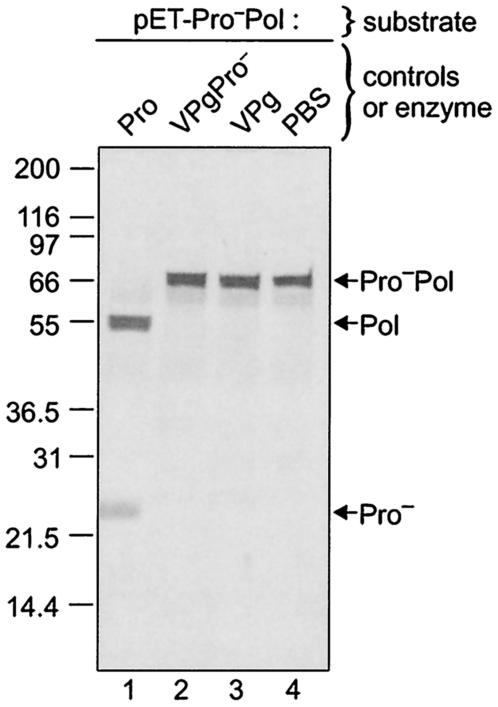
The mature MD145-12 proteinase was expressed in bacteria using the pET system (Fig. 2A, lane 9). In addition, the VPg alone (Fig. 2A, lane 7) and a precursor protein consisting of VPg and inactivated Pro (VPgPro−) (Fig. 2A, lane 3) were expressed in bacteria and included as controls for the detection of nonspecific bacterial protease activity. A nonradiolabeled total bacterial cell lysate containing each of these expressed proteins was incubated with the radiolabeled Pro−Pol substrate overnight at 30°C. The proteinase encoded by pET-Pro was able to cleave the Pro−Pol substrate into the 57-kDa Pol and the 22-kDa His-tagged Pro (Fig. 5, lane 1). No cleavage was observed when the Pro−Pol substrate was incubated with bacterial lysates containing either VPgPro− or VPg (Fig. 5, lanes 2 and 3, respectively) or with PBS alone (Fig. 5, lane 4). These data demonstrated that our mature bacteria-expressed recombinant proteinase was enzymatically active and that it was able to mediate cleavage in trans.
FIG. 5.
Development of a trans cleavage assay for detection of active forms of the viral proteinase. The radiolabeled Pro−Pol precursor protein was generated from the pET-Pro−Pol construct by TNT. Five microliters was used as a substrate in the trans cleavage assay. The enzymatic activity present in the Pro, VPgPro−, and VPg bacterial total cell lysates was assayed. Following overnight incubation at 30°C, the reaction assays were resolved in a 10 to 20% Tris-Gly polyacrylamide gel, and the 35S-labeled translation products were detected by autoradiography. The negative control (lane 4) consisted of 5 μl of the in vitro translation reaction mixture incubated with 5 μl of PBS (pH 7.4) overnight at 30°C. The cleavage products Pol and Pro− as well as the uncleaved Pro−Pol complex are indicated by arrows on the right.
The enzymatically active mature recombinant proteinase (designated rPro and containing an engineered His tag) was purified from bacteria by Ni-NTA affinity chromatography (Fig. 6A, lane 2) in order to further characterize its trans cleavage activity on selected MD145-12 ORF1 polyprotein substrates. Our data thus far had shown evidence for the production of at least six mature proteins from the ORF1 polyprotein: Nterm, NTPase, p20, VPg, Pro, and Pol. We examined whether rPro could mediate processing of these proteins from the ORF1 polyprotein in trans. The purified rPro enzyme (2 μg) was incubated with a 200-kDa full-length radiolabeled MD145-12 ORF1 polyprotein (Pro−-cORF1) that contained intact cleavage sites but an inactivated proteinase. The rPro was able to cleave the polyprotein into the mature nonstructural proteins (Fig. 6B, lane 2). For comparison, rPro was incubated with the TNT products from an ORF1 construct (cORF1) encoding an active proteinase. Before the addition of rPro, the TNT products contained both precursors and fully processed proteins (Fig. 6B, lane 3). The addition of rPro to this reaction resulted in the additional cleavage of precursors such as p20VPgProPol, ProPol, and p20VPgPro into their fully processed forms (Fig. 6B, lane 4).
FIG. 6.
trans cleavage activity of purified recombinant proteinase on native or variant forms of the ORF1 polyprotein. (A) SDS-PAGE analysis of the MD145-12 rPro before (lane 1) and after (lane 2) Ni-NTA purification. Proteins were stained with Coomassie blue, and the proteinase is indicated by an arrow. Lane M contains the protein molecular weight marker. (B) TNT reactions using the constructs (lanes 1 to 11) indicated above each lane were performed for 1.5 h at 30°C in the presence of [35S]Met. Five microliters of the TNT reaction mixture was incubated with either 5 μl of PBS (minus sign) or 5 μl of PBS containing 2 μg of purified rPro (plus sign). In lane 7, the TNT reaction mixture was incubated for 1.5 h at 30°C and immediately stored at −20°C without proteinase treatment. The negative control (neg) was a TNT reaction mixture containing pSPORT1. trans cleavage assays were analyzed by electrophoresis in a 12% Tris-Gly polyacrylamide gel prior to autoradiography. The proteins of interest are marked by arrows on the right. The asterisk in lane 9 indicates the putative NTPasep20 protein. (C) TNT reactions using the constructs (lanes 1 to 5) indicated above each lane were performed overnight at 30°C in the presence of [35S]Met. The NV FL101 plasmid (lanes 4 and 5) is a full-length cDNA clone of Norwalk virus (NV) (GI/1). Five microliters of the TNT reaction mixture was incubated with either 5 μl of PBS at pH 7.4 (minus sign) or 5 μl of PBS containing 2 μg of MD145-12 rPro (plus sign). The negative control (neg, lane 6) was a TNT reaction mixture containing pSPORT1. trans cleavage assays were analyzed by electrophoresis in a 12% Tris-Gly polyacrylamide gel prior to autoradiography. The proteins of interest for MD145-12 and NV are indicated on the left and right of the gel, respectively.
We used the trans cleavage assay to examine whether the ablation of certain cleavage sites would cause the utilization of alternative cleavage sites by the proteinase. We first examined the 24 h TNT products from the Q330A/ORF1 construct, in which cleavage between the Nterm and NTPase proteins was abolished to form the 80-kDa NtermNTPase complex (Fig. 6B, lane 5). Although the 80-kDa protein was predominant, faint bands corresponding to the possible Nterm and the NTPase proteins, likely produced by cleavage at an alternative site, were observed (Fig. 6B, lane 5). Addition of the rPro to this reaction showed evidence for processing of precursors such as p20VPgProPol and ProPol into their mature forms (Fig. 6B, lane 6). However, the 80-kDa protein and the possible Nterm and NTPase proteins produced by alternative cleavage remained at the same intensity. This observation indicates that the dipeptide Q330/G331 is the preferred accessible cleavage site for the proteinase between the Nterm and NTPase proteins.
The Q696A/ORF1 construct contains a mutated cleavage site between the NTPase and p20. Following incubation of the TNT reaction mixture for 1.5 h, two proteins corresponding in mass to the Nterm protein and the NTPasep20VPgProPol complex were observed (Fig. 6B, lane 7). However, after 24 h of incubation, the band corresponding to the NTPasep20VPgProPol decreased, while the intensity of bands consistent with the p20VPgProPol, Pol, and NTPase proteins increased (Fig. 6B, lane 8). The release of the NTPase indicated that an alternative cleavage site might be utilized near Q696/G697. The rPro was added to the Q696A/ORF1 TNT reaction (Fig. 6B, lane 9). Proteins consistent with the cleaved NTPase and p20 proteins were observed, indicating that proteolytic processing had occurred even in the presence of the mutated cleavage site between them. However, the new cleavage was less efficient since a unique protein consistent with the noncleaved NTPasep20 was still present after proteinase treatment (Fig. 6B, lane 9, asterisk). Because this second alternative cleavage site was not recognized in trans by the proteinase, it must likely be recognized only in cis when the preferred cleavage site is not available.
The TNT products of the E875,1008,1189A/ORF1 construct (which contains mutated cleavage sites in the p20VPgProPol precursor) were treated with 2 μg of rPro (Fig. 6B, lane 11). Before and after treatment, the predominant proteins were the p20VPgProPol precursor, NTPase, and Nterm (Fig. 6B, lanes 10 and 11, respectively). This experiment indicates that the cleavage sites E875/G876, E1008/A1009 and E1189/G1190 are unique, because evidence for alternative cleavage was not observed. Moreover, without the addition of rPro, the ORF1 polyprotein was cleaved to generate the NTPase and Nterm proteins, suggesting that the proteinase is active in the form of p20VPgPro and possibly also in the p20VPgProPol precursor proteins.
Previous studies of the GI Southampton and Norwalk virus ORF1 polyproteins reported evidence for little or no processing of the p113 protein (analogous to the MD145-12 p20VPgProPol precursor) in TNT reactions (10, 16). We examined whether the MD145-12 proteinase could process this Norwalk virus precursor in trans. The NV FL101 plasmid (which contains a cloned cDNA consensus sequence of the Norwalk virus genome) was used as the template in a TNT reaction mixture that was incubated 24 h in the absence or presence of 2 μg of MD145-12 rPro (Fig. 6C, lanes 4 and 5, respectively). Consistent with a previous study (10), prolonged incubation of the Norwalk virus TNT reaction mixture showed processing of the ORF1 polyprotein into the three major products, p113 (p20VPgProPol), p48 (Nterm), and p40 (NTPase) (Fig. 6C, lane 4). We found evidence for additional processing of the Norwalk virus polyprotein into p74, p57, and p19 proteins under these conditions (Fig. 6C, lane 4) consistent in observed mass to MD145-12 ProPol, Pol, and Pro, respectively (Fig. 6C, lane 1). Addition of the MD145-12 rPro to the Norwalk virus TNT products resulted in apparent further processing of the p113 and other precursor proteins such as ProPol into p57, p19, and p16 (Fig. 6C, lane 5) that corresponded in mass to the predicted Norwalk virus cleavage products of Pol, Pro, and VPg, respectively. The Norwalk virus VPg (p16) has one methionine residue, in contrast to the VPg of MD145-12, which allowed its detection in this experiment. It should be noted that a product, here designated p30, was also observed in the Norwalk virus trans cleavage assay (Fig. 6C, lane 5). It is likely that this protein corresponds to the predicted 22-kDa protein immediately upstream of the VPg that is analogous to the p20 of MD145-12. The Norwalk virus “p22” protein likely migrates more slowly, similar to the p20 of MD145-12 (which migrates as a 24-kDa protein). A 66-kDa protein observed in the Norwalk virus trans cleavage experiment may be the equivalent of the MD145-12 p20VPgPro, but this will need to be verified in future studies. Taken together, the in vitro proteolytic processing profiles of these GI and GII norovirus strains appear similar. In addition, the GII proteinase can recognize several GI cleavage sites in trans.
Effects of mutations in and near the ProPol dipeptide cleavage site on trans cleavage efficiency.
The time course experiment of ORF1 proteolytic processing in TNT (Fig. 3A) showed evidence for efficient early cis cleavage of the 200-kDa polyprotein by the proteinase, at dipeptides Q330/G331 and Q696/G697, to release the Nterm, NTPase, and p20VPgProPol proteins. The p20VPgProPol precursor was processed gradually over a 24-h incubation period, indicating that cleavages at the dipeptides E875/G876, E1008/A1009, and E1189/G1190 occurred more slowly. We examined whether the difference in the cleavage rate could be explained by the nature of the amino acid residue in the P1 position of the dipeptides. The two “early” cleavages contained a Gln in the P1 position, while the three cleavage sites of the precursor p20VPgProPol contained Glu. To test this hypothesis in the trans cleavage assay, we engineered a series of cleavage site mutations between Pro and Pol, using the pET-ProPol construct (with an active 3CL Pro) as diagrammed in Fig. 7A. For each construct, the corresponding TNT products were incubated overnight at 30°C in the presence or absence of 2 μg of purified MD145-12 rPro (Fig. 7B). The cleavage efficiency was estimated by quantitation of the signals captured with a phosphorimager (Fig. 7C). cis cleavage was not observed in any of the ProPol constructs, including the control with the wild-type E1189/G1190 dipeptide cleavage site (Fig. 7B, odd-numbered lanes). It was of interest that the ProPol construct with an engineered QG was not cleaved in cis, since the QG dipeptides at positions 330 and 331 and 696 and 697 were efficient cleavage sites. Addition of exogenous proteinase was used to determine whether the modified dipeptide cleavage sites were cleavable in trans. The pET-ProPol translation product was used as a positive control and was cleaved in trans to yield two proteins, the 57-kDa polymerase and the 19-kDa Pro (Fig. 7B, lane 2). For the constructs in which the Glu residue in the P1 position was replaced by an Ala, Gly, or Gln residue, cleavage in trans was not observed (Fig. 7B, lanes 4, 6, and 8). Of note, the Q1189/G1190 cleavage site sequence was identical to that of the efficient Q330/G331 and Q696/G697 sites, indicating that the primary sequence of the cleavage site was not the only determinant for trans cleavage by the proteinase.
The effects of changes in other residues in or near the dipeptide cleavage site were examined. In the construct G1190Q/ProPol, the Gly at the P1′ position was changed to a Gln residue (generating a E/Q site). The trans cleavage activity was reduced (Fig. 7B, lane 10), resulting in 68% efficiency in cleavage of the ProPol complex (Fig. 7C).
The effects of changing amino acid residues at the P2 and P2′ positions were examined by using the L1188A/ProPol and G1191A/ProPol constructs, respectively. We observed incomplete cleavage in trans by the proteinase when the Leu in the P2 position was replaced by an Ala (Fig. 7B, lane 12). In this experiment, 85% of the ProPol complex was cleaved (Fig. 7C). For the P2′ position construct (G1191A/ProPol), the ProPol complex was entirely cleaved (Fig. 7B, lane 14, and 7C).
Taken together, at least three amino acid substitutions in the P1 position of the ProPol cleavage site could not be tolerated, and we could not transport an efficient upstream dipeptide cleavage site (Q/G) into this position. However, the efficiency of the ProPol cleavage site recognition by the proteinase could be altered by substitutions in the P1′ and P2 positions. These data suggest that the primary sequence of the dipeptide cleavage site and its conformation when presented to the proteinase are important determinants of cleavage efficiency. Moreover, the observed stability of the pET-ProPol TNT product incubated overnight suggests that there is little or no cis cleavage of the free ProPol complex during the processing of the ORF1 polyprotein (Fig. 7B, lane 1).
trans cleavage of the ORF1 polyprotein by the MD145-12 ProPol precursor.
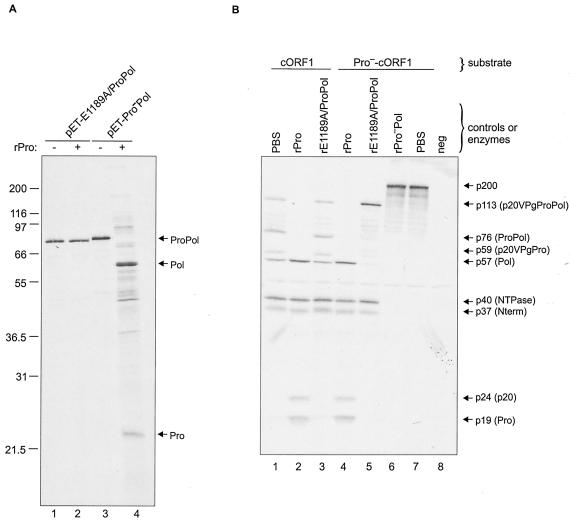
Because the cysteine proteinase of poliovirus is known to be active when it is part of the 3CD complex (13, 35), we examined whether the proteinase of MD145-12 is active when part of the ProPol complex. Two forms of recombinant ProPol were engineered to address this question. The rProPol derived from the construct pET-E1189A/ProPol contained an inactivated cleavage site between Pro and Pol and an active proteinase domain, and that which was derived from the construct pET-Pro−Pol contained an intact cleavage site between Pro and Pol and an inactivated proteinase. The constructs encoding these proteins were analyzed in TNT and, as expected, no cis cleavage was observed (Fig. 8A, lanes 1 and 3, respectively). Incubation of rPro with the Pro−Pol protein resulted in trans cleavage between Pro and Pol (Fig. 8A, lane 4), and incubation of rPro with the E1189A/ProPol protein did not (Fig. 8A, lane 2).
FIG. 8.
Proteinase activity of the ProPol complex. (A) Five microliters of the TNT products derived from constructs pET-E1189A/ProPol (lane 1) and pET-Pro−Pol (lane 3) was incubated 24 h with 5 μl of PBS to monitor for cis cleavage activity. The TNT products were also incubated with 2 μg of rPro to detect trans cleavage (lanes 2 and 4, plus signs). ProPol, Pol, and Pro proteins are indicated by arrows. (B) The cORF1 (lanes 1, 2, and 3) and Pro−-cORF1 constructs (lanes 4 to 7) were translated in vitro for 1.5 h at 30°C. trans cleavage assays were performed 24 h at 30°C. For each construct, an aliquot of 5 μl was incubated with either 5 μl of PBS (lanes 1 and 7) or 2 μg of rPro (lanes 2 and 4). The cORF1 and Pro−-cORF1 translation products were also incubated with 2 μg of bacterially expressed and partially purified rE1189A/ProPol (lanes 3 and 5) or 2 μg of rPro−Pol (lane 6). A TNT reaction mixture containing pSPORT1 was included as a control (lane 8). The cleavage assays were analyzed in a 12% Tris-Gly polyacrylamide gel prior to autoradiography. The proteins of interest are indicated with arrows.
The two forms of recombinant ProPol (rPro−Pol and rE1189A/ProPol) were then expressed in bacteria and partially purified on an Ni-NTA affinity column (data not shown). The ProPol proteins were examined for their proteinase activity in a trans cleavage assay using the ORF1 polyprotein templates encoded by cORF1 and Pro−-cORF1. As controls, the purified rPro−Pol or PBS were incubated overnight with the Pro−-cORF1 polyprotein substrate, and no cleavage was observed (Fig. 8B, lanes 6 and 7, respectively). Other controls for this experiment included the incubation of the cORF1 TNT products in the absence or presence of 2 μg of purified MD145-12 rPro (Fig. 8B, lanes 1 and 2, respectively). In the absence of proteinase, the proteins p20VPgProPol, ProPol, p20VPgPro, Pol, NTPase, and Nterm were observed (Fig. 8B, lane 1). The addition of rPro resulted in additional processing, with cleavage of the larger precursors and the appearance of p20 and Pro (Fig. 8B, lane 2). When the rE1189A/ProPol protein was added to the cORF1 translation products, little evidence for additional processing of the C-terminal p113 precursor was detected (Fig. 8B, lane 3). This observation indicated that rE1189A/ProPol could not efficiently process the cleavage sites within the p20VPgProPol complex in trans.
The incubation of the Pro−-cORF1 200-kDa translation product with 2 μg of rE1189A/ProPol protein yielded three proteins, p20VPgProPol, NTPase, and Nterm (Fig. 8B, lane 5). Of note, the rE1189A/ProPol protein again did not show efficient processing of the p20VPgProPol precursor into the ProPol, Pol, p20, and Pro proteins that were observed in the rPro reaction mixture containing the 200-kDa polyprotein as a substrate (Fig. 8B, lane 4). These results indicate that the ProPol complex has activity as a proteinase but that it differs from the mature proteinase in its ability to function in trans on the p20VPgProPol precursor.
DISCUSSION
Five cleavage sites recognized by the MD145-12 virus-encoded 3CL cysteine proteinase were mapped by using a combination of biochemical and genetic techniques. Cleavages at the dipeptides Q330/G331, Q696/G697, E875/G876, E1008/A1009, and E1189/G1190 defined the borders of six mature nonstructural proteins designated Nterm, NTPase, p20, VPg, Pro, and Pol. The MD145-12 cleavage map is consistent with that reported for the Southampton virus (GI/2) (16, 18) and the various cleavage sites reported for the Camberwell (also GII/4), Norwalk (GI/1), and Chiba (GI/4) noroviruses (10, 27, 28).
Kinetic analysis of in vitro polyprotein proteolytic processing showed that the MD145-12 ORF1 polyprotein is rapidly cleaved in a TNT reaction into three products, Nterm (37 kDa), NTPase (40 kDa), and the p20VPgProPol complex (113 kDa), similar to those reported previously for Southampton virus and other noroviruses (6, 10, 27). Prolonging the TNT incubation time to 24 h allowed us to detect further proteolytic processing of the MD145-12 p20VPgProPol complex into additional precursors and products, which have not been observed in such studies of other norovirus strains (10, 16). Prolonging the TNT incubation time also resulted in evidence for additional proteolytic processing of the Norwalk virus polyprotein encoded in a full-length consensus cDNA clone of the Norwalk virus genome. We identified the additional MD145-12 cleavage products, by using a combination of several techniques, as ProPol, p20VPgPro, Pol, p20VPg, VPgPro, p20, and Pro (designated p76, p59, p57, p38, p37*, p20, and p19, respectively). Synthesis of the Camberwell ORF1 polyprotein in COS cells yielded several protein products not readily detected in TNT, including ProPol, Pol, and Pro, which led to the suggestion that cellular factors not present in the TNT might be required to achieve full processing of the ORF1 polyprotein (16, 27). Cellular factors have been implicated in enhancing the cleavage efficiency of the poliovirus 3CD protein (2). However, our ability to identify the fully processed Pol, Pro, and p20 proteins indicates that complete cleavage of the p20VPgProPol complex by the MD145-12 proteinase can be achieved in TNT; but it must be given sufficient time.
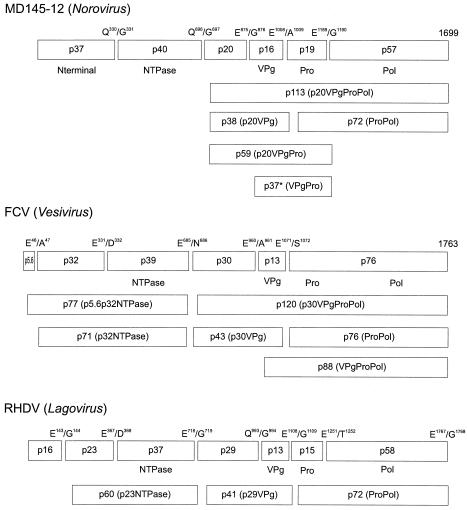
Comparison of the norovirus nonstructural proteins and their precursors with those of RHDV and FCV, analyzed in cell culture systems, reveals certain similarities and differences (Fig. 9). All caliciviruses encode an N-terminal protein of unknown function, an NTPase, a 20- to 30-kDa protein of unknown function, the VPg, a 3CL cysteine proteinase, and an RNA-dependent RNA polymerase. Although some variation has been observed among caliciviruses in their processing strategies to release the final mature nonstructural proteins (14, 19, 30, 32, 34), the accumulation of the stable MD145-12 precursor proteins p20VPg and ProPol, similar to those identified in cell culture studies of FCV and RHDV, suggests that these proteins are important in the calicivirus replication strategy. Of interest, these precursor proteins are likely analogous to the picornavirus 3AB and 3CD proteins, respectively (14, 30, 32, 33). One striking difference between the noroviruses in comparison with FCV and RHDV is the absence of an intermediate precursor containing part of the N-terminal protein (possibly the picornavirus 2B equivalent) complexed with the NTPase (p71 for FCV and p60 for RHDV) (19, 30). The p71 and p60 precursors are detected in FCV- and RHDV-infected cells, respectively, and further processing of these precursors releases the N terminus of the NTPase. The high efficiency of the cleavage between the Nterm and NTPase proteins for MD145-12 and other noroviruses (10, 16, 27) argues against the production of a stable NtermNTPase precursor during a norovirus infection.
FIG. 9.
Comparison of the nonstructural polyprotein cleavage maps of MD145-12 (Norovirus), feline calicivirus (FCV) (Vesivirus), and rabbit hemorrhagic disease virus (RHDV) (Lagovirus) (14, 19) and their known processing intermediates. Each cleavage product is designated by the letter “p” followed by its molecular mass. Functional domains and cleavage sites are indicated.
The establishment of the MD145-12 cleavage map allowed us to bacterially express and purify the mature viral proteinase in order to further characterize its substrate specificity. It was shown previously that the norovirus proteinase was enzymatically active when expressed in bacteria (17, 28). The norovirus proteinase resembles the 3C proteinase of the picornaviruses (4, 28). However, unlike most picornavirus 3C proteinases, the norovirus proteinase recognizes not only the QG dipeptide but also dipeptides with a Glu in the P1 position. We established a trans cleavage assay with an ORF1 polyprotein template as the substrate and the purified rPro as the enzyme and confirmed that the mature proteinase can cleave each of the five mapped cleavage sites in trans. Moreover, our data showed that the GII MD145-12 proteinase has the ability to mediate trans processing of cleavage sites in the polyprotein of a GI norovirus. The trans cleavage assays on polyprotein substrates with mutated cleavage sites were useful in the verification of several sites in the MD145-12 cleavage map. Indeed, the change of the P1 residue to an Ala residue either abolished or greatly reduced trans as well as cis cleavage on the polyprotein substrate. These data demonstrated that Q330/G331, Q696/G697, E875/G876, E1008/A1009, and E1189/G1190 are the preferred cleavage sites accessible to the proteinase in the ORF1 polyprotein, even in the presence of several additional QG, EG, and EA dipeptides in the ORF1 amino acid sequence. It should be noted that suppression of the Q696/G697 (and possibly Q330/G331) cleavage sites resulted in the apparent utilization of a nearby alternative cleavage site in cis by the proteinase, but we did not map this minor product. The biological relevance of alternative cleavage sites mapped in in vitro studies must await the development of cell culture or replicon systems for this virus group.
It has been shown for the picornaviruses (11, 21) and the caliciviruses (10, 31, 34) that certain amino acid residues in the P1 and P4 positions of the dipeptide cleavage site are essential for recognition by the proteinase. Our data obtained with the ProPol precursor as the substrate and the purified MD145-12 rPro as the enzyme in a trans cleavage assay confirmed the importance of the P1 position. Furthermore, the P1′ and P2 positions also proved important in that cleavage efficiency could be reduced by amino acid substitutions in these positions. We examined whether we could improve the in vitro efficiency of cleavage by changing the E1189/G1190 cleavage site within the ProPol substrate precursor to Q1189/G1190, the latter of which was an efficient upstream cleavage site. However, the presence of a Gln (Q) in the P1 position in the ProPol cleavage site abolished trans cleavage by exogenous rPro and did not result in the acquisition of autocatalytic cis cleavage mediated by the ProPol precursor. These data further support the role of both primary and conformational requirements for recognition and cleavage by the MD145-12 proteinase. Our data were in agreement with those of Hardy et al. (10), in which cis cleavage efficiency was not improved in the Norwalk virus polyprotein by changing the ProPol cleavage site from E/G to Q/G. These observations are important for future studies aimed at generating chimeric caliciviruses with nonstructural protein exchanges. The dipeptide cleavage site sequence as well as its conformation will need to be considered in order to achieve efficient cleavage by a homologous or nonhomologous proteinase.
For certain picornaviruses such as encephalomyocarditis virus, trans proteinase activity has been demonstrated for precursor proteins such as 3ABC, 3CD, and P3 (22), corresponding in norovirus to p20VPgPro, ProPol, and p20VPgProPol, respectively. Our data suggest that these norovirus precursors may have proteinase activity. The experiment depicted in Fig. 6 (lane 10) indicated that the proteinase was active when it was part of the noncleaved p20VPgPro (and likely the p20VPgProPol) precursor because upstream cleavage occurred between the Nterm and NTPase even when cis cleavage within p20VPgProPol was abolished by mutation of its cleavage sites. Mutation of E875/G876 and E1008/A1009 (Fig. 4, lanes 2 and 3, respectively) indicated that the proteinase is active also when it is part of VPgPro and, again, p20VPgPro. Because ProPol is a common calicivirus precursor, we examined whether the ProPol from MD145-12 could function as a proteinase. We expressed the MD145-12 rProPol in bacteria (with cleavage between the two proteins inactivated by mutagenesis of the cleavage site) and showed that it was active in trans on an ORF1 polyprotein substrate. Of interest, only the Q330/G331 and Q696/G697 dipeptides were processed efficiently by the rProPol. The nature of the surrounding residues at these QG cleavage sites may give insight into the cleavage requirement for ProPol in that the cleavage sites are part of a conserved LXXYELQG motif. Future studies will be needed to examine the essential residues in this motif. The observation that only the efficient early cleavages of the polyprotein can be mediated by the ProPol precursor suggests that its role, if any, in proteolytic processing may be limited to certain cleavage events, as described previously for the poliovirus 3CD protein (13, 35). The poliovirus 3CD protein apparently cleaves only the QG dipeptides between VP0, VP3, and VP1, and the residues present at the P1 and P4 positions of these cleavage sites are essential for their recognition by 3CD (3). Because the 3CD of poliovirus (25) and ProPol of FCV (33) are likely multifunctional proteins, it will be interesting to examine whether norovirus ProPol precursors might have multiple functions during replication.
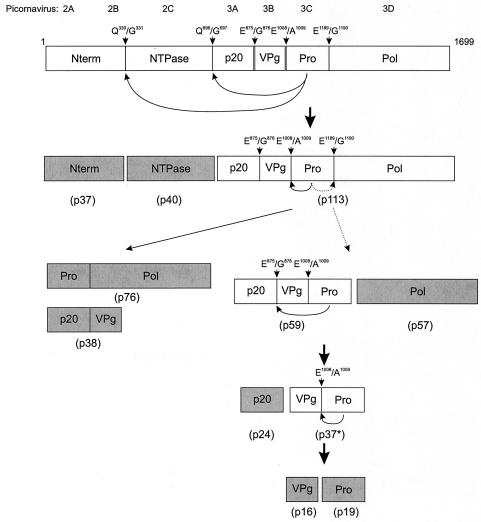
Taken together, the data in this study allow us to propose a model for proteolytic processing of the MD145-12 ORF1 polyprotein (Fig. 10). The kinetic experiment indicated that cleavage of the two QG dipeptides (Q330/G331 and Q696/G697), which resulted in the release of the proteins Nterm, NTPase, and p20VPgProPol, occurs rapidly as a cotranslational cleavage event as proposed for Southampton virus (16). Our new data indicate that proteolytic processing of the p20VPgProPol then occurs to release several precursors and products in two possible secondary cleavage pathways. In one pathway, the cleavage following the release of the Nterm and NTPase takes place at the E1008/A1009 dipeptide to release p20VPg and ProPol. The ProPol precursor accumulates throughout the 24-h time course, indicating that it is unable to mediate efficient autocatalytic cis cleavage at its E1189/G1190 dipeptide cleavage site, consistent with observations by Someya et al. (28). This is supported by our in vitro studies of ProPol in which prolonged incubation of this precursor did not result in cis cleavage (Fig. 7B, lane 1). In a second pathway, a secondary cleavage occurs directly at the E1189/G1190 dipeptide to release p20VPgPro and Pol. The evidence for these two secondary cleavage events is the accumulation of p20VPg, ProPol, p20VPgPro, and Pol over time as the amount of p20VPgProPol complex decreases. Following the secondary cleavages, further proteolytic processing releases the mature proteins. At 8 h, a decrease in the p20VPgPro precursor was observed that was likely due to cis or trans cleavage to release p20 and VPgPro. The VPgPro may then be cleaved to release VPg and Pro. These cleavages were supported by our immunoprecipitation data (Fig. 3B) demonstrating the presence of VPgPro and the absence of cleaved Pro at the 1.5 h time point, followed by a decrease in VPgPro and an increase in cleaved Pro at the 24 h time point. In our model, certain proteins (indicated in gray in Fig. 10) appeared to be the final stable products of cleavage that might be present in a norovirus infection. These are Nterm (although further processing cannot be ruled out), NTPase, p20, VPg, Pro, Pol, p20VPg, and ProPol. Other proteins (indicated in white in Fig. 10) represent either precursors that were never detected in this study (the intact polyprotein) or that decreased over time, including the p20VPgProPol, p20VPgPro, and VPgPro.
FIG. 10.
Model for temporal proteolytic processing of the MD145-12 ORF1 polyprotein by the viral proteinase. Active dipeptide cleavage sites and their locations are indicated. Primary cleavages at Q330/G331 and Q696/G697 release the N-terminal and NTPase proteins. Two possible secondary cleavage pathways involve processing of the p20VPgProPol precursor. In one secondary pathway, the proteinase may cleave at E1008/A1009 to release p20VPg and ProPol. In the other pathway (shown with the dotted arrows), the proteinase may cleave at E1189/G1190 to release p20VPgPro and Pol. Additional processing of the released precursors occurs over time. Likely cis cleavages executed by the proteinase are shown immediately below the indicated precursor. White boxes indicate proteins in which the observed levels decrease over time. Gray boxes indicate proteins that remain present or accumulate over time. The designation for each protein based on molecular mass is indicated in parentheses.
Although cell culture systems or replicon systems are not yet available for the noroviruses, knowledge of the MD145-12 cleavage precursors and products identified in this work may facilitate the development of systems for the study of norovirus replication modeled by the caliciviruses that are characterized in cell culture systems. The processing map of the MD145-12 ORF1 polyprotein is similar to that of FCV, which has been verified by reverse genetics (30), and in addition, many of the processing events we have identified for our norovirus strain in vitro are consistent with those observed in FCV-infected cells (30). The slower proteolytic processing of the MD145-12 p20VPgProPol precursor suggests the existence of a strategy in norovirus replication to control the synthesis of each cleavage product and processing intermediate. Such a strategy has been proposed for the picornaviruses, in which both precursors and products have defined roles in replication as summarized by Racaniello (25). The definition of the borders of the mature MD145-12 nonstructural proteins and the stable precursors will also facilitate studies of their structure and function, and this information may be useful in the development of prevention and treatment strategies for these ubiquitous pathogens.
Acknowledgments
We thank Bob Schackmann (University of Utah) and John Shannon (University of Virginia) for assistance with the protein sequence analysis and Kyeong-Ok Chang (NIAID, NIH) for helpful discussions. We extend our appreciation to Albert Z. Kapikian (NIAID, NIH) for critical review of the manuscript and support of this project.
REFERENCES
- 1.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. [DOI] [PubMed] [Google Scholar]
- 2.Blair, W. S., X. Li, and B. L. Semler. 1993. A cellular cofactor facilitates efficient 3CD cleavage of the poliovirus P1 precursor. J. Virol. 67:2336-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blair, W. S., and B. L. Semler. 1991. Role for the P4 amino acid residue in substrate utilization by the poliovirus 3CD proteinase. J. Virol. 65:6111-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boniotti, B., C. Wirblich, M. Sibilia, G. Meyers, H. J. Thiel, and C. Rossi. 1994. Identification and characterization of a 3C-like protease from rabbit hemorrhagic disease virus, a calicivirus. J. Virol. 68:6487-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Clarke, I. N., and P. R. Lambden. 2001. The molecular biology of human caliciviruses. Novartis Found. Symp. 238:180-191. [DOI] [PubMed] [Google Scholar]
- 7.Glass, P. J., L. J. White, J. M. Ball, I. Leparc-Goffart, M. E. Hardy, and M. K. Estes. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 74:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green, K. Y., G. Belliot, J. L. Taylor, J. Valdesuso, J. F. Lew, A. Z. Kapikian, and F. Y. Lin. 2002. A predominant role for Norwalk-like viruses as agents of epidemic gastroenteritis in Maryland nursing homes for the elderly. J. Infect. Dis. 185:133-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, K. Y., R. M. Chanock, and A. Z. Kapikian. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 10.Hardy, M. E., T. J. Crone, J. E. Brower, and K. Ettayebi. 2002. Substrate specificity of the Norwalk virus 3C-like proteinase. Virus Res. 89:29-39. [DOI] [PubMed] [Google Scholar]
- 11.Jewell, D. A., W. Swietnicki, B. M. Dunn, and B. A. Malcom. 1992. Hepatitis A virus 3C proteinase substrate specificity. Biochemistry 31:7862-7869. [DOI] [PubMed] [Google Scholar]
- 12.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 13.Jore, J., B. De Geus, R. J. Jackson, P. H. Pouwels, and B. E. Enger-Valk. 1988. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J. Gen. Virol. 69:1627-1636. [DOI] [PubMed] [Google Scholar]
- 14.König, M., H. J. Thiel, and G. Meyers. 1998. Detection of viral proteins after infection of cultured hepatocytes with rabbit hemorrhagic disease virus. J. Virol. 72:4492-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambden, P. R., E. O. Caul, C. R. Ashley, and I. N. Clarke. 1993. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 259:516-519. [DOI] [PubMed] [Google Scholar]
- 16.Liu, B., I. N. Clarke, and P. R. Lambden. 1996. Polyprotein processing in Southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J. Virol. 70:2605-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, B. L., P. R. Lambden, H. Gunther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine enteric calicivirus: relationship to the Norwalk-like viruses. J. Virol. 73:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, B. L., G. J. Viljoen, I. N. Clarke, and P. R. Lambden. 1999. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J. Gen. Virol. 80:291-296. [DOI] [PubMed] [Google Scholar]
- 18a.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed]
- 19.Meyers, G., C. Wirblich, H. J. Thiel, and J. O. Thumfart. 2000. Rabbit hemorrhagic disease virus: genome organization and polyprotein processing of a calicivirus studied after transient expression of cDNA constructs. Virology 276:349-363. [DOI] [PubMed] [Google Scholar]
- 20.Noel, J. S., R. L. Fankhauser, T. Ando, S. S. Monroe, and R. I. Glass. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 179:1334-1344. [DOI] [PubMed] [Google Scholar]
- 21.Pallai, P. V., F. Burkhardt, M. Skoog, K. Schreiner, P. Bax, K. A. Cohen, G. Hansen, D. E. H. Palladino, K. S. Harris, M. J. Nicklin, and E. Wimmer. 1989. Cleavage of synthetic peptides by purified poliovirus 3C proteinase. J. Biol. Chem. 264:9738-9741. [PubMed] [Google Scholar]
- 22.Parks, G. D., J. C. Baker, and A. C. Palmenberg. 1989. Proteolytic cleavage of encephalomyocarditis virus capsid region substrates by precursors to the 3C enzyme. J. Virol. 63:1054-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfister, T., and E. Wimmer. 2001. Polypeptide p41 of a Norwalk-like virus is a nucleic acid-independent nucleoside triphosphatase. J. Virol. 75:1611-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pletneva, M. A., S. V. Sosnovtsev, and K. Y. Green. 2001. The genome of Hawaii virus and its relationship with other members of the caliciviridae. Virus Genes 23:5-16. [DOI] [PubMed] [Google Scholar]
- 25.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 26.Schechter, I., and A. Berger. 1967. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27:157-162. [DOI] [PubMed] [Google Scholar]
- 27.Seah, E. L., J. A. Marshall, and P. J. Wright. 1999. Open reading frame 1 of the Norwalk-like virus Camberwell: completion of sequence and expression in mammalian cells. J. Virol. 73:10531-10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Someya, Y., N. Takeda, and T. Miyamura. 2000. Complete nucleotide sequence of the Chiba virus genome and functional expression of the 3C-like protease in Escherichia coli. Virology 278:490-500. [DOI] [PubMed] [Google Scholar]
- 29.Someya, Y., N. Takeda, and T. Miyamura. 2002. Identification of active-site amino acid residues in the Chiba virus 3C-like protease. J. Virol. 76:5949-5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sosnovtsev, S. V., M. Garfield, and K. Y. Green. 2002. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 76:7060-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sosnovtsev, S. V., S. A. Sosnovtseva, and K. Y. Green. 1998. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 72:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sosnovtseva, S. A., S. V. Sosnovtsev, and K. Y. Green. 1999. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J. Virol. 73:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei, L., J. S. Huhn, A. Mory, H. B. Pathak, S. V. Sosnovtsev, K. Y. Green, and C. E. Cameron. 2001. Proteinase-polymerase precursor as the active form of feline calicivirus RNA-dependent RNA polymerase. J. Virol. 75:1211-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wirblich, C., M. Sibilia, M. B. Boniotti, C. Rossi, H. J. Thiel, and G. Meyers. 1995. 3C-like protease of rabbit hemorrhagic disease virus: identification of cleavage sites in the ORF1 polyprotein and analysis of cleavage specificity. J. Virol. 69:7159-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ypma-Wong, M. F., P. G. Dewalt, V. H. Johnson, J. G. Lamb, and B. L. Semler. 1988. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology 166:265-270. [DOI] [PubMed] [Google Scholar]