Abstract
A process that we refer to as control by epistasy of synthesis (CES process) occurs during chloroplast protein biogenesis in Chlamydomonas reinhardtii: the synthesis of some chloroplast-encoded subunits, the CES subunits, is strongly attenuated when some other subunits from the same complex, the dominant subunits, are missing. Herein we investigate the molecular basis of the CES process for the biogenesis of the cytochrome b6f complex and show that negative autoregulation of cytochrome f translation occurs in the absence of other complex subunits. This autoregulation is mediated by an interaction, either direct or indirect, between the 5′ untranslated region of petA mRNA, which encodes cytochrome f, and the C-terminal domain of the unassembled protein. This model for the regulation of cytochrome f translation explains both the decreased rate of cytochrome f synthesis in vivo in the absence of its assembly partners and its increase in synthesis when significant accumulation of the C-terminal domain of the protein is prevented. When expressed from a chimeric mRNA containing the atpA 5′ untranslated region, cytochrome f no longer showed an assembly-dependent regulation of translation. Conversely, the level of antibiotic resistance conferred by a chimeric petA-aadA-rbcL gene was shown to depend on the state of assembly of cytochrome b6f complexes and on the accumulation of the C-terminal domain of cytochrome f. We discuss the possible ubiquity of the CES process in organellar protein biogenesis.
Keywords: chloroplast gene expression, cytochrome b6f complex assembly, translational autoregulation, epistasy, Chlamydomonas reinhardtii
The major oligomeric protein complexes found in organellar energy transducing membranes display a concerted accumulation of their constitutive subunits: a number of yeast respiratory mutants and Chlamydomonas photosynthetic mutants display a pleiotropic loss of most subunits from a given complex, although their mutations were characterized as affecting primarily only a single subunit (1–7). In most cases, these phenotypes result from a posttranslational degradation of the unassembled subunits, with no alteration in their rates of synthesis (8–10).
Studies of Chlamydomonas mutants have revealed another contribution to the stoichiometric accumulation of subunits during chloroplast protein biogenesis that we have referred to as a control by epistasy of synthesis (CES; refs. 11 and 12). Some chloroplast-encoded subunits display a much lower rate of synthesis in the absence of their assembly partners, as detailed in Table 1 (6, 13–17). Thus, chloroplast protein synthesis is to some extent hierarchical. We define CES subunits as subunits whose rate of synthesis appears assembly-dependent and define dominant subunits as those whose absence results in reduced synthesis of CES proteins. A variety of mechanisms can account for these observations. The employed mechanism may actually differ from one CES subunit to another.
Table 1.
The CES subunits in C. reinhardtii
| Photosynthetic protein complex | Subunits
|
Ref. | |
|---|---|---|---|
| Dominant | CES | ||
| Cytochrome b6f | SUIV | cyt. f | 6 |
| Photosystem I | PsaB | PsaA | 15 |
| Photosystem II | D2 | D1 | 14 |
| D1 | apoCP47 | 13 | |
| ATP synthase | Su. β | Su. α | 16 |
| RUBP | SS | LS | 17 |
Absence of the indicated dominant subunits results in attenuated synthesis of the corresponding CES subunit.
RUBP, ribulose bisphosphate carboxylase; cyt. f, cytochrome f; Su., subunit; SS, small subunit; LS, large subunit.
Herein we address the molecular basis of the CES process for the biogenesis of the cytochrome b6f complex. Mutant strains lacking cytochrome b6 or subunit IV of the cytochrome b6f complex (SUIV), two major chloroplast-encoded subunits of this complex, show an apparent 90% decrease in the rate of cytochrome f synthesis, which is not accompanied by a decreased half-life of the protein. Therefore, a 10% accumulation of unassembled cytochrome f occurs in the membrane. In contrast, SUIV and cytochrome b6 are rapidly degraded in the absence of cytochrome f, with no alteration of their synthesis rate (6). SUIV and cytochrome b6, therefore, are dominant over cytochrome f in the hierarchical organization of subunit synthesis for cytochrome b6f complex biogenesis.
Cytochrome f is encoded by the chloroplast petA gene. It is a c-type cytochrome, synthesized as a precursor protein with a lumen-targeting peptide that drives the translocation of most of the apoprotein through the thylakoid membrane. Preapocytochrome f then matures to holocytochrome f on the luminal side of the thylakoid membrane, by covalent heme attachment and cleavage of the targeting peptide (for reviews, see refs. 18 and 19). Mature cytochrome f remains membrane-associated through a C-terminal-located α-helix. Interestingly, two mutant strains in which a cytochrome f variant was obtained by site-directed mutagenesis showed a 3-fold increase in the rate of cytochrome f synthesis. In one case, the protein, truncated at its C terminus, lacked its transmembrane anchor and its stromal extension of 15 residues. The resulting mutant overexpressed a soluble form of cytochrome f that accumulated in the thylakoid lumen (20). In the other case, a highly protease-sensitive polypeptide was created by substituting the cysteinyl residues involved in the covalent binding of the c-type heme. This hemeless cytochrome f was synthesized at a much higher rate than wild-type cytochrome f, although the modified polypeptide accumulated to less than 0.5% of the wild-type level (21).
Herein, we provide conclusive evidence that the contrasting effects on cytochrome f synthesis—underexpression in absence of SUIV versus overexpression in strains lacking significant accumulation of the C-terminal (Cter) domain of the protein—originate from a common molecular mechanism: a negative feedback regulation of cytochrome f translation, governed by the steady-state concentration of a structural protein motif present in the Cter domain of the unassembled protein (12); this motif, which is shielded upon assembly but totally or virtually absent in the truncated or hemeless cytochrome f mutants, is readily accumulated and highly exposed in mutants failing to synthesize an assembly partner such as cytochrome b6 or SUIV. We show that the petA 5′ untranslated region (UTR) contains all of the target information required for this regulation, thus ruling out posttranslational regulation and arguing for a control at the level of translation initiation.
MATERIALS AND METHODS
Strains, Media, and Genetic Methods.
A wild-type Chlamydomonas reinhardtii strain (mt+) derived from strain 137c and a petA (mt+) deletion strain (6) were used for chloroplast transformation experiments. The other mutant strains used in this study were the deletion strain ΔpetD (mt+), previously named ΔQ in ref. 6, the nuclear mutant strains mcd1-F16 (mt−) (22, 23) and ccs1-ac206 (mt−) (24), and the F52L-55V (mt+) transformant (21). Wild-type and mutant strains were grown on Tris–acetate–phosphate (TAP) medium (pH 7.2) at 25°C under dim light (5–6 μE per m2 per s) (25). Crosses were performed as described (25). Characterization of cytochrome b6f mutant phenotypes was carried out by using their fluorescence induction kinetics (26, 27).
Antibiotic-Resistance Tests.
Cells initially grown on TAP medium were transferred to selective medium by streaking them continuously with a platinum loop from a zone at high concentration down to a zone where individual clones could be recovered. After 8–10 days, their sensitivity to antibiotics was detected by the simultaneous observation of two phenomena: a change from a dark-green to a yellow-brown color in the region of high cell density and the absence of division figures from individual cells in the region of low cell density, as determined by light microscopy.
Nucleic Acid Manipulations.
Plasmid pF52L-55VΔpetD with the heme-attachment-defective cytochrome f sequence, a deletion of the petD gene, and the aadA cassette (conferring spectinomycin and streptomycin resistance) was constructed by replacing the entire petA-petD intergenic region located downstream of the aadA cassette in plasmid pAF52L-55V (21) with the 3′ UTR of the petD gene, using the strategy as described (20). Previous experiments have shown that this replacement does not modify the expression of the pet genes (ref. 28 and R.K., unpublished results).
Oligonucleotides FMETDIR (5′-TCGCGACATGTCCTAACCAAGTATTTACTACT-3′) and FSTOPINV (5′-ACGGCTGCAGTTAGAAGTTCATTTCTGCTA-3′) were used as primers with plasmid piWF (6) as a template to amplify by PCR the 950-bp cytochrome f coding sequence. The amplified fragment, after digestion with AflIII and PstI (two restriction sites underlined in the sequences of the oligonucleotides above), was cloned into the vector pdFBE (6) that had been digested with NcoI and PstI to create plasmid pAFRF. This substituted the aadA coding region from plasmid pdFBE with the petA coding region fused in-frame to the first 25 amino acids of the atpA gene, which were present in plasmid pdFBE (6, 29, 30). pAFRF was then digested with AccI and ScaI, and the resulting 3.7-kbp fragment was ligated to the 4.5-kbp fragment obtained from digestion of piWF with the same enzymes to yield plasmid pAFFF.
Oligonucleotides PETAPROM (5′-GCGAATTCGCAGGCAGTGGCGGTACC-3′) and PETAATG (5′-GCGGATCCATGGACATAATTTTATTAATCTTAAAAC-3′) were used as primers to amplify the 690-bp 5′ region of the petA gene containing the petA promoter and the petA 5′ UTR, including the methionine initiation codon and the second amino acid (serine) of cytochrome f. The amplified fragment was digested with EcoRI and BamHI (two restriction sites underlined in the sequence of the oligonucleotides above) and cloned into the vector pBKS− to yield plasmid p5F. p5F was digested with NcoI (boldface type in the oligonucleotide sequence above) and AlwNI, which cleave immediately downstream of the petA initiation codon and at the ori locus of the pBKS− vector, respectively. The resulting 1,550-bp fragment was ligated to the 3.1-kbp fragment obtained by digesting PUC-ATPX-AAD (29) with the same enzymes to create plasmid pFKR. The cassette FKR was then removed from the plasmid by digestion with EcoRV and SmaI and cloned into the unique StuI site of the plasmid pR12–23 (31) to create plasmid pFKR12.
Transformation of C. reinhardtii.
Wild-type and ΔpetA cells were transformed by tungsten-particle bombardment as described (6). Phototrophic transformants were selected on minimum medium at 5–6 μE per m2 per s. Transformants containing the aadA cassette were selected on TAP/spectinomycin (100 μg/ml)-containing plates. Resistant clones were then screened by fluorescence for defective cytochrome b6f activity and subcloned on spectinomycin-containing plates until they reached homoplasmy, as determined by DNA filter hybridizations. At least three transformants were analyzed for each construct. Pulse-labeling experiments, protein isolation, separation, and analysis were carried out as in ref. 6.
RESULTS
Strains That Express a Short-Lived Form of Cytochrome f Escape the CES Process.
Fig. 1 shows a pulse-labeling study comparing strains that underexpress (ΔpetD) or overexpress (F52L-F55V) cytochrome f as compared with the wild-type control. The ΔpetD strain lacks the chloroplast petD gene encoding SUIV of the cytochrome b6f complex. Consequently, cytochrome f, being a CES subunit, displays a marked decrease in its rate of synthesis, resulting in the accumulation of only 10% of the wild-type level (8) but all in an unassembled configuration. The F52L-55V strain, because of the substitution of the two cysteinyl residues involved in covalent heme binding to apocytochrome f, expresses a highly protease-sensitive variant of cytochrome f that does not accumulate in vivo (<0.5% of the wild-type level of cytochrome f). The rate of synthesis of hemeless cytochrome f is approximately 3-fold higher than that of wild-type cytochrome f (2).
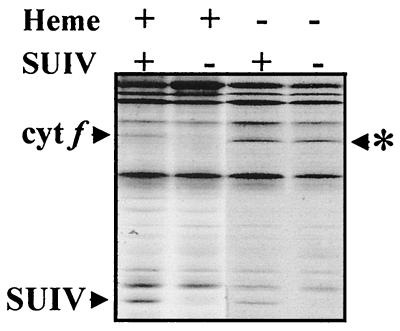
Figure 1.
Synthesis of hemeless cytochrome f escapes the CES process. Chloroplast translates from wild-type, ΔpetD, F52L-55V, and F52L-55VΔpetD strains (lanes from left to right). Whole-cell polypeptides were pulse-labeled for 5 min with [14C]acetate in the presence of an inhibitor of cytoplasmic translation and separated in SDS/12–18% polyacrylamide gels in the presence of 8 M urea. Arrowhead, positions of cytochrome f and SUIV deduced from comparison with known polypeptide patterns; ∗, position of apocytochrome f, which migrates slightly faster than holocytochrome f (20).
We wished to determine whether the increased rate of cytochrome f synthesis in F52L-55V reflected a regulatory mechanism that was similar to that responsible for the decreased rate of cytochrome f synthesis in ΔpetD, namely, a feedback mechanism governed by the concentration of the Cter domain of unassembled cytochrome f. Strains expressing the unstable hemeless cytochrome f lack significant accumulation of the Cter domain and should bypass the CES process: thus, synthesis of hemeless cytochrome f should be insensitive to the presence or absence of its assembly partners. We therefore transformed the chloroplast genome of wild-type C. reinhardtii with plasmid pF52L-55VΔpetD carrying the F52L-55V mutations altering heme binding to cytochrome f, a deletion of the petD gene to prevent SUIV synthesis and a aadA cassette conferring spectinomycin resistance to allow selection of the desired double mutants.
We used pulse labeling to determine the rate of synthesis of hemeless cytochrome f in these transformants, which lack SUIV. Fig. 1 shows that its rate of synthesis was 3-fold higher than that of wild-type cytochrome f, whether SUIV was made (Fig. 1, F52L-55V) or not (Fig. 1, F52L-55VΔpetD). Therefore, this strain, which fails to accumulate the Cter domain of the protein in vivo, no longer exhibits the CES process.
The petA 5′ UTR Is Required for the CES Process.
The decrease of cytochrome f synthesis in the absence of its assembly partners could operate at the level of translation or by the susceptibility of newly synthesized polypeptide chains to proteolytic degradation. To discriminate between these hypotheses, we replaced the petA promoter and the petA 5′ UTR, where cis-acting signals for translation initiation are located, by those of the atpA gene (Fig. 2A). The atpA gene encodes the α subunit of the ATP synthase complex, whose expression is independent of the expression of the pet genes. The resulting plasmid pAFFF was introduced by biolistic transformation into the chloroplast of the ΔpetA deletion strain. Phototrophic transformants, hereafter termed AFFF, were recovered on minimal medium, demonstrating that the promoter and 5′ UTR of atpA were able to drive cytochrome f synthesis at rates high enough to sustain phototrophic growth.
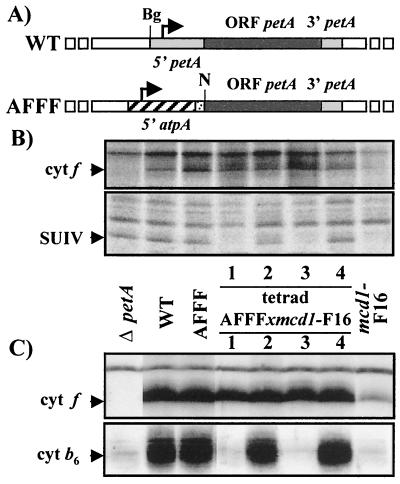
Figure 2.
Cytochrome f synthesis under control of the atpA 5′ UTR escapes the CES process. (A) Maps of the petA gene in wild-type and AFFF strains (Bg, BglII; N, NcoI). The heavily hatched box in the 5′ region of atpA denotes the sequence encoding the 25 first amino acids of the α subunit of the ATP synthase complex. (B) Newly synthesized cytochrome f and SUIV detected by pulse-labeling experiments in ΔpetA, wild-type, parental strains, and progeny of a representative tetrad from the cross AFFF × mcd1-F16. (C) Accumulation of cytochromes f and b6, detected with specific antibodies in the same strains.
We next investigated whether the synthesis of cytochrome f, now under the control of the atpA 5′ UTR, was still regulated by the presence of its assembly partners. To this end, we crossed AFFF (mt+) with the nuclear mutant mcd1-F16 (mt−), a strain that does not synthesize SUIV because petD mRNA is unstable (23). In this cross, all the daughter cells of each tetrad inherited the chimeric chloroplast gene AFFF, uniparently transmitted by the mt+ parent. In contrast, only two members of the tetrad inherited the nuclear mcd1-F16 mutant allele transmitted by the mt− parent, the two other members having a wild-type nuclear genome. We analyzed the expression of the pet genes in tetrad progeny by protein pulse labeling (Fig. 2B) or by immunoblotting using specific antibodies raised against cytochrome f or cytochrome b6 (Fig. 2C).The mcd1-F16 mutant members (the first and third members of the representative tetrad shown Fig. 2) failed to synthesize SUIV (Fig. 2B) and, consequently, also failed to accumulate cytochrome b6, as did the parental mcd1-F16 strain (Fig. 2C). The two other members synthesized SUIV (Fig. 2B) and accumulated wild-type levels of cytochrome b6 (Fig. 2C).
As shown in Fig. 2B, the rate of synthesis of cytochrome f was similar in the four daughter cells of the tetrad, irrespective of the presence of SUIV. Therefore, at variance with cytochrome f expressed from the wild-type petA gene, whose synthesis and accumulation decreased in the absence of SUIV in the parental mcd1-F16 strain (Fig. 2, compare lanes mcd1-F16 and WT), cytochrome f expressed from the chimeric petA gene was no longer regulated by the presence of its assembly partners. Accordingly, cytochrome f, being a stable protein, accumulated to the wild-type level in the four daughter cells, even though those two bearing the mcd1-F16 mutation lacked SUIV and cytochrome b6. No changes in petA mRNA levels were detected among the tetrad progeny by RNA filter hybridizations (data not shown). In summary, we conclude that translation initiation of cytochrome f, rather than posttranslational degradation of the polypeptide, is involved in this CES process.
A Reporter Gene, Driven by the petA 5′ UTR, Mimics the CES Behavior of Cytochrome f.
If the petA 5′ UTR contains all the target information required for the autoregulation of cytochrome f synthesis, it should confer the same regulatory properties to a reporter gene translated under its control. The product of such a reporter gene should be underexpressed in an assembly-deficient strain lacking SUIV and overexpressed in strains unable to accumulate cytochrome f, for example, those defective in heme binding to cytochrome f.
We introduced the chimeric gene petA-5′-UTR-aadA-rbcL-3′-UTR, hereafter referred to as FKR into a neutral site of the chloroplast genome (Fig. 3A). The rbcl 3′ UTR confers stability to chimeric RNAs. As expected, the FKR cassette conferred spectinomycin and streptomycin resistance to the resulting FKR12(mt+) transformants. They were subsequently crossed to the two mt− nuclear mutants, mcd1-F16 (see above) and ccs1-ac206, which is blocked at the level of apo- to holocytochrome f conversion (24) and overexpresses cytochrome f at the level of translation, as does the F52L-55V strain (data not shown). We identified the mutant progeny in the crosses by monitoring their fluorescence induction kinetics.
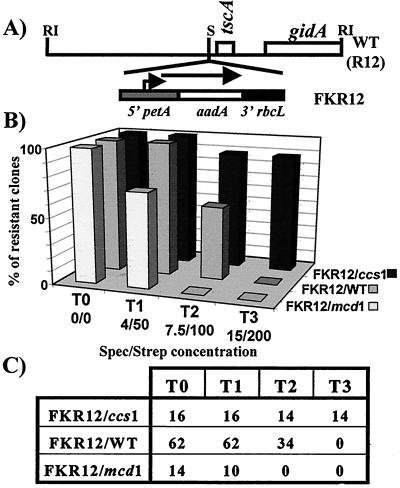
Figure 3.
petA 5′ UTR confers CES behavior to the aadA reporter gene product. (A) Map of the R12 fragment in wild-type and FKR12 strains. The positions of known genes and relevant restriction sites are indicated (S, StuI; RI, EcoRI) (31). (B) Percentage of strains with FKR12/mcd1, FKR12/WT, and FKR12/ccs1 genotypes, as indicated, growing on TAP (T0), T1, T2, or T3 medium, as indicated. Absolute numbers of growing clones (resistant clones) are indicated in C. The 62 FKR12/WT strains originated from three crosses—32 from FKR12 × WT, 14 from FKR12 × mcd1-F16, and 16 from FKR12 × ccs1-ac206—that yielded the same ratio of about 50% resistant clones on T2.
Because the aadA gene product cannot be detected in pulse-labeling experiments, the expression of the FKR cassette in the progeny of these crosses was monitored by testing their level of antibiotic resistance. To minimize aadA-independent background variations in their sensitivity to either spectinomycin or streptomycin, we used a combination of these two antibiotics. Growth of the progeny from the crosses was then assayed on solid TAP medium supplemented with increasing concentrations of antibiotics (streptomycin/spectinomycin, respectively): from 0 (as a growth control, T0), to 4/50 (T1), 7.5/100 (T2), or 15/200 (T3) μg/ml (Fig. 3B). FKR12 was also crossed to a mt− wild-type strain to control for any unknown modifiers of antibiotic resistance.
The correlation between the genotype of the strains and their antibiotic resistance phenotype was drawn from analysis of at least seven tetrads for each cross. Growth data for all the clones that we tested are summarized in Fig. 3 B and C. All members of tetrads derived from the cross between FKR12 and wild-type, as well as the daughter cells from crosses with the mcd1 and ccs1 parental strains having the wild-type nuclear alleles (FKR12/WT), grew on T1 medium and died on T3 medium. Remarkably, about half of the FKR12/WT clones, whether analyzed in one batch or in three separate batches corresponding to each of the three crosses, grew on T2 medium whereas the other half did not, suggesting individual variation, most likely of nuclear origin, in susceptibility to the combined antibiotics.
In contrast to these progeny with a wild-type nucleus, all but two ccs1 mutant progeny were able to grow on T2 and T3 media (FKR12/ccs1). This demonstrates that the aadA reporter gene is, at least, 2-fold overexpressed in a ccs1 relative to a wild-type nuclear context, in agreement with the 3-fold overexpression of cytochrome f in the ccs1 strain.
In contrast, all mcd1 mutant progeny died on either T2 or T3 medium. Moreover, 4 of these mutant products, of 14, were unable to grow on T1 medium (FKR12/mcd1). The aadA cassette, therefore, is underexpressed in a mcd1 relative to a wild-type nuclear context. Indeed, the higher antibiotic susceptibility of the FKR12/mcd1 progeny does not simply result from the loss of photosynthetic capability, because, in the FKR12 × ccs1 cross, the more resistant progeny were the nonphotosynthetic ones. The decrease in antibiotic resistance was less than expected from the 10-fold decrease of cytochrome f synthesis in the mcd1 strain. However, the physiology of antibiotic resistance in Chlamydomonas is poorly understood and it may not respond in a linear way to the accumulation of the AadA protein.
RNA filter hybridizations, using a probe specific for the aadA coding region, showed no correlation between the level of resistance to the antibiotics and the steady-state level of aadA mRNA. In fact, FKR12/WT strains contained less aadA mRNA than FKR12/mcd1 strains but more than FKR12/ccs1 strains (data not shown). This suggests that the chimeric transcript is degraded upon translation, a situation already observed for some aadA-containing cassettes (32).
DISCUSSION
How the CES Process Operates During Cytochrome b6f Complex Biogenesis: Toward a Molecular Mechanism.
Insight into the molecular basis for the regulation of cytochrome f synthesis can be drawn from the similar behavior of hemeless and Cter-truncated cytochromes f, which are both expressed at high and SUIV-independent rates (ref. 20 and Fig. 1). The increased rates of synthesis cannot be explained by ribosome pausing during the translation of the Cter domain, because this domain is still present in the hemeless mutant. One cannot invoke a rate-limiting step associated with the ligation of heme, because this would be bypassed in the strain expressing hemeless cytochrome f but not in the Cter-truncated protein.
The up-regulation of cytochrome f synthesis has to be attributed to a common feature of both strains, the absence of unassembled Cter domain, whose concentration is negligible in both cases. It is deleted in the Cter-truncated mutant, which nevertheless accumulates the remainder of the protein in the lumen (20). It does not accumulate in the hemeless cytochrome f mutant, because the apocytochrome f is rapidly degraded (21). In contrast, unassembled Cter domain readily accumulates in mutants expressing the wild-type version of cytochrome f but lacking SUIV or cytochrome b6. These strains show very low rates of cytochrome f synthesis (6). We propose that, because most of the Cter domain is shielded into assembled cytochrome b6f complexes, wild-type cells display intermediate rates of cytochrome f synthesis controlled by the steady-state concentration of unassembled cytochrome f, which in turn depends both on the rates of assembly and dissociation constant of cytochrome b6f complexes.
Translation initiation is, most likely, the key step in the regulation of cytochrome f synthesis. We can exclude other potential target steps, such as decreased rates of elongation during cytochrome f translation, decreased rates of cytochrome f translocation across the thylakoid membrane, or cotranslational degradation of the nascent polypeptide. In the strain AFFF, where cytochrome f synthesis is driven by the atpA promoter and 5′ UTR, we observed normal processing and thylakoid insertion (Fig. 2); yet the CES process is not observed. Furthermore, experiments with the FKR cassette demonstrate that the petA 5′ UTR is able to confer CES behavior to the aadA reporter gene. Because the protein translated from this gene is completely different from that of cytochrome f (Fig. 3), the coding sequence per se cannot have a regulatory role. Thus, we conclude that translation initiation of cytochrome f is autoregulated via the petA 5′ UTR through a protein motif shielded upon assembly, most likely within the Cter domain of unassembled cytochrome f, as illustrated in Fig. 4. The precise characterization of the regulatory residues is currently in progress.
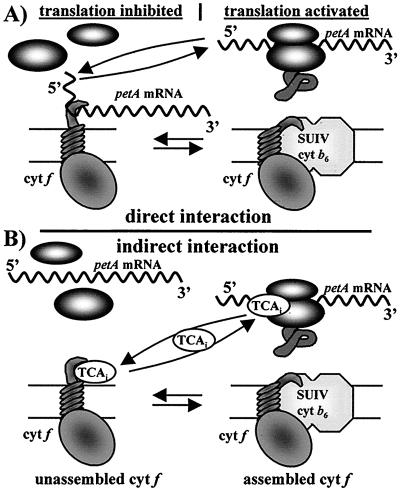
Figure 4.
Hypothetical mechanisms for the CES behavior of cytochrome f. (A) Direct interaction between the Cter domain of cytochrome f and the petA 5′ UTR. (B) Indirect interaction which relies on a putative ternary effector, TCAi, an activator of cytochrome f translation. (Left) Inhibition of cytochrome f synthesis caused by the accumulation of unassembled cytochrome f, as observed in strains lacking SUIV, but not in strains lacking accumulation of the Cter domain of cytochrome f. (Right) Activation of cytochrome f synthesis, the petA messenger (A) or the TCAi factor (B) being made available upon assembly. The intermediate rate of synthesis observed in wild-type cells results from an equilibrium between these situations.
Autoregulation of Cytochrome f Synthesis.
How the Cter domain of cytochrome f exerts a feedback control on translation initiation remains presently unknown. One possibility, illustrated in Fig. 4A, corresponds to the mechanism prevailing in most translational autoregulation processes described so far in both eukaryotes and prokaryotes. The proteins involved contain known or proposed RNA-binding domains. Typically, the RNA-binding motif of the protein under translational autoregulation interacts with its own mRNA to modify some posttranscriptional step—maturation, processing, or translation—that ultimately down-regulates protein expression (for review, see ref. 33). This is the case of certain Escherichia coli and yeast ribosomal proteins (34–36); the bacteriophage T4 gene 32 and 43 products (37, 38); the E. coli SecA, poly nucleotide phosphorlase, and poly(A) binding protein (39–41); the tumor suppressor p53 (42); and the yeast RNA helicase Dpb2p (43). Cytochrome f, however, has no reported RNA binding activity nor does its Cter domain contain any typical RNA binding motif.
The regulation of cytochrome f expression is also distinct from that of β-tubulin, another non-RNA binding protein showing an assembly-dependent autoregulation of gene expression in animal cells. Unassembled β-tubulin prompts the degradation of polysome-bound tubulin mRNA (44), whereas the petA mRNA levels remain constant and show no correlation with cytochrome f translation rates.
We hypothesize therefore that there is an indirect interaction between the Cter motif, firmly bound to the membrane, and the 5′ UTR of the petA mRNA. This autocontrol would implicate a ternary effector, a translational activator capable of competitive binding to the Cter motif of cytochrome f and to the petA 5′ UTR. As illustrated Fig. 4B, the activator would be trapped by the Cter domain of cytochrome f until this subunit assembles into a cytochrome b6f complex. Upon assembly, the activator is released from the Cter domain and becomes available for interaction with the 5′ UTR of petA mRNA, where it mediates translation initiation. Such an effector may be defective in several nuclear mutants we have isolated that are specifically defective in cytochrome f translation: these mutants define a single nuclear locus, TCA1 (22). We are currently examining the possible participation of TCA1 as an effector of the autocontrol.
Generality of the CES Process in Organelle Protein Synthesis.
Apart from cytochrome f, CES subunits are found in all major thylakoid complexes in C. reinhardtii (refs. 6 and 13–16, see Table 1). An additional and recent example is that of the chloroplast-encoded large subunit (LS) of ribulose bisphosphate carboxylase, whose synthesis is down-regulated in the absence of the nuclear-encoded small subunit (17).
Most interestingly, several recent observations suggest that the same chloroplast CES subunits are conserved in higher plants. In the maize nuclear mutant crp1, which is primarily impaired in the processing of the petB-petD cotranscript and, therefore, lacks SUIV synthesis, the translation of cytochrome f is reduced (45). Failure to synthesize both apoCP47 and D1 was observed in the barley vir-115 mutant, which has a primary defect in D1 expression (46, 47). Although one cannot exclude a dual effect of the crp1 and vir-115 mutations, these observations are easily understood if cytochrome f and apoCP47 were CES subunits in maize and barley as they are in Chlamydomonas. Also, the expression of large subunit of the ribulose bisphosphate carboxylase is decreased in tobacco antisense plants that underexpress the nuclear-encoded small subunit (48).
A CES process may also contribute to the biogenesis of mitochondrial complexes. In the yeast Saccharomyces cerevisiae, for example, the mitochondrion-encoded subunits 6 and 8 of the ATP synthase complex show reduced synthesis in a mutant affecting subunit 9 (2). Similarly, the rate of synthesis of cytochrome oxidase (COX) I, a protein centrally located in the cytochrome oxidase complex and encoded by a mitochondrial gene, is reduced in yeast strains deficient in other COX subunits (8, 49–52), including mitochondrion-encoded COXII and COXIII. Furthermore, COXI, although synthesized in reduced amount in a strain deficient in COX7, is stable, and the COXII and COXIII subunits are synthesized at wild-type levels but rapidly degraded (52), a hierarchical situation strikingly similar to the one we described for the cytochrome b6f complex in C. reinhardtii (6).
Whether the CES process occurs through a unique molecular mechanism in organellar protein synthesis is not yet known. Stampacchia et al. (53) failed to observe an involvement of the psaA 5′ UTR in the regulation of synthesis of the CES subunit PsaA, one of the major photosystem I reaction center subunits in C. reinhardtii. In contrast, the CES process also relies on the regulation of translation initiation of the α subunit in the biogenesis of the chloroplast ATP synthase from C. reinhardtii (B. Rimbault, D. Drapier, J.G.-B., and F.-A.W., unpublished results). Similarly, the rbcL mRNA level is unaffected by the decrease accumulation of the small subunit of ribulose bisphosphate carboxylase, whereas its binding to polysomes is reduced, suggesting a specific decrease in the rate of translation initiation of large subunit (48).
The mechanism for the CES process in cytochrome b6f biogenesis in C. reinhardtii chloroplasts could thus reflect a rather ubiquitous phenomenon in the biogenesis of organellar proteins. A number of nuclear factors acting specifically on the translation of a particular organellar gene, as does the TCA1 factor discussed above, have been identified in C. reinhardtii (for review, see ref. 54) and in yeast (for review, see ref. 55). These factors could turn out to be essential components of the regulatory circuitry resulting in the control of protein assembly through translational autoregulation in organelles.
The ubiquitous CES process in organelles is probably not an energy-saving mechanism aimed at limiting production of unneeded polypeptides. It concerns only a restricted subset of the organelle-encoded polypeptides that are protease-resistant but show limited accumulation when unassembled. Rather, the unique properties of the CES subunits offer a means to catalyze multiple-subunit assembly. The various protease-susceptible subunits arrive at their assembly site from the nucleo-cytosol and organelle compartments in a stochastic way. Instead of having a high probability of being recognized as substrates for degradation because they are transiently unassembled, they would be rapidly titrated by the protease-resistant CES subunits acting as anchors and shelters for assembly.
Acknowledgments
We thank D. Drapier, C. de Vitry, and F. Zito for critical reading of the manuscript and stimulating discussions. This work was supported by the Centre National de la Recherche Scientifique, Unité Propre de Recherche 9072, the Collège de France and National Science Foundation/Centre National de la Recherche Scientifique Grant INT-9603351 (to D.B.S. and F.A.W.). R.K. was a recipient of a doctoral fellowship from the Ministère de la Recherche et de l’Enseignement supérieur. D.B.S. was a recipient of a Guggenheim Fellowship and the Georges Morel Prize from the Institut National de la Recherche Agronomique.
ABBREVIATIONS
- Cter
C-terminal domain of cytochrome f
- SUIV
subunit IV of the cytochrome b6f complex
- UTR
untranslated region
- CES
control by epistasy of synthesis
- COX
cytochrome oxidase
References
- 1.Dowhan W, Bibus C R, Schatz G. EMBO J. 1985;4:179–184. doi: 10.1002/j.1460-2075.1985.tb02334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ooi B G, Lukins H B, Linnana A W, Nagley P. Nucleic Acids Res. 1987;15:1965–1977. doi: 10.1093/nar/15.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delepelaire P. EMBO J. 1984;3:701–706. doi: 10.1002/j.1460-2075.1984.tb01872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemaire C, Wollman F-A. J Biol Chem. 1989;264:10228–10234. [PubMed] [Google Scholar]
- 5.Girard-Bascou J, Chua N H, Bennoun P, Schmidt G, Delosme M. Curr Genet. 1980;2:215–221. doi: 10.1007/BF00435689. [DOI] [PubMed] [Google Scholar]
- 6.Kuras R, Wollman F-A. EMBO J. 1994;13:1019–1027. doi: 10.1002/j.1460-2075.1994.tb06350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemaire C, Girard-Bascou J, Wollman F-A, Bennoun P. Biochim Biophys Acta. 1986;851:229–238. [Google Scholar]
- 8.Nakai T, Yasuhara T, Fujiki Y, Oshashi A. Mol Cell Biol. 1995;15:4441–4452. doi: 10.1128/mcb.15.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearce D A, Sherman F. J Biol Chem. 1995;270:20879–20882. doi: 10.1074/jbc.270.36.20879. [DOI] [PubMed] [Google Scholar]
- 10.De Vitry C, Olive J, Drapier D, Recouvreur M, Wollman F-A. J Cell Biol. 1989;109:991–1006. doi: 10.1083/jcb.109.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wollman F-A, Girard-Bascou J. Médecine/Science. 1994;11:I–XIV. [Google Scholar]
- 12.Wollman F-A, Kuras R, Choquet Y. In: Photosynthesis: From Light to Biosphere. Mathis P, editor. III. Dordrecht, the Netherlands: Kluwer; 1995. pp. 737–742. [Google Scholar]
- 13.Bennoun P, Spierer-Herz M, Erickson J, Girard-Bascou J, Pierre Y, Delosme M, Rochaix J-D. Plant Mol Biol. 1986;6:151–160. doi: 10.1007/BF00021484. [DOI] [PubMed] [Google Scholar]
- 14.Erickson J M, Rahire M, Malnoe P, Girard-Bascou J, Pierre Y, Bennoun P, Rochaix J-D. EMBO J. 1986;8:1745–1754. doi: 10.1002/j.1460-2075.1986.tb04422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girard-Bascou J, Choquet Y, Schneider M, Delosme M, Dron M. Curr Genet. 1987;12:489–495. doi: 10.1007/BF00419557. [DOI] [PubMed] [Google Scholar]
- 16.Drapier D, Girard-Bascou J, Wollman F-A. Plant Cell. 1992;4:283–295. doi: 10.1105/tpc.4.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khrebtukova I, Spreitzer R J. Proc Natl Acad Sci USA. 1996;93:13689–13693. doi: 10.1073/pnas.93.24.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howe G, Merchant S. Photosynth Res. 1992;40:147–165. doi: 10.1007/BF00019332. [DOI] [PubMed] [Google Scholar]
- 19.Wollman F-A. In: Molecular Biology of Chlamydomonas: Chloroplasts and Mitochondria. Rochaix J-D, Goldschmidt-Clermont M, Merchant S, editors. Dordrecht, the Netherlands: Kluwer; 1998. , in press. [Google Scholar]
- 20.Kuras R, Wollman F-A, Joliot P. Biochemistry. 1995;34:7468–7475. doi: 10.1021/bi00022a021. [DOI] [PubMed] [Google Scholar]
- 21.Kuras R, Büschlen S, Wollman F-A. J Biol Chem. 1995;270:27797–27803. doi: 10.1074/jbc.270.46.27797. [DOI] [PubMed] [Google Scholar]
- 22.Girard-Bascou J, Choquet Y, Gumpel N, Culler D, Purton S, Merchant S, Laquerrière F, Wollman F-A. In: Photosynthesis: From Light to Biosphere. Mathis P, editor. III. Dordrecht, the Netherlands: Kluwer; 1995. pp. 683–686. [Google Scholar]
- 23.Drager R G, Girard-Bascou J, Choquet Y, Kindle K L, Stern D B. Plant J. 1998;13:85–96. doi: 10.1046/j.1365-313x.1998.00016.x. [DOI] [PubMed] [Google Scholar]
- 24.Howe G, Merchant S. EMBO J. 1992;11:2789–2801. doi: 10.1002/j.1460-2075.1992.tb05346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris E H. The Chlamydomonas Source Book. San Diego: Academic; 1989. [Google Scholar]
- 26.Bennoun P, Delepelaire P. In: Methods in Chloroplast Molecular Biology. Edelman M, Hallick R B, Chua N-H, editors. Amsterdam: Elsevier; 1982. pp. 25–38. [Google Scholar]
- 27.Zito F, Kuras R, Choquet Y, Kössel H, Wollman F-A. Plant Mol Biol. 1997;33:79–86. doi: 10.1023/a:1005734809834. [DOI] [PubMed] [Google Scholar]
- 28.Sturm N R, Kuras R, Büschlen S, Sakamoto W, Kindle K L, Stern D B, Wollman F-A. Mol Cell Biol. 1994;14:6171–6179. doi: 10.1128/mcb.14.9.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldschmidt-Clermont M. Nucleic Acids Res. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiedler H R, Schmidt R, Leu S, Shavit N, Strotmann H. FEBS Lett. 1995;377:163–166. doi: 10.1016/0014-5793(95)01332-6. [DOI] [PubMed] [Google Scholar]
- 31.Choquet Y, Rahire M, Girard-Bascou J, Erickson J, Rochaix J-D. EMBO J. 1992;11:1697–1704. doi: 10.1002/j.1460-2075.1992.tb05220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zerges W, Rochaix J-D. Mol Cell Biol. 1994;14:5268–5277. doi: 10.1128/mcb.14.8.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Standart N, Jackson R J. Biochimie. 1994;76:867–879. doi: 10.1016/0300-9084(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 34.Yates J L, Nomura M. Cell. 1980;21:517–522. doi: 10.1016/0092-8674(80)90489-4. [DOI] [PubMed] [Google Scholar]
- 35.Benard L, Ehresmann P, Ehresmann C, Portier C. Biochimie. 1996;78:568–576. doi: 10.1016/S0300-9084(96)80003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Villardel J, Warner J R. Proc Natl Acad Sci USA. 1996;93:1596–1600. doi: 10.1073/pnas.93.4.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuerk C, Eddy S, Parma D, Gold L. J Mol Biol. 1990;213:749–761. doi: 10.1016/S0022-2836(05)80261-X. [DOI] [PubMed] [Google Scholar]
- 38.Gold L, O’Farrel P, Russel M. J Biol Chem. 1990;213:749–761. [Google Scholar]
- 39.Salavatti R, Oliver D. RNA. 1995;1:745–753. [PMC free article] [PubMed] [Google Scholar]
- 40.Robert-le Meur M, Portier C. EMBO J. 1992;11:2633–2641. doi: 10.1002/j.1460-2075.1992.tb05329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bag J, Wu J. Eur J Biochem. 1996;251:7251–7262. [Google Scholar]
- 42.Mosner J, Munnenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W. EMBO J. 1995;14:4442–4449. doi: 10.1002/j.1460-2075.1995.tb00123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barta I, Iggo R. EMBO J. 1995;14:3800–3808. doi: 10.1002/j.1460-2075.1995.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gay D, Sisodia S S, Cleveland D W. Proc Natl Acad Sci USA. 1989;86:5763–5767. doi: 10.1073/pnas.86.15.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barkan A, Walker M, Nolasco M, Johnson D. EMBO J. 1994;13:3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gamble P E, Mullet J E. J Biol Chem. 1989;264:7236–7243. [PubMed] [Google Scholar]
- 47.Kim J, Gamble-Klein P, Mullet J E. Plant Mol Biol. 1994;25:459–467. doi: 10.1007/BF00043874. [DOI] [PubMed] [Google Scholar]
- 48.Rodermel S, Haley J, Jiang C Z, Tsai C H, Bogorad L. Proc Natl Acad Sci USA. 1996;93:3881–3885. doi: 10.1073/pnas.93.9.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poutre C, Fox T D. Genetics. 1987;115:637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cabral F, Schatz G. J Biol Chem. 1978;253:4396–4401. [PubMed] [Google Scholar]
- 51.Cabral F, Solioz M, Rudin Y, Schatz G. J Biol Chem. 1978;253:297–304. [PubMed] [Google Scholar]
- 52.Calder K M, McEwen J E. Mol Microbiol. 1991;5:1769–1777. doi: 10.1111/j.1365-2958.1991.tb01926.x. [DOI] [PubMed] [Google Scholar]
- 53.Stampacchia O, Girard-Bascou J, Zanasco J L, Zerges W, Bennoun P, Rochaix J-D. Plant Cell. 1997;9:773–782. doi: 10.1105/tpc.9.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rochaix J-D. Plant Mol Biol. 1996;32:327–341. doi: 10.1007/BF00039389. [DOI] [PubMed] [Google Scholar]
- 55.Grivell L A. Eur J Biochem. 1989;182:477–493. doi: 10.1111/j.1432-1033.1989.tb14854.x. [DOI] [PubMed] [Google Scholar]