Abstract
The surface of the bluetongue virus core forms a T=13 quasiequivalent icosahedral protein shell with 260 trimers of a single gene product: VP7 protein. Underneath is a smooth layer, made up of VP3 protein, which appears to guide and nucleate the assembly of VP7 trimers. The contacts between the two shells are extensive but nonspecific, and construction of the T=13 icosahedral shell requires polymorphism in the association of the VP7 subunits, each of which has two domains that contribute to trimer formation. We used structural and relative sequence information to guide an investigation of how such a complex structure is achieved during virus assembly and what residues are required to form a stable capsid. Fifteen single or multiple site-specific substitution mutations were introduced into the helical domain of VP7, which is closely associated with the VP3 layer, and the effects on capsid assembly were analyzed. Our data show that both the position and the nature of single residues are critical for the attachment of VP7 to VP3 and that formation of a stable VP7 lattice is not the automatic consequence of trimer formation.
One of the largest multilayered viruses to have detailed structural information available is bluetongue virus (BTV), the type member of the Orbivirus family. The BTV virion is composed of three layers from the innermost VP3 layer, through intermediate VP7 layer, to the outer layer composed of proteins VP5 and VP2 (23, 24). Available biochemical information suggests that the outer layer is required for entry into the mammalian cells (10), which ultimately denudes the virion of both VP2 and VP5 to leave a transcriptionally active core composed of VP7 and VP3 in the cytoplasm. Structurally similar cores can be isolated from mature virion preparations by chymotrypsin and have proved suitable for crystallization (8).
The core particle contains 780 molecules of the 38-kDa VP7 protein that encapsidate and interact with 120 molecules of the 100-kDa VP3 protein to form an icosahedral structure (700 Å in diameter). The 780 copies of VP7 are arranged as trimers on a T=13 icosahedral lattice and form the outer surface of the core. In such an icosahedral lattice, there are five quasiequivalent trimers: P, Q, R, S, and T (8, 9). The precise juxtaposition of these trimers varies slightly in accordance with the requirements of quasiequivalence. The 120 VP3 copies form an innermost layer that acts as scaffolding for VP7 deposition (8). The mismatch between the number of subunits in the VP3 and VP7 layers poses an interesting problem as to how these layers reconcile to form an intact icosahedral structure. Although the X-ray structure of the BTV core has provided a wealth of information on how 260 VP7 trimers are organized on the VP3 layer with 120 subunits and how these two protein layers interact with one another, the process by which the VP7 trimers are assembled on the VP3 layer in the infected cells is not clear.
Interestingly, in recent years structural studies on representatives of a number of genera within the Reoviridae family have provided data on the inner shell protein layers and have revealed that the surface protein shell has either an incomplete T=13 organization, such as in orthoreoviruses (4, 5, 22), aquareoviruses (27), oryzaviruses (17), and fijiviruses, or a complete T=13 architecture, such as in orbiviruses (8, 21, 26), rotaviruses (20), phytoreoviruses, and coltiviruses, and these are attached to the surface of an underlying protein shell composed of 120 copies of a capsid protein. Whereas in the first group of viruses the T=2 layer has prominent turrets that protrude into the incomplete T=13 layers at the fivefold axes, the overall structure in the second group is almost spherical, reinforcing the shape of the subcore underneath. Remarkably, unlike the T=13 layer, the T=2 layer with 120 subunits is shared by most viruses with segmented double-stranded RNA genomes that have been studied to date, including bacteriophage phi6 (3). Such architecture is likely to be preserved for essential biological functions such as keeping the transcriptional complex and genome in the correct configurations across the different viruses. In contrast, during evolution some changes must be permitted in outer layers from virus to virus, since this layer is akin to the outer capsid, the protein molecules of which are responsible for the different tropism exhibited for different types of host cell.
For BTV the interactions between the T=13 layer of VP7 and the T=2 layer of VP3 are intimate and extensive. In solution, VP7 forms hexamers and pentamers of trimers, and it has been crystallized and studied at high resolution (7). Each VP7 has two domains: an upper domain with a β-sandwich fold and a lower helical domain. Although both domains contribute to the formation of the trimer, it is the lower domain that exclusively controls both the hexameric and pentameric association of the trimers on the T=13 icosahedral lattice and their interactions with the scaffolding layer of VP3. The X-ray crystallization data have shown that the VP7 trimers contact the underlying VP3 lattice through a range of hydrophobic residues that occur on the relatively flat underside of the trimers (8). However, the precise contribution of key residues and the extent to which the formation of the trimer is necessary for VP3 interaction remain of unclear.
In the present study, we expanded upon the findings of a preliminary investigation (15) of the role of key VP7 residues in trimer formation to assess the role of VP7 helices 2 and 9 in virus core formation. Based on the three-dimensional structure of the core, a series of substitutions of amino acid residues were introduced, and the mutated VP7 was expressed in insect cells by using recombinant baculoviruses. The effects of each mutation on the overall VP7 structure, its oligomerization, and the interaction with VP3 to form the core were analyzed as described previously (6, 15). The structure of the core was also examined by electron microscopy (EM). Our data identify the protein-protein interactions between VP7 trimers and between VP7 and VP3 that are necessary for successful assembly and suggest a pathway of assembly for this complex layered viral core.
MATERIALS AND METHODS
Viruses and cells.
Spodoptera frugiperda 9 (Sf9) cells were grown in suspension or in monolayer cultures at 28°C in SF900 II (Gibco-BRL). Derivatives of Autographa californica nuclear polyhedrosis virus containing the wild-type BTV-10 VP7 mutants were plaque purified and propagated as described previously (15).
Site-directed mutagenesis, construction of recombinant transfer vectors, and isolation of recombinant baculoviruses expressing mutant VP7 proteins.
Using the single-strand capacity of the baculovirus transfer vector pAcCL29 (16), synthetic oligonucleotides and wild-type BTV-10 VP7 DNA were used to prepare VP7 mutants essentially as described previously (15). The mutated sequences in the recombinant plasmids were identified by sequence analyses (25). Mutant DNA preparations were used to generate recombinant baculoviruses as described elsewhere (12).
Purification of CLPs and analysis of proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Baculovirus-expressed core-like particles (CLPs) were purified from recombinant virus infected cell lysates as described previously (6, 15). In brief, Sf9 cells were coinfected in suspension culture with recombinant baculovirus, Ac17BTV3 expressing VP3 (11), together with either one of the mutant VP7 baculoviruses or recombinant baculovirus Ac10BTVP7 expressing wild-type VP7 (19). After incubation at 28°C for 48 h, infected cells were harvested, lysed by Dounce homogenization in TNN buffer (200 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% [vol/vol] Nonidet P-40).
Assembled CLPs were purified from the soluble fraction by centrifugation on a 35% CsCl gradient or by a discontinuous sucrose step gradient (66% [wt/vol] and 40% [wt/vol] sucrose in 200 mM Tris-HCl [pH 8.0]-150 mM NaCl) as described previously (6, 15). The presence of VP3 and VP7 proteins was analyzed by SDS-PAGE and Western blotting with anti-BTV-10 polyclonal antibodies as described previously (15). For comparison of each expressed protein, the multiplicity of infection was always fixed, and within each experiment a control (green fluorescent protein expression) was always included to provide a normalizing factor.
To obtain the ratios between VP7 and VP3 molecules, each gel was scanned and the ratio was calculated by densitometry.
Purification of recombinant VP7.
The wild-type and mutant BTV-10 VP7 proteins were purified as described previously (2, 15). The extract containing soluble VP7 was stored at −20°C prior to analysis.
Velocity sedimentation analysis.
Purified VP7 protein was applied onto 15 to 30% (vol/vol) glycerol gradients made in 20 mM Tris (pH 8.0)-100 mM NaCl-1 mM dithiothreitol-0.5 mM EDTA and sedimented at 60,000 rpm in a Beckman Vti 65 rotor at 4°C for 90 min. Molecular mass makers in the gradients were provided by high-molecular-mass calibration kits (Pharmacia). The molecular mass standard consisted of the following proteins: catalase, 4 × 58 kDa = 232 kDa; lactate dehydrogenase, 4 × 36 kDa = 140 kDa; and serum albumin, 67 kDa. Gradients were fractioned from the bottom, and VP7 molecules were detected by SDS-PAGE and Western blotting.
EM.
Purified wild-type and mutant CLPs were resuspended in water, and 10-μl portions of CLPs were absorbed onto carbon-coated copper 400 mesh EM grids for 15 min, washed with water, and negatively stained with 1% (wt/vol) uranyl acetate. Grids were examined in a Hitachi H-7000 electron microscope at 75 kV.
RESULTS
Exchange of AHSV and BTV VP7 sequences and their influence on VP7-VP3 interaction.
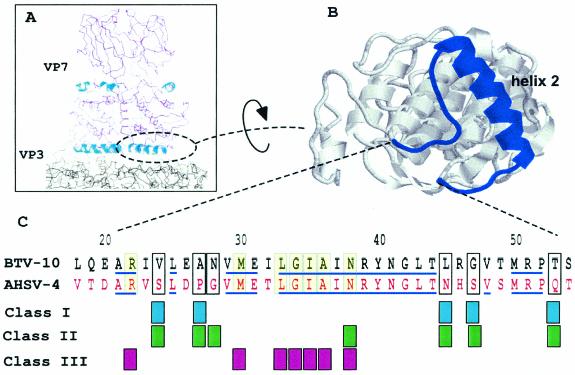
The flat base of VP7 trimers that interact with VP3 are formed largely by the three copies of α-helix 2 (of the total of nine helices) and the segments of the chain (amino acids [aa] 22 to 26 and aa 44 to 54) that lead into and out of the helix. To assess the role of the residues responsible for such interaction, we targeted a cluster of amino acids within helix 2 to replace with sequences from African horse sicknesses virus (AHSV), a close relative of BTV. A complete sequence alignment of BTV and AHSV VP7 together with the positions of known VP7 secondary structures have been reported previously (15). Within the α-helix 2 (aa 27 to 44) all residues are identical except aa 27 to 28 (Fig. 1). Within the turn that leads to this helix, only one of five residues (aa 24) is dissimilar, whereas two (aa 22 and 25) are identical and one is similar (aa 22). Of the 10 residues at the end of α-helix 2, the chain that leads to the next helix shares only 4 identical residues (aa 46 and 51 to 53) with AHSV. We opined that the difference of sequence in this region of VP7 would indicate residues tolerated for maintenance of a functional interaction with VP3, as suggested by the comparative X-ray structures of BTV and AHSV VP7 proteins (2, 7).
FIG. 1.
Sites of VP7 mutations in VP7 helix 2. A cross section through the structure of the BTV core is shown with a VP7 trimer above the smooth VP3 subcore (above left). Helix 2 is indicated and shown in more detail viewed from the underside (above right). In the lower part of the the figure is an alignment of BTV and AHSV helix 2 residues, with grouping of the different mutations used to assess helix 2 function. Class I was derived from consideration of the homology; class II is equivalent to class I but with a deliberate change of residue character; class III shows a change of character at residues homologous between BTV and AHSV. Identical sequences are indicated by lines.
Selected BTV residues V24, A27, L45, G47, and T53 likely to interact with the VP3 scaffold were mutated to the equivalent AHSV residues, either individually or in pairs: V24S/A27P, L45N/G47S, and T53Q (Fig. 1, class I). Changes were designed not to affect local interactions and not to disturb the overall folding pattern of the molecule. In addition, four mutants were generated (Fig. 1, class II) that incorporated more drastic changes (e.g., hydrophobic to charged residues) to assess the tolerance of the region to sequence changes not observed in natural isolates. V24S/A27P was mutated either to V24D/A27P or together with an additional substitution at N28E, creating a triple-site mutation, V24D/A27P/N28E. Both L45N/G47S and T53Q were also mutated to charged residues, creating L45K/G47D and T53R, respectively.
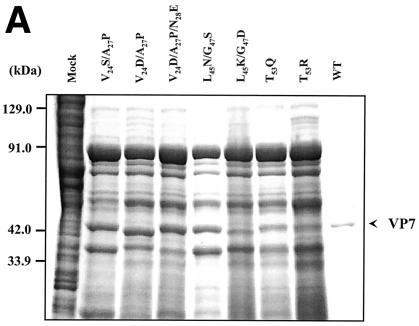
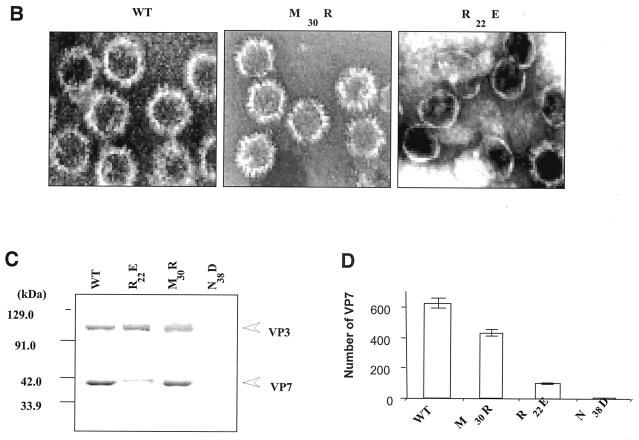
Prior to assessment of the role of these residues on VP7 trimer formation and on the formation of the core, each mutant protein was expressed singly using recombinant baculoviruses, and the effect on protein stability was examined by SDS-PAGE, followed by Coomassie blue staining (Fig. 2A). All mutant proteins were expressed at a high level in soluble form, similar to the wild-type recombinant protein, demonstrating that no individual or combination of mutation led to misfolding, aggregation, or degradation.
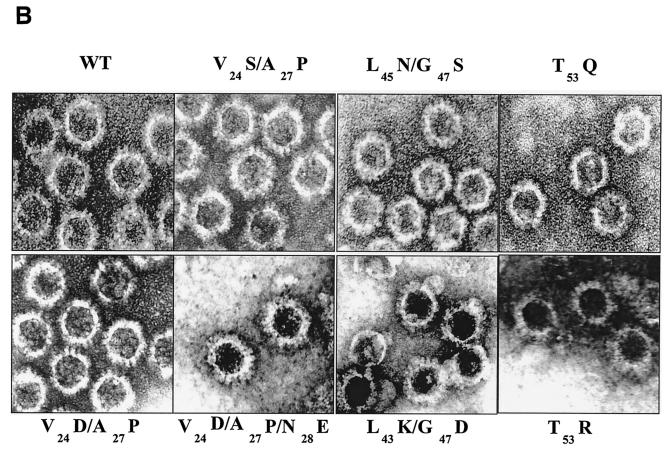
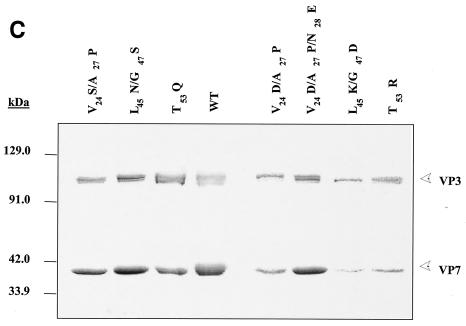
FIG. 2.
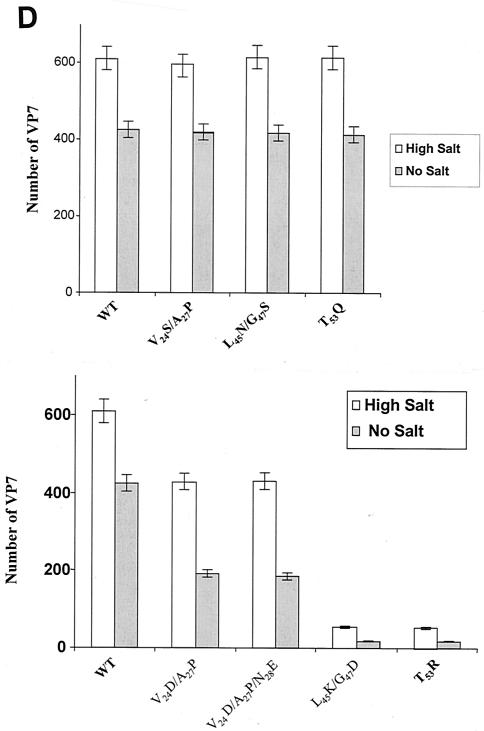
Analysis of VP7 class I and class II mutant protein interaction with BTV VP3 protein. Sf9 cells were coinfected with recombinant baculoviruses expressing BTV VP3 and either wild-type VP7 (WT) or various VP7 mutants (V24S/A27P, L45N/G47S, T53Q, V24D/A27P/N28E, V24D/A27P, L45K/G47D, and T53R, as indicated). The resulting CLPs were purified on CsC1 gradients and then analyzed. (A) Analysis of infected cell lysates by SDS-10% PAGE, followed by Coomassie blue staining. Note that VP3 is occasionally visualized as a doublet due to cleavage by resident protease of Sf9 cells. (B) EM images of CLPs formed by wtVP3+wtVP7 or wtVP3 with differ ent mutant VP7s (upper panel, class I; lower panel, class II) as indicated. (C) CLPs were analyzed by SDS-PAGE and stained with Coomassie brilliant blue. The positions of VP3 and VP7 are indicated. The CLP identity is indicated above each lane. (D) Number of VP7 molecules per CLP. Sucrose gradient (for low salt)- or CsCl gradient-purified CLPs were resolved by SDS-PAGE, and the image of the gel was captured and analyzed by using AlphaImager 2000 v.3.2 software (Alpha Innotech Co.). The number of VP7 molecules per CLP was calculated by determining the ratio of the density of VP7 to VP3, with VP3 as the denominator. The data shown are representative of three experiments, the value of which varied by no more than 10%. The number of VP7 molecules per CLP is listed under the conditions indicated.
The role of mutated residues on capsid assembly was assessed after coexpression of each VP7 derivative with a recombinant virus expressing wild-type VP3 protein in insect cells. Coexpression of VP7 and VP3 results in the synthesis of BTV CLPs that can be isolated by cesium chloride density gradients and exhibit a morphology by EM typical of BTV cores (6). All mutants showed the typical CLP morphology that appeared to indicate assembly with the VP3 scaffold; however, some assembled particles had visible differences in the thickness of the VP7 layer, as indicated by arrowheads in the lower panel of Fig. 2B. Assembled VP3 particles formed in the absence of VP7 are too unstable to isolate on a density gradient, and only in the presence VP7 could the CLPs be purified (28). Purified particles were also analyzed by SDS-PAGE; in these analyses it was observed that, although all mutant proteins were attached to VP3 layers, variable levels of VP7 to VP3 were apparent for a number of mutants (see Fig. 2C). The three substitution mutants of VP7 of BTV with AHSV sequences (shown in the upper panel of Fig. 2B) appeared not to alter CLP morphology or the VP3/VP7 ratio substantially (Fig. 2C), but all four mutants that substituted non-AHSV sequences had some effects on CLP morphology and showed less VP7 per VP3 than did wild-type CLPs (Fig. 2B and C).
Quantitation of helix 2 mutant stability in assembled CLP particles.
Since both EM and SDS-PAGE analysis indicated that some VP7 mutants were unstably attached to the VP3 scaffold and lost upon purification, the relative efficiency of CLP layer interaction was assessed. To do this, we used an established assay in which each mutant CLP preparation was purified on sucrose gradients in the presence or absence of 250 mM salt or, alternatively, first purified by using CsCl gradients (high salt) and then analyzed for stability in low salt, as reported previously (15). After isolation, the protein content of each mutant CLP preparation was analyzed by SDS-PAGE and, after a staining step, the relative amounts of VP3 and VP7 were assessed by densitometry. Purified wild-type CLPs usually lack 60 “P” trimers, i.e., the less well bonded of the VP7 trimers (8, 9). Therefore, 200 trimers of VP7 per CLP (i.e., 600 VP7 molecules per 120 molecules of VP3) were considered a full complement (B. V. Prasad, unpublished data). In the presence of a high salt concentration (250 mM), the level of VP7 trimers in each type of mutant CLP and associated with VP3 for mutants V24S/A27P, L45N/G47S, and T53Q were the same as for native CLPs (Fig. 2B and C). Each type of mutant CLPs retained ∼600 VP7 molecules per particle in the presence of a high salt concentration but, like the wild-type CLP, had only ∼420 VP7 molecules per particle in the absence of salt when ionic interactions displaced a further subset of VP7 trimers (Fig. 2D). However, the VP7 content of mutant CLPs was notably low where these residues were mutated to non-AHSV residues, with the maximum number of VP7 molecules being only ∼420 even in high-salt conditions. The difference between the mutants is more striking when mutant CLPs were isolated wholly in the absence of salt when, for some of these mutants, only a very low number of VP7 trimers remained attached to the VP3 layer. The first two mutants, V24D/A27P and V24D/A27P/N28E, in which an additional change was introduced at residue 28, behaved similarly, both mutants having only ca. ∼240 VP7 molecules attached to VP3 in absence of salt but up to ∼420 VP7 molecules in its presence (Fig. 2D). It is likely that not only the most distant P trimers but also the adjacent Q trimers were absent from these preparations. For the other two mutants that introduced two charged residues (L45K/G47D and T53R) very few, if any (<20 per CLP), VP7 molecules were present on CLPs in low-salt conditions and, unlike the other mutants, the presence of a high salt concentration did not improve the attachment of VP7 trimers on VP3 layer beyond about one-tenth (∼60 per CLP) of normal VP7/VP3 ratio (Fig. 2B to D and Table 1).
TABLE 1.
Mutations in the VP7 gene and their effects on trimer and CLP stability
| Helix, class, and no. | VP7 mutant | CLP stabilitya in:
|
No. of VP7/CLP (high salt) | Trimer forma- tion | |
|---|---|---|---|---|---|
| No salt | High salt | ||||
| Helix 2 | |||||
| Class I | |||||
| I | V24S/A27P | +++ | ++++ | 600 | Yes |
| II | L45N/G47S | +++ | ++++ | 600 | Yes |
| III | T53Q | +++ | ++++ | 600 | Yes |
| Class II | |||||
| IV | V24D/A27P | + | +++ | 420 | Yes |
| V | V24D/A27P/N28E | ++ | +++ | 420 | Yes |
| VI | L45K/G47D | − | + | 60 | Yes |
| VII | T53R | − | + | 60 | Yes |
| Class III | |||||
| VIII | R22E | + | + | 60 | Yes |
| IX | M30R | ++ | +++ | 420 | Yes |
| X | N38D | − | − | Yes | |
| XI | L33S/G34S/I35S/A36S | − | − | ||
| Helix 9 | |||||
| XII | G337Q | +++ | ++++ | 600 | Yes |
| XIII | P338Q | + | ++ | 420 | Yes |
| XIV | A342R | +++ | ++++ | 600 | Yes |
| XV | A346R | + | ++ | 420 | Yes |
| XVI | G337Q/P338Q | − | + | 60 | Yes |
| XVII | A342R/A346R | − | + | 60 | Yes |
Mutations at conserved residues of VP7 have a drastic effect on capsid assembly.
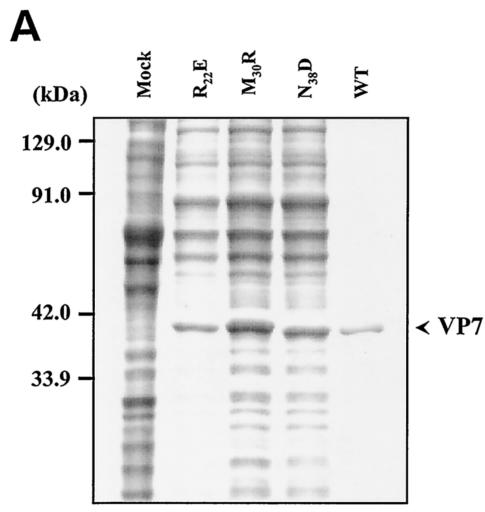
As indicated above, the residues of VP7 helix 2 are highly conserved among three gnat-transmitted orbiviruses (BTV, epizootic hemorrhagic disease of deer virus [EHDV], and AHSV), which indicates their functional importance (15). To investigate this further, we identified selected highly accessible residues based on the sequences that are shared by all three viruses and that are at the juxtaposition with the VP3 layer. Such highly conserved sequences in this region of VP7 would indicate conservation of functional interaction with VP3. For example, M30 and N38 on helix 2 and the residue R22 (Fig. 1, class III) are highly conserved and therefore were replaced with residues with similar side chain size, but different in polarity such as R to E, M to R, and N to D. Such changes were expected not to disturb the folding pattern of the molecule but to interfere with the protein-protein contacts. In addition, a strong hydrophobic region encompassing four residues, LGIA (aa 33 to 36), which is shared by all three VP7 molecules and thus indicates its functional importance, was also mutated by replacing each of the four residues with polar serine residue to avoid a strong effect on overall helix 2 structure. Each mutant was expressed by using recombinant baculoviruses as before, and their effects on overall protein folding, solubility, and CLP formation were again examined. While the four-serine-substitution mutant perturbed the overall structure of the VP7, producing an insoluble protein molecule (data not shown), all three single mutants were expressed as soluble proteins (Fig. 3A) that differed in their ability to form CLP (Fig. 3B and C). Substitution of asparagine by aspartate (at residue 38) had a severe effect on particle assembly and perturbed the formation of the VP7 lattice on the VP3 layer. Similarly, R22E formed few particles in which only very few (∼60) VP7 molecules were attached to the VP3 layer even when stabilized by the presence of high salt and exhibited various particle dimensions (Fig. 3D). Only mutation M30R retained a high number of VP7 trimers (∼420 molecules) in the presence of a high salt concentration. Nonetheless, in conditions of low salt, attachment of this mutant VP7 to VP3 was much less stable than the wild-type CLPs, indicating that all three targeted residues are involved in interaction with VP3 lattice.
FIG. 3.
Mutation at conserved VP7 residues within alpha helix 2 and its effects on CLP formation. Wild type (WT) and VP7 mutants R22E, M30R, and N38D were analyzed as described for Fig. 2A. (A) Analysis of mock-infected (Mock) and infected cell lysates was done by SDS-10% PAGE, followed by Coomassie blue staining. (B) EM analysis of CLPs formed by wild-type (WT) VP3 plus wild-type VP7 or by wild-type VP3 with different VP7 mutants as indicated. (C) CLPs were analyzed by SDS-PAGE and stained with Coomassie brilliant blue. The positions of VP3 and VP7 are indicated; the CLP identity is indicated above each lane. (D) The number of VP7 molecules per CLP under high-salt conditions was calculated as described in Fig. 2 legend. The data shown are representatives of three experiments, the value of which varied by no more than 10%.
Mapping the trimer-trimer interacting sites in capsid assembly.
The sideways interactions between the VP7 trimers themselves must be structurally variable in order to form a complex T=13 lattice on the surface of the VP3 subcore. The asymmetric unit in the icosahedral lattice contains the five quasiequivalent trimers P, Q, R, S, and T, with P being the closest to the icosahedral fivefold axis and the others situated farther outward in order. The S trimer is thus adjacent to the icosahedral twofold axis, and the T trimer is found on the icosahedral threefold axis. Remarkably, the various trimers in the assembled core remain virtually identical, while making such different and relatively strong contacts with VP3 and with each other (8).
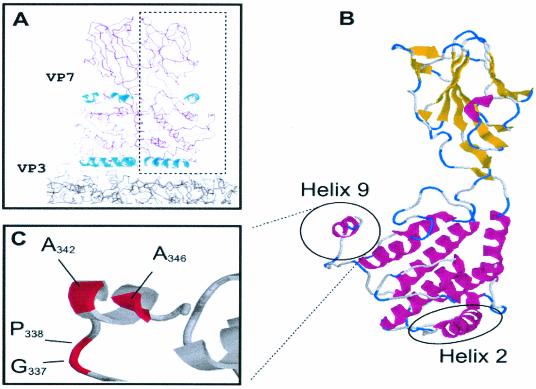
Our initial mutagenesis studies demonstrated that deletion or extension of residues at the C terminus of VP7 abolished core formation (14). Subsequent crystallographic data revealed that this region consists of a single helix (helix 9) formed from residues 341 to 347 and is on the outside of the trimer (Fig. 4). Preceding this helix is a highly hydrophobic and particularly flexible loop (sequence NPMPGPL), suggesting that this part of the structure may be able to adopt a number of conformations to provide the varied stabilizing side-to-side contacts required by the different trimer-trimer interfaces. A study specifically substituting these hydrophobic and flexible residues for arginine/glutamine was therefore undertaken to investigate putative trimer-trimer interactions. For example, G337Q was constructed to make a long polar side chain (Fig. 4). Since P338 is also shared by another closely related orbivirus, EHDV VP7 (but not by AHSV), this residue was also substituted by glutamine (P338Q) to assess its effect on CLP formation. A third mutation was generated in which both residues were substituted, creating G337Q/P338Q. It is noteworthy that both VP3 and VP7 can be exchanged between BTV and EHDV still result in CLP formation (13). Similarly, substitution mutations in residues of the highly hydrophobic helix 9 were also generated. In this helix A342 and A346 are highly conserved and exposed on the surface of the molecule, implying an involvement in intermolecular contacts. Perturbation of these putative contacts was achieved by substitution to arginine, generating two single substitution mutations (A342R and A346R) and one double mutation (A342R/A346R).
FIG. 4.
VP7 trimer highlighting helix 9. (A) Cross section through the structure of VP7 above the smooth VP3 subcore. Helix 9 is highlighted in the context of the VP7 trimer and shown in more detail in relation to a single monomer in panel B. (C) The relative positions of the residues described in the text for assessment of the role of helix 9 in trimer-trimer interaction are shown.
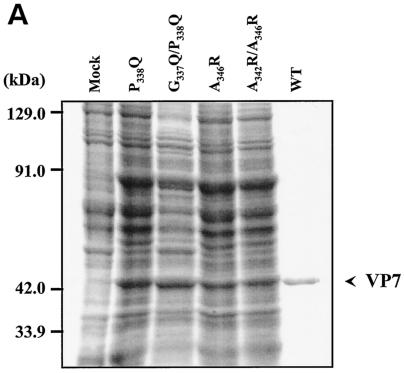
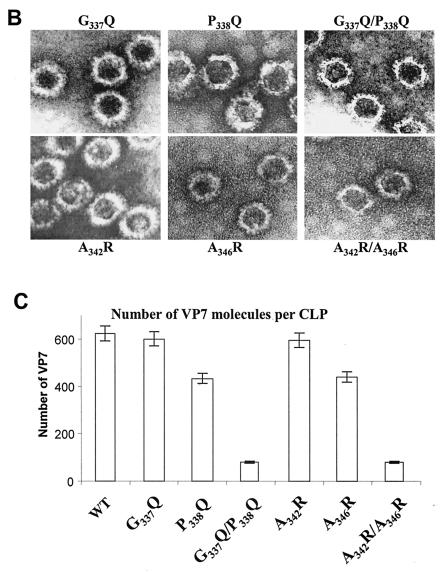
After expression with recombinant baculoviruses (Fig. 5A), the structural and functional effects of all six mutants targeting the C terminus on interaction with VP3 were investigated through coexpression experiments as before. Both G337Q and A342R still formed CLPs with a full VP7 complement (200 trimers) on the VP3 subcore (Fig. 5B and C). In contrast, the two other single mutants, P338Q and A346R, were deleterious for CLP assembly, and the attachment of mutant VP7 trimers was less stable since vary few particles could be seen by EM. Both the double mutants G337Q/P338Q and A342R/A346R caused drastic reduction of CLP stability, suggesting that both P338 and A346 are critical for trimer-trimer interaction during core assembly.
FIG. 5.
Location and effects of mutations at conserved VP7 residues within alpha helix 9. (A) Analysis of infected cell lysates by four (P338Q, G337Q/P338Q, A346R, and A342R/A346R) of the six mutant viruses by SDS-10% PAGE, followed by Coomassie blue staining. Analysis of coexpression of VP3 with wild-type VP7 (WT) or with mutants (G337Q, P338Q, A342R, A346R, G337Q/P338Q, and A342R/A346R) was done as indicated. (B) EM analysis of CLPs formed by native VP3 protein and native VP7 or different mutant VP7 as indicated. (C) The numbers of VP7 molecules per CLP under high-salt conditions and calculated as described for Fig. 2 are given. The data shown are representatives of three experiments, the value of which varied by no more than 10%.
Failure to attach to the VP3 subcore is not the result of poor VP7 trimer formation.
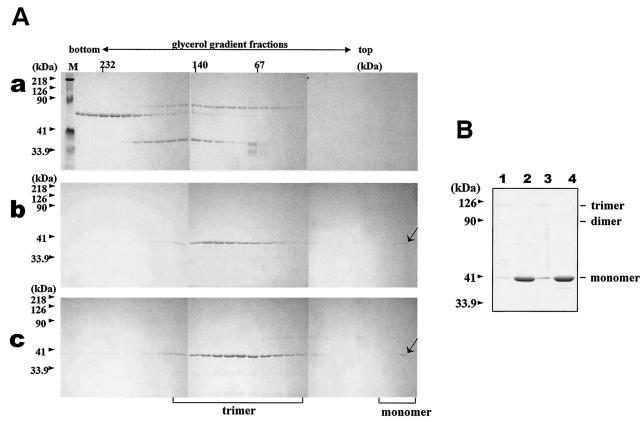
A clear corollary to our use of the CLP assembly assay to assess the interaction of the VP7 trimer with the VP3 subcore is that poor VP7 attachment is the result of a failure of VP7 mutants to trimerize rather than a lack of interaction with VP3 per se. The trimeric nature of mutants that had profound effects on CLP assembly was therefore examined individually. Each helix 2 (e.g., L45K/G47D, T53R/V24D, A27P/R22E, and N38D) and helix 9 (A342R/A346R and G337Q/G338Q) mutant was purified to homogeneity after expression in the absence of VP3, as described previously (2). Since isolated VP7 forms trimers readily in solution, the oligomeric nature of each purified VP7 preparation was determined by two different methods: first, by SDS-PAGE analysis of unboiled samples (previously established to detect VP7 oligomers [15]) and, second, by sedimentation of the oligomers through glycerol gradients. When analyzed by SDS-PAGE, all VP7 mutants were shown to form trimers that were as clearly detected as those of the wild-type VP7 (data not shown but a summary is presented in Table 1). Glycerol gradient sedimentation confirmed that each mutant existed stably as a trimer (demonstration data obtained with mutant N38 shown in Fig. 6). Failure to form the flat underside of the VP7 trimer, which is necessary for VP3 interaction, is not the cause of mutant failure to associate with VP3. Thus, the CLP assembly assay properly measures the local side chain requirements for an effective interaction. Trimerization alone is therefore not sufficient to drive the assembly of the VP7 lattice onto the VP3 subcore.
FIG. 6.
Trimerization assay of the wild-type and mutated BTV-10 VP7. Recombinant wild-type and mutant N38D VP7 was purified and assayed for trimer formation as described in Materials and Methods. (A) Sedimentation profiles of soluble VP7 protein on glycerol gradients. After purification, soluble VP7 mutants were subjected to velocity sedimentation analysis on 15 to 30% glycerol gradients to separate VP7 trimers. Fractions from the bottom to the top (left to right) were subjected to SDS-PAGE and stained with Coomassie brilliant blue. An arrow marks the position of VP7. (a) High-molecular-weight calibration markers (in kilodaltons), catalase (4 × 58 kDa = 232 kDa), lactate dehydrogenase (4 × 36 kDa = 140 kDa), and serum albumin (67 kDa); (b) wild-type VP7; (c) mutant N38D VP7. Lane M shows molecular weight markers (Bio-Rad) for SDS-PAGE. (B) Coomassie blue-stained SDS-PAGE profiles of wild-type (lanes 1 and 2) and N38D VP7 samples (lanes 2 and 4) either boiled at 100°C for 5 min or incubated at 37°C for 5 min (lanes 1 and 3) prior to SDS-PAGE analysis. The positions of the VP7 monomer, dimer, and trimer are indicated.
DISCUSSION
The structure of the assembled BTV core has received much attention; however, there has been relatively little study of the role of specific residues in the protein-protein interactions necessary to preserve the stable capsid structure or of the intermediates of the assembly pathway that achieve the final spherical icosahedral structure. We have addressed these issues here by a designed mutagenesis of VP7 and the assessment of its influence on interactions with VP3 and the subsequent overall effects on capsid assembly.
We previously established a useful model system in which self-assembly of VP3 and VP7 proteins into empty CLPs was achieved by coexpression of their respective genes in insect cells (6). Although during gradient purification CLPs appear to lose readily one set of VP7 trimers (i.e., P trimers), as seen by cryo-EM analysis (Fig. 7), the system nevertheless provides a tool for examining the protein-protein interactions that may occur during virus assembly. In a first series of mutations, we targeted the regions of the flat undersurface of VP7 trimers that adhere closely to the VP3 surface; we examined, in particular, helix 2 and its associated loops. Mutations consisted of either substitution by using related orbivirus (AHSV) sequences or nonrelated sequences, predominantly substituting hydrophobic to charged residues. Another group of mutations targeted certain sequences within this region that are highly conserved within the gnat-transmitted orbivirus. In the first group (V24S/A27P, L45N/G47S, and T53Q), some mutations included hydrophobic to polar and/or more rigid residues. However, when CLP assembly was examined with each of these VP7 mutants, all those that mimicked AHSV sequence behaved much like BTV and had no detectable effect on overall folding or the molar ratio of VP7 to VP3 in CLPs. This is particularly notable for T53Q, for which threonine with the aliphatic hydroxyl side chain was replaced by acidic glutamate. Thus, the data confirmed our previous structural data that revealed that VP7 structures of these two orbiviruses are highly conserved (1). These data support our previous research wherein it was demonstrated that certain domain-switched mutants of VP7 between AHSV and BTV were compatible with CLP assembly (18).
FIG. 7.
Cryo-EM three-dimensional reconstruction of CLP. A surface representation of CsCl gradient-purified CLP (wild-type VP3 plus wild-type VP7) shows the trimers of VP7 in the protomeric unit of the core and shows the loss of the P trimer.
Unlike the homologous changes, however, any combination of mutation to AHSV sequence, together with mutations that changed the character of the residue from hydrophobic to charged (V24D/A27P, L45K/G47D, and T53R), produced instability, sometimes severe, on CLP assembly. CLPs were more stable when isolated in high salt, suggesting that solvation of the charged groups can prevent a substantial mismatch between the VP7 and VP3 proteins, whereas the assembly is driven by the hydrophobic interactions. Interestingly, when V24D/A27P was mutated further to V24D/A27P/N28E with an additional change introduced at the next position (N28), by substitution of a glutamic acid residue, very little difference in instability of CLPs between these two mutants could be detected.
The effects of a second group of mutations on CLP assembly, where the changes were introduced at conserved residues, were more profound. All three mutants M30R, R22E, and N38D have affected the stable attachment of VP7 on the VP3 subcore, retaining fewer VP7 molecules than the wild-type CLPs. In particular, a single change, asparagine at residue 38 to aspartate, abolished CLP assembly. It may be explained by the long side chain of aspartate hindering the contact between the VP3 layer and VP7 trimers.
VP7 molecules oligomerize into trimers when expressed alone, in the absence of VP3 protein. It can therefore be speculated that attachment of preformed trimers onto the VP3 subcore and subsequent formation of VP7 layer would be directed by side-to-side interaction of adjacent trimers. Interactions between neighboring trimers at the twofold axis are through a thin band around the lower domains and thus avoid the need for different conformations of the VP7 trimers. The role of the VP7 trimer in assembly was therefore investigated through site-directed changes in helix 9, a prominent feature on the side of the VP7 lower domain involved in trimer packing. Of the various mutations at different sites, the importance of at least two particular residues, P338 and A346, to CLP assembly was shown after biochemical and morphological assay. While the single-site mutants P338Q and A346R could still form reasonably stable CLPs, the double mutations G337Q/P338Q and A342R/A346R resulted in poor CLP morphology, with only ∼60 VP7 molecules retained by the VP3 surface of each CLP isolated in the presence of high salt.
Since helix 9 was not involved in direct interaction with the VP3 subcore, the effect of these mutations is indirect and relates to the lateral packing of the VP7 trimers on the VP3 surface. Our data are in line with structural data that showed that the residues 336 to 338 have high B-factors and have the ability to adjust to various quasiequivalent environments in the T=13 lattice (8).
Although these changes confirmed the role of the helix in trimer-trimer interaction, trimer formation per se was not altered, suggesting that trimer assembly alone is not sufficient for completion of VP7 attachment to the subcore. In terms of the overall assembly pathway of the BTV core, cryo-EM and X-ray structures revealed that, of the 13 unique contacts made between the VP3 and VP7 shells, the contact that aligns the VP7 trimer axis with the icosahedral threefold axis of the VP3 layer is the strongest. This suggests it may nucleate the assembly of the VP7 lattice on the VP3 subcore once the first trimer is anchored (8, 9). It has been postulated that preformed hexamers of VP7 may propagate around the initial VP7 trimer, forming a sheet that loosely enwraps the VP3 layer. The data obtained here did not support a gradient of trimer association from a single nucleation site since mutations that destabilize the CLP particle still allow assembly of some VP7 lattice on the VP3 shell. An alternate model for assembly suggests that multiple sheets of VP7 may form at different nucleation sites. Our data are consistent with a pathway of core assembly that involves multiple preferred initiation sites for VP7 assembly on VP3 (as illustrated in Fig. 8) and a second set of weaker interactions that then “fill the gaps.” The distinction between VP7 trimers that first occupy the subcore and those that follow is intriguingly subtle since it must be sufficient to allow variation in packing and yet not prevent the overall biological purpose of virus assembly.
FIG. 8.
Diagram of the juxtaposition of VP7 trimers in relation to VP3 and the likely VP7 assembly pathway. Trimers of VP7 adopt subtly different conformations in relation to each other in order to accommodate the spatial variation required for completion of a T=13 shell. (A) The trimers are labeled at P, Q, R, S, and T, with P providing the center of the group located on the fivefold axis. (B) A suggested order of assembly filling the trimer positions sequentially and consistent with partially filled shells associated with the mutants analyzed here.
Acknowledgments
We thank B. V. Prasad (Baylor College, Houston, Tex.) for providing us with the cryo-EM image of CLP and D. Stuart and J. Grimes (University of Oxford) for designing the mutations. We are very grateful to Ian M. Jones (University of Reading) and Vijay Reddy (The Scripps Research Institute, La Jolla, Calif.) for very fruitful discussions during preparation of the manuscript.
This work was supported by grants from the National Institutes of Health (United States) and the BBSRC (United Kingdom).
REFERENCES
- 1.Basak, A. K., P. Gouet, J. Grimes, P. Roy, and D. Stuart. 1996. Crystal structure of the top domain of African horse sickness virus VP7: comparisons with bluetongue virus VP7. J. Virol. 70:3797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basak, A. K., D. I. Stuart, and P. Roy. 1992. Preliminary crystallographic study of bluetongue virus capsid protein, VP7. J. Mol. Biol. 228:687-689. [DOI] [PubMed] [Google Scholar]
- 3.Butcher, S. J., T. Dokland, P. M. Ojala, D. H. Bamford, and S. D. Fuller. 1997. Intermediates in the assembly pathway of the double-stranded RNA virus phi6. EMBO J. 16:4477-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dryden, K. A., D. L. Farsetta, G. Wang, J. M. Keegan, B. N. Fields, T. S. Baker, and M. L. Nibert. 1998. Internal/structures containing transcriptase-related proteins in top component particles of mammalian orthoreovirus. Virology 245:33-46. [DOI] [PubMed] [Google Scholar]
- 5.Dryden, K. A., G. Wang, M. Yeager, M. L. Nibert, K. M. Coombs, D. B. Furlong, B. N. Fields, and T. S. Baker. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 122:1023-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.French, T. J., and P. Roy. 1990. Synthesis of bluetongue virus (BTV) corelike particles by a recombinant baculovirus expressing the two major structural core proteins of BTV. J. Virol. 64:1530-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimes, J., A. K. Basak, P. Roy, and D. Stuart. 1995. The crystal structure of bluetongue virus VP7. Nature 373:167-170. [DOI] [PubMed] [Google Scholar]
- 8.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. C. Mertens, and D. I. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 9.Grimes, J. M., J. Jakana, M. Ghosh, A. K. Basak, P. Roy, W. Chiu, D. I. Stuart, and B. V. Prasad. 1997. An atomic model of the outer layer of the bluetongue virus core derived from X-ray crystallography and electron cryomicroscopy. Structure 5:885-893. [DOI] [PubMed] [Google Scholar]
- 10.Hassan, S. H., and P. Roy. 1999. Expression and functional characterization of bluetongue virus VP2 protein: role in cell entry. J. Virol. 73:9832-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inumaru, S., H. Ghiasi, and P. Roy. 1987. Expression of bluetongue virus group-specific antigen VP3 in insect cells by a baculovirus vector: its use for the detection of bluetongue virus antibodies. J. Gen. Virol. 68:1627-1635. [DOI] [PubMed] [Google Scholar]
- 12.King, L. A., and R. D. Possee (ed.). 1992. The baculovirus expression system: a laboratory guide. Chapman and Hall, London, England.
- 13.Le Blois, H., B. Fayard, T. Urakawa, and P. Roy. 1991. Synthesis and characterization of chimaeric particles between epizootic haemorrhagic disease virus and bluetongue virus: functional domains are conserved on the VP3 protein. J. Virol. 65:4821-4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeBlois, H., and P. Roy. 1993. A single point mutation in the VP7 major core protein of bluetongue virus prevents the formation of core-like particles. J. Virol. 67:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limn, C.-H., N. Staeuber, K. Monastyrskaya, P. Gouet, and P. Roy. 2000. Functional dissection of the major structural protein of bluetongue virus: identification of key residues within VP7 essential for capsid assembly. J. Virol. 74:8658-8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livingstone, C., and I. Jones. 1989. Baculovirus expression vectors with single strand capability. Nucleic Acids Res. 17:2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, G., Z. H. Zhou, M. L. Baker, J. Jakana, D. Cai, X. Wei, S. Chen, X. Gu, and W. Chiu. 1998. Structure of double-shelled rice dwarf virus. J. Virol. 72:8541-8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monastyrskaya, K., N. Staeuber, G. Sutton, and P. Roy. 1997. Effects of domain-switching and site-directed mutagenesis on the properties and functions of the VP7 proteins of two orbiviruses. Virology 237:217-227. [DOI] [PubMed] [Google Scholar]
- 19.Oldfield, S., A. Adachi, T. Urakawa, T. Hirasawa, and P. Roy. 1990. Purification and characterization of the major group-specific core antigen VP7 of bluetongue virus synthesized by a recombinant baculovirus. J. Gen. Virol. 71:2649-2656. [DOI] [PubMed] [Google Scholar]
- 20.Prasad, B. V., G. J. Wang, J. P. Clerx, and W. Chiu. 1988. Three-dimensional structure of rotavirus. J. Mol. Biol. 199:269-275. [DOI] [PubMed] [Google Scholar]
- 21.Prasad, B. V., S. Yamaguchi, and P. Roy. 1992. Three-dimensional structure of single-shelled bluetongue virus. J. Virol. 66:2135-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 23.Roy, P. 2000. Orbiviruses and their replication, p. 1835-1869. In B. N. Fields (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 24.Roy, P. 1996. Orbiviruses and their replication, p. 1709-1734. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 25.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoehn, G., S. R. Moss, P. A. Nuttall, and E. A. Hewat. 1997. Structure of Broadhaven virus by cryoelectron microscopy: correlation of structural and antigenic properties of Broadhaven virus and bluetongue virus outer capsid proteins. Virology 235:191-200. [DOI] [PubMed] [Google Scholar]
- 27.Shaw, A. L., S. K. Samal, K. Subramanian, and B. V. Prasad. 1996. The structure of aquareovirus shows how the different geometries of the two layers of the capsid are reconciled to provide symmetrical interactions and stabilization. Structure 4:957-967. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka, S., and P. Roy. 1994. Identification of domains in bluetongue virus VP3 molecules essential for the assembly of virus cores. J. Virol. 68:2795-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]