Abstract
G protein-coupled receptors (GPCRs) constitute a large superfamily involved in various types of signal transduction pathways triggered by hormones, odorants, peptides, proteins, and other types of ligands. The superfamily is so diverse that many members lack sequence similarity, although they all span the cell membrane seven times with an extracellular N and a cytosolic C terminus. We analyzed a divergent set of GPCRs and found distinct loop length patterns and differences in amino acid composition between cytosolic loops, extracellular loops, and membrane regions. We configured GPCRHMM, a hidden Markov model, to fit those features and trained it on a large dataset representing the entire superfamily. GPCRHMM was benchmarked to profile HMMs and generic transmembrane detectors on sets of known GPCRs and non-GPCRs. In a cross-validation procedure, profile HMMs produced an error rate nearly twice as high as GPCRHMM. In a sensitivity-selectivity test, GPCRHMM’s sensitivity was about 15% higher than that of the best transmembrane predictors, at comparable false positive rates. We used GPCRHMM to search for novel members of the GPCR superfamily in five proteomes. All in all we detected 120 sequences that lacked annotation and are potentially novel GPCRs. Out of those 102 were found in Caenorhabditis elegans, four in human, and seven in mouse. Many predictions (65) belonged to Pfam domains of unknown function. GPCRHMM strongly rejected a family of arthropod-specific odorant receptors believed to be GPCRs. A detailed analysis showed that these sequences are indeed very different from other GPCRs. GPCRHMM is available at http://gpcrhmm.cgb.ki.se.
Keywords: Hidden Markov model, G protein-coupled receptors, protein classification
The G protein-coupled receptors (GPCRs) make up a large and diverse superfamily of transmembrane (TM) proteins involved in signal transduction pathways. All known examples share a seven-TM helix topology with an extracellular N terminus. The extracellular signal is invariably transduced to a cytosolic heterotrimeric G protein complex. Despite this common overall architecture and mechanism, GPCRs can be divided into families with a striking lack of common sequence motifs (Bockaert and Pin 1999; Fredriksson et al. 2003). This is reflected by the vast number of ligands that activate the receptors, which range from neurotransmitters, hormones, and peptides to external stimuli such as light and odors.
Although large numbers of GPCR genes have been reported in several proteomes, there may be unidentified GPCRs that cannot be detected by ordinary sequence similarity searches. Such searches have an intrinsic limitation in that they cannot detect very distant homologies or sequences belonging to entirely new GPCR families. For example, the Pfam database contains a number of GPCR families, each one modeled by a separate profile HMM. As these families are distinct enough to not overlap, they would also be unlikely to recognize a novel GPCR family.
Alternative strategies for GPCR identification have targeted the seven-TM topology that is conserved across all members. In the simplest form these strategies use hydropathy-curve algorithms to detect proteins with seven hydrophobic stretches (Gao and Chess 1999). A more sophisticated approach is to use general-purpose TM topology predictors such as HMMTOP (Tusnady and Simon 1998) or Phobius (Käll et al. 2004), which are computational models trained to detect TM helices and their orientation in the membrane. The most straightforward way to employ a TM topology predictor for GPCR identification is to scan databases for proteins predicted to have seven TM helices and an extracellular N terminus. For example, Takeda and colleagues detected a large number of novel GPCRs when searching the human proteome for proteins predicted to have 6–8 TM helices (Takeda et al. 2002). The predictions thus obtained included a large number of false positives, but were further refined using additional computational screens.
Kim and colleagues developed a different approach called QFC for “Quasi-periodic Feature Classifier” (Kim et al. 2000). In QFC, each sequence is characterized by a set of physicochemical features. A hyperplane in the feature space was optimized to separate GPCRs from other sequences. QFC was applied to the Drosophilia melanogaster proteome and contributed to the identification of a putative new GPCR family (7tm_6/PF02949 in Pfam) (Clyne et al. 1999). Inoue and colleagues developed a combined classification and identification tool based on the loop length pattern of GPCRs (Inoue et al. 2004). This is a stepwise rule-based algorithm that, given a topology prediction, classifies sequences into GPCR or non-GPCR, and further into GPCR subclasses. The rules are binary (1 for long and 0 for short loops), and were set from observations about typical loop lengths. The algorithm is dependent on a topology prediction so for GPCR identification it would essentially be a way to remove false positives.
7TMHMM by Möller and colleagues (Möller 2001) is another GPCR specific predictor. The model is derived from TMHMM (Krogh et al. 2001), and contains sub-models for the seven TM helices and the cytosolic and extracellular loops, but is not trained on GPCR sequences. 7TMHMM is not primarily designed to identify novel GPCRs, but to map the cytosolic loops of known GPCRs for prediction of G protein specificity (Möller et al. 2001).
Because the topology of GPCRs is more conserved than the primary sequence, topology-based detection methods make a lot of sense. However, no method exists that is based on a comprehensive analysis of GPCR sequences, and that is explicitly trained to identify GPCRs. In this paper we address this issue and present an analysis of sequence features in the GPCR superfamily. We found no sequence motifs that are conserved across all families, yet TM topology-related features such as loop lengths and amino acid compositions are relatively conserved. This leads to the notion that proper GPCR function depends on appropriate loop lengths and amino acid composition in the different regions of the GPCR.
We used the results of this analysis to construct GPCRHMM, a hidden Markov Model that specifically recognizes GPCRs based on TM topology-related features. GPCRHMM was benchmarked on three test sets, and its performance was compared to other GPCR detection strategies. GPCRHMM proved to be very sensitive yet much more specific than other methods.
GPCRHMM is a method for identification of GPCR sequences and families that lack sequence similarity to known examples, and therefore escape detection by homology searching. We searched five complete proteomes with GPCRHMM and present novel GPCR findings in the proteomes of Caenorhabditis elegans, D. melanogaster, Fugu rubripes, mouse, and human.
Results
Analysis of GPCR families
In order to model general aspects of G protein-coupled receptors we needed a diverse set of training sequences, but at the same time we had to avoid including false positives as these would corrupt the model. Many sequences are annotated as putative GPCRs based on sequence similarity to known members or because they have a probable seven-TM topology. We initially extracted 13 Pfam families that are classified as verified or putative GPCR families by the specialized database GPCRDB (Horn et al. 2001) (Table 1), and analyzed them to reveal families that could be incorrectly annotated as GPCR. For comparison, we also included two families known not to be GPCRs in the analysis: the bacteriorhodopsin family (Bac_rhodopsin/PF01036), which is structurally similar to rhodopsin but not G protein-coupled, and the protein kinase family (Pkinase/PF00069), which is completely unrelated.
Table 1.
GPCRHMM model training included sequences from 11 Pfam GPCR protein families
| No. of sequences in | ||||
| Pfam family | Description | Training set | Pfam full set | Max. sequence identity within training set |
| 7tm_1 | Rhodopsin family | 64 | 6486 | 60% |
| 7tm_2 | Secretin family | 53 | 410 | 40% |
| 7tm_3 | Metabotropic glutamate family | 31 | 219 | 40% |
| Frizzled | Frizzled/Smoothened family membrane region | 19 | 122 | 60% |
| STE3 | Pheromone A receptor | 18 | 38 | 60% |
| STE2 | Fungal pheromone mating factor STE2 GPCR | 3 | 9 | 60% |
| Dicty_CAR | Slime mold cyclic AMP receptor | 3 | 5 | 60% |
| 7tm_4 | C. elegans chemoreceptor | 35 | 268 | 30% |
| 7tm_5 | C. elegans chemoreceptor | 26 | 280 | 30% |
| 7tm_6 | D. melanogaster odorant receptor | 0 | 82 | — |
| Mlo | Plant putative GPCR | 0 | 55 | — |
| V1R | Vomeronasal organ pheromone receptor family | 33 | 171 | 60% |
| TAS2R | Mammalian taste receptor protein | 26 | 59 | 60% |
Two putative GPCR families, 7tm_6 and Mlo, were excluded from the training set based on literature and sequence analysis. Sequence redundancy was reduced to avoid data biases (see Materials and Methods). All in all, 311 sequences were used for training, partitioned into the 11 families as given in the third column.
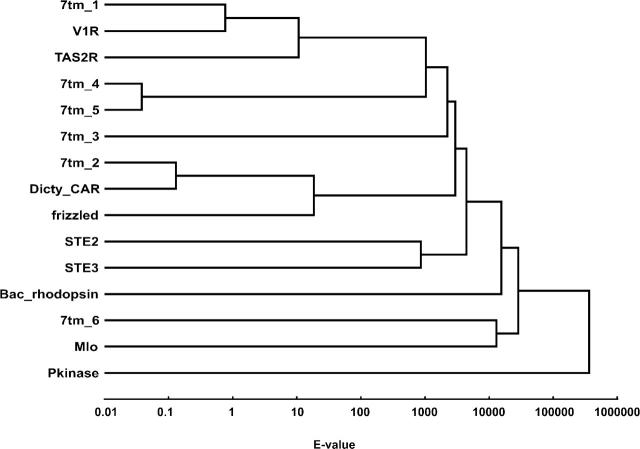
To investigate sequence similarity between these families, we used the profile HMMs provided by Pfam to cross-match the families. The full-length members in each Pfam family were scored against the HMMs of all the other families. If two families were remotely homologous, we would expect most members of one family to get a relatively high score to the other family’s HMM. To display the resulting matrix of pairwise relationships, we clustered all the families into a tree with the UPGMA algorithm (Fig. 1). As distance measure between two families we used the best (lowest) of the two median E-values generated in the searches.
Figure 1.
Similarity-based relationship tree of 13 confirmed or putative GPCR families and two families that are known not to be GPCRs: the bacteriorhodopsin (a proton pump) and the protein kinase families. Distances between families were obtained as follows. The Pfam HMM (“glocal” model) representing each of the family was used to score the full-length Pfam sequences of all other families. The logarithm of the lowest of the two median E-values from each reciprocal search was used as distance measure in the UPGMA algorithm. The database size for the HMM searches was set to 106 sequences. To avoid negative distances, a constant was added to all values in the distance matrix but this was compensated for on the X-axis scale. The tree places bacteriorhodopsin between the confirmed GPCR families and Mlo and 7tm_6, suggesting that the latter are not GPCRs.
Two families emerged as outliers in the cross-match analysis: the fly-specific odorant receptor family 7tm_6, and the plant family Mlo (Mlo/PF03094). These two families are placed on a branch of their own that is distant from confirmed GPCR families. The Bacteriorhodopsin family (which are proton pumps) clusters between the known GPCRs and the two families. This indicates that the 7tm_6 and Mlo families are either highly divergent GPCRs that would be important to include in the training set, or incorrectly annotated, which would contaminate the training set. We therefore analyzed these two families further (see following section), which led us to exclude them from model training.
Analysis of the 7tm_6 and Mlo families
The 7tm_6 family is composed of arthropod-specific odorant receptors (Hallem and Carlson 2004) with high sequence divergence. The response to odors has been well characterized (Hallem et al. 2004), but to our knowledge, nobody has experimentally confirmed the proteins’ membrane spanning topology or the proposed coupling to G proteins.
We ran the TM topology predictors Phobius, HMMTOP, and TMHMM on the 40 7tm_6 sequences in the Pfam seed, and only nine of the 120 predictions agreed with a GPCR topology. The most often predicted topology was an inverted GPCR topology, i.e., seven TM segments but with a cytosolic N terminus. Contrary to these predictions, the UniProt annotation of the sequences corresponds to a normal GPCR TM topology. We noticed that the UniProt topology annotation strongly disagrees with the “positive inside rule.” This rule states that positively charged amino acids (Arg and Lys) are mainly located on the cytosolic side of the membrane (von Heijne 1986). We counted the balance of positive amino acid residues for the UniProt topology of the 40 7tm_6 proteins. On average, they had 20 more positive amino acids on the extracellular side than on the cytosolic side of the membrane (26 vs. 6). If only the 10 residues closest to the membrane were taken into account, the excess on the extracellular side was 7 (12 vs. 5) on average. The latter calculation on 7tm_1, 7tm_2, and 7tm_3 yielded between 7 and 10 fewer amino acids on the extracellular side compared to the cytosolic side.
The plant Mlo family is also an outlier in the GPCR family tree in Figure 1. A seven-TM topology has been confirmed experimentally for one of the family members (Devoto et al. 2003), but there is no evidence for interaction with G proteins. On the contrary, Mlo seems to function independently of G proteins (Kim et al. 2002). Since the G protein repertoire of plants consists of only one or two heterotrimeric G protein complexes (Jones 2002), one can expect the number of GPCRs to be limited in plants. The A. thaliana genome encodes another transmembrane protein, GCR1, with recognizable sequence similarity to other GPCRs (Josefsson and Rask 1997). GCR1 has been coimmunoprecipitated with the Gα subunit GPA1 (Pandey and Assmann 2004), which indicates G protein coupling. Although the activating ligand remains to be identified, this finding has turned GCR1 into a more likely GPCR candidate than Mlo.
Analysis of the training set
We compiled a redundancy-reduced (see Materials and Methods) training set of 311 GPCRs from 11 families (i.e., excluding 7tm_6 and Mlo) and created a multiple alignment of all sequences. Consistent with earlier reports (Bockaert and Pin 1999; Gether 2000; Fredriksson et al. 2003), visual inspection of the alignment did not reveal any universally conserved residues or motifs. We looked in particular for two common GPCR features: the DRY-motif (the residues [ED]RY in the interface between the third TM helix and the second cytosolic loop), and a cysteine in each of the first and second extracellular loops. The DRY-motif is primarily found in the 7tm_1 family sequences and is not conserved in other families. The cysteines were found in a majority of the sequences but not as a rule. Although no multiple alignment program can guarantee a correct alignment from very divergent sequences, these results strongly suggest that conserved sequence motifs cannot form the basis for a general GPCR predictor.
It should be mentioned here that interfamily sequence similarities have been reported in a study of TM regions in human GPCR sequences (Fredriksson et al. 2003). The degree of similarity is suggestive of a common ancestry but not enough for a motif-based classifier, even though the study only included human sequences.
Because our goal was to train a model based on universal GPCR features, we analyzed the length and amino acid distributions of individual TM helices and loop regions. This is not a trivial task because the exact start and end of the membrane spanning regions is unknown for most of the sequences. We used a combination of UniProt annotation, TM prediction tools, and profile HMMs to locate the membrane and loop regions (see Materials and Methods).
Length analysis
For each of the seven TM helices, we calculated the mean length in the training set but found no systematic difference between them. All seven helices had a median length between 22 and 24 amino acids. In contrast, the loops displayed substantially more length diversity (Fig. 2). The first and the last loops are short (median 11 and 9 amino acids), while the four central loops were on average 20–27 amino acids long. It is important to note that the length distribution of the first cytosolic loop is much tighter than all the other loops, suggesting that its length is biologically more important. Although the third extracellular loop had a shorter median length it shows much more variation in length. The N- and C-terminal segments had much broader length distributions (data not shown), and are therefore of less interest in this work. Our results on median length and length distributions are essentially in agreement with earlier studies (Otaki and Firestein 2001; Inoue et al. 2004).
Figure 2.
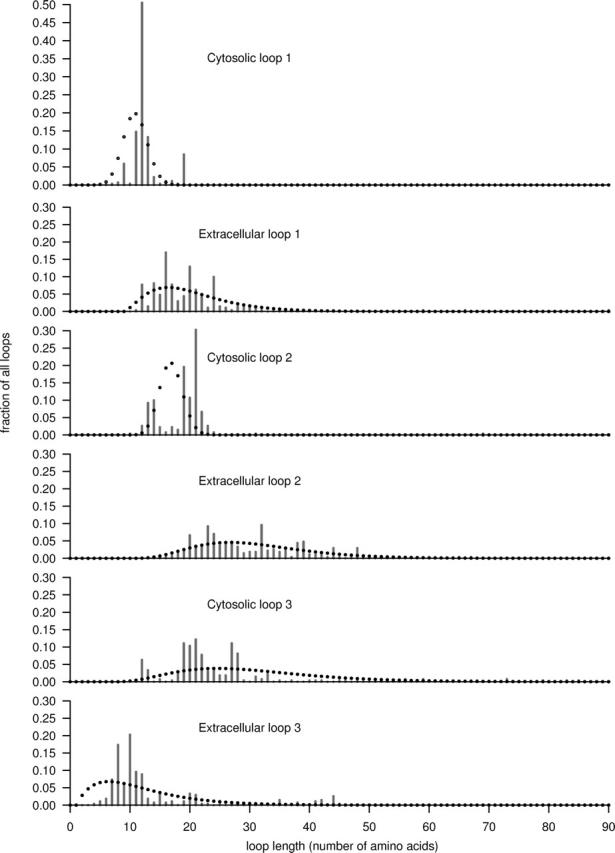
Loop length distributions of the training set sequences (bars) and modeled length distributions (dots). The observed lengths: most notable is the conserved and short length of the first cytosolic loop. Also, the second cytosolic loop has a narrow length distribution. In contrast, the first extracellular loop includes a number of long examples. The second extracellular and the third cytosolic loops have wide length distributions and long median lengths. The third extracellular loop is often short but has a wide length distribution. Modeled length distributions: the data was fitted to binomial distributions (cytosolic loop 1 and 2) or to negative binomial distributions (the remaining loops). The estimated distributions follow the observed data reasonably well given the trade-off between modeling quality and the risk of overtraining on imperfect data.
Amino acid composition analysis
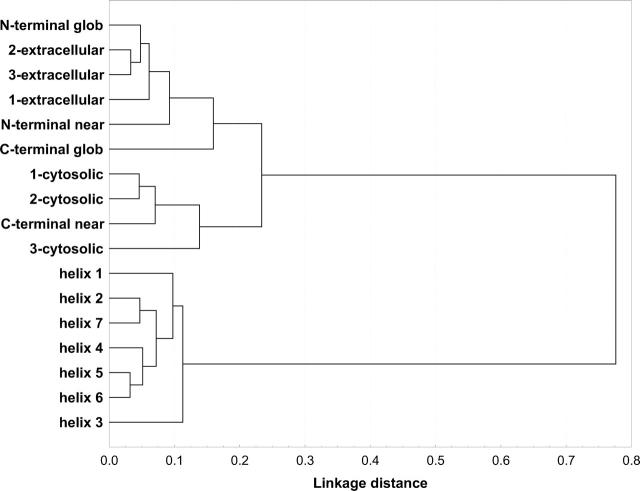
Amino acid frequencies were derived for each region using all sequences. A measure based on relative entropy (see Materials and Methods) was used to derive distances between amino acid distributions. These were used to generate a UPGMA tree to show the relation of the different regions (Fig. 3). Not surprisingly, amino acid distributions of TM and soluble regions form two distinct groups. Extracellular regions are more similar to each other than to cytosolic regions and vice versa. For the analysis we chose to divide the N- and C-terminal regions into two regions each, as we suspected that the region “close” to the membrane could have a composition different from the rest of the terminus. We arbitrarily defined “close” to be within 15 amino acids of the membrane. Figure 3 shows that the C-terminal region closest to the membrane (“C-terminal near”) is indeed clustering with the cytosolic loop regions, and that it is quite distinct from the “globular” C-terminal region.
Figure 3.
An amino acid composition-based relationship tree of the different topological regions in the training set GPCRs. A distance measure based on relative entropy was used (see Materials and Methods). Terminology: the numbering is from the N terminus to the C terminus. “1-extracellular” is the first extracellular loop, “1-cytosolic” is the first cytosolic loop, and so forth. “N/C-terminal near” corresponds to the 15 residues closest to the membrane in the N/C-terminal soluble regions, while “N/C-terminal glob” represents the remaining residues to the respective termini.
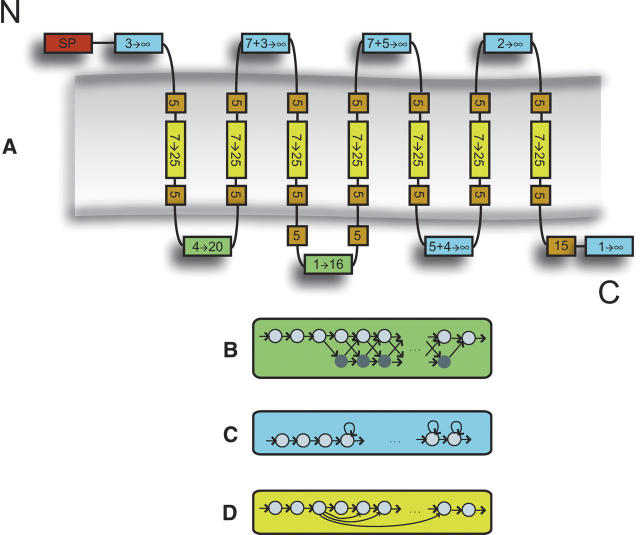
From the observed length and amino acid composition features we constructed a hidden Markov model with an architecture that is natural to GPCR sequences (see Fig. 4). We refer to the model as “compartmentalized” as each TM helix and loop region is modeled by a separate compartment with a specific amino acid probability distribution and length model. Each compartment is composed of a series of connected states with tied (identical) amino acid emission probabilities, and the architecture of connections gives rise to different length distributions.
Figure 4.
(A) Overview of the GPCRHMM architecture. A box where the possible length interval is indicated represents each model compartment. To model different types of sequence lengths data we have used three sets of connectivity layouts that correspond to different distributions. See Materials and Methods for a description of the signal peptide (SP) compartment. (B) In this connectivity layout the emitting states are accompanied by “silent” states that do not emit amino acids. This generates a distribution with a limited maximum length, and was used to model the first and second cytosolic loops. (C) Here, the states have a self-transition and a transition to the next state. All self-transitions are given the same probability. This generates a length distribution with unlimited maximum length, which was used for other remaining loops. The notation x + y → ∞ means that the compartment has a fixed length region of x states followed by a region of y states allowing lengths of y → ∞. (D) This layout of forward connected emitting states was used to model the core of a TM helix.
We used two types of architectures to model loop lengths. Loops that were highly conserved in length (cytosolic loop 1 and 2) were modeled in a way that restricts the length to a finite maximum value, while the other loops were allowed to be infinitely long with a decreasing probability. The encoded length distribution is determined by the transition probabilities between the states in a compartment, and these were estimated from the training set. By tying the transition probabilities the number of free transition parameters is reduced to only two per compartment. The training procedure and details of the model are described in Materials and Methods.
Comparison to other methods
We compared GPCRHMM to other possible strategies for GPCR detection: Pfam profile HMMs, two general TM topology predictors (Phobius and HMMTOP), and two specialized GPCR predictors (7TMHMM and QFC). Pfam is a sensitive resource for sequence similarity searches and is frequently employed for genome annotation. We wanted to know whether the individual Pfam GPCR families can be used to train a compartmentalized HMM capable of detecting remote homologs that the family profile HMMs would not find.
GPCRHMM and Pfam accuracies were tested using cross-validation such that one family at the time was taken out and buried in a set of 1071 non-GPCR sequences. The other families were used to recognize the taken out family by means of training a GPCRHMM model, or in the case of Pfam, by building profile HMMs. The accuracy was measured as Minimum Error Rate (MER), which is the lowest number of false positive and false negative classifications (see Materials and Methods). GPCRHMM produced an MER of 50 while the Pfam searches gave a considerably higher (worse) MER of 92. This indicates that GPCRHMM would have a higher probability of detecting a previously unknown family of GPCRs than the collection of GPCR profile HMMs in Pfam.
GPCRHMM was also compared to four topology predictor methods. These methods could not be subjected to a cross-validation test; hence, we compared them to a GPCRHMM model trained on all sequences in the training set. All methods were tested for sensitivity and selectivity on a homology-reduced version of GPCRDB and the negative test set used above. If we require the general-purpose TM predictors Phobius and HMMTOP to predict the topology correctly, i.e., seven TM regions and an extracellular N terminus, these methods record a relatively low sensitivity of 79.6% and 79.3% (Table 2). In comparison, GPCRHMM with a global score cutoff of −15 and a local score cutoff of 0 produced about 15% higher sensitivity at about the same false positive rate (~1%). By raising the cutoff in GPCRHMM, the false positive rate was eliminated with only 1% loss in sensitivity.
Table 2.
Benchmarking GPCRHMM against other methods
| Sensitivity (on 1706 positives) | False positive rate (on 1071 negatives, soluble and other TM proteins) | False positive rate | |
| GPCRHMM | |||
| global score >−15 | 94.4% | 0.93% | 6.86% |
| global score >−5 | 93.7% | 0.28% | 2.55% |
| global score >0 | 92.8% | 0.00% | 1.18% |
| HMMTOP | |||
| ø7i | 79.3% | 1.11% | |
| 6–8 TM | 95.4% | 8.87% | |
| Phobius | |||
| ør7i | 79.6% | 1.21% | |
| 6–8 TM | 94.8% | 9.80% | |
| QFC | 95.5% | 11.4% | 81.2% |
| 7TMHMM | 93.5% | 10.0% | 93.3% |
At a false positive rate of ~1%, GPCRHMM has ~15% higher sensitivity than HMMTOP and Phobius. The only way for other methods to achieve a similar sensitivity as GPCRHMM’s is to accept a 10-fold higher false positive rate on the first negative set and even higher on the bacterial negative set. For comparison, at a false positive rate of 8.87%, GPCRHMM reached a sensitivity of 98.0%. Sensitivity was measured on a nonredundant subset of GPCRDB, and the false positive rate on a set of TM (non-GPCRs) and soluble sequences as well as on E. coli transmembrane proteins. GPCRHMM was run with a local score cutoff of 0 in all three cases.
The general TM predictors reach increased sensitivity levels if we accept all topologies predicted with six to eight TM regions as GPCRs. This setting improved the sensitivity of Phobius and HMMTOP to about the same as GPCRHMM (global score cutoff −15) but their false positive rate was increased about 10-fold. By lowering the GPCRHMM global score cutoff to −53 and not using a local cutoff, it reached the same false positive rate as HMMTOP 6–8 TM (8.87%), but at a sensitivity of 98.0% compared to HMMTOP’s 95.4%. Thus, at comparable false positive rates, GPCRHMM is a more sensitive method than Phobius and HMMTOP.
7TMHMM and QFC are dedicated GPCR predictors. Both methods, however, perform roughly as Phobius and HMMTOP in increased sensitivity mode, i.e., with acceptable sensitivity but with a high false positive rate. We also tested the performance on another negative set: all Escherichia coli proteins predicted by Phobius to have five or more TM helices. Since prokaryotes lack GPCRs, this dataset is a true negative set even though it may contain seven TM proteins. Here, QFC and 7TMHMM had false positive rates of 81.2% and 93.3%, while GPCRHMM only predicted 1%–7% depending on the cutoff. It is therefore fair to say that although QFC and 7TMHMM can detect multispanning proteins they are not GPCR-specific.
Whole proteome searches
We applied GPCRHMM to five complete proteomes: Human, Mouse, C. elegans, D. melanogaster, and F. rubripes (Table 3; Supplemental Tables S1–S5). The highest number of hits was detected in mouse, followed by C. elegans and human. This is consistent with earlier research, and reflects comparatively large numbers of receptors for environment sensing: olfactory receptors in mouse (Young et al. 2002; Zhang and Firestein 2002) and chemoreceptors in C. elegans (Robertson 1998, 2000; Mombaerts 1999; Remm and Sonnhammer 2000).
Table 3.
GPCRHMM predictions of GPCRs in five proteomes
| Full name | Pfam ID | Human | Mouse | C. elegans | Drosophila | Fugu |
| Predictions with Pfam GPCR support | ||||||
| Rhodopsin-like family | 7tm_1 | 785 | 1412 | 122 | 78 | 354 |
| Secretin-like family | 7tm_2 | 78 | 49 | 6 | 25 | 84 |
| Metabotropic glutamate family | 7tm_3 | 35 | 171 | 5 | 11 | 62 |
| C. elegans chemoreceptor family | 7tm_4 | 248 | ||||
| C. elegans chemoreceptor family | 7tm_5 | 281 | ||||
| C. elegans Sra, Srb, Sre, Srg | Sr[a,b,e,g] | 101 | ||||
| Frizzled/Smoothened family | Frizzled | 11 | 14 | 5 | 5 | 8 |
| Vomeronasal pheromone receptor | VIR | 5 | 121 | |||
| Mammalian taste receptor | TAS2R | 28 | 19 | |||
| Ocular albinism proteins | Ocular_alb | 1 | 1 | 1 | ||
| Lung seven transmembrane receptor | Lung_7-TM_R | 1 | 1 | 1 | 2 | |
| Predictions with other GPCR support | ||||||
| InterPro rhodopsin-like | 5 | 20 | 81 | 1 | 6 | |
| Other GPCR families | 7 | 5 | 11 | 4 | 4 | |
| Serpentine receptor annotation | 442 | |||||
| Novel GPCR predictions | ||||||
| No annotation | 3 | 6 | 42 | 3 | 1 | |
| Pfam DUF1171 | 1 | 1 | 1 | 2 | 1 | |
| Pfam DUF40 | 44 | |||||
| Pfam DUF286 | 7 | |||||
| Pfam DUF621 | 5 | |||||
| Pfam DUF1182 | 2 | |||||
| Pfam DUF32 | 1 | |||||
| Suspected false positives (non-GPCR annotation or low sequence complexity) | 58 | 42 | 48 | 41 | 100 | |
| Total predictions (Fraction of proteome) | 1018 (3.4%) | 1862 (5.8%) | 1453 (6.5%) | 172 (1.2%) | 621 (1.9%) |
The predictions were divided into the groups Pfam support, other support, or novel predictions. The “other support” section includes predictions that did not match Pfam but have GPCR support from InterPro, UniProt, WormBase, or FlyBase annotation. Suspected false positives are either sequences with a proven alternative annotation, such as ion channels, or sequences with repeating patterns of low complexity. Low complexity was particularly frequent in Fugu rubripes, where ~45% of the false positives contained low complexity regions. A large fraction of the C. elegans predictions belong to families specific to worm, and many of these contain Pfam domains of unknown function (DUF).
Most of the GPCR predictions matched one of the existing Pfam GPCR families. Of these, the 7tm_1 family (PF00001) is the most common except for in C. elegans where chemoreceptor families dominate. In C. elegans, 7tm_1 only stands for 8% of all predicted GPCRs, whereas in human and mouse, this family alone stands for about 75%. In D. melanogaster and F. rubripes, the 7tm_1 fraction is close to 50%.
For several proteins that did not belong to a Pfam family, we were able to find other sources of annotation, such as InterPro, UniProt, Wormbase (Harris et al. 2004) and FlyBase (FlyBase Consortium 2003), that supported a function as GPCR. The majority of the proteins in this category are proteins that belong to the “Rhodopsin-like GPCR superfamily” in InterPro (IPR000276). Here, the similarity was too weak for detection by the corresponding Pfam 7tm_1 model but was possible to detect using other InterPro protein family databases such as PRINTS, Prosite or SMART. GPCRHMM predicted 113 (81 in C. elegans) proteins that had rhodopsin-like InterPro annotation but were not classified as 7tm_1 in Pfam. Another 31 GPCRHMM predicted proteins were not members of a Pfam GPCR family, but were annotated as GPCRs by one of the other databases. This includes the intimal thickness-related receptors (Tsukada et al. 2003) and members of the transmembrane seven superfamily (Spangenberg et al. 1998). In C. elegans, 442 of the GPCRHMM predictions were annotated as “serpentine receptor,” supporting a GPCR function. Most of these (413) were of other types than Sr[a,b,e,g], which exist in Pfam.
The remaining predictions can be regarded as new discoveries of GPCRs. GPCRHMM detected 55 proteins in the five proteomes with no annotation at all. The majority (42) of those proteins were found in the C. elegans proteome. Of the 55 predictions with no annotation, only nine show homology to clearly annotated GPCRs, as detected by Blastp searches against UniProt (E-value cutoff = 10−5). Other novel predictions belonged to Pfam families that do not indicate a GPCR function. Also here C. elegans yielded most predictions, 60 of 65. One of the families was Pfam DUF1171 (PF06664), which was found in all five proteomes. The DUF1171 domain, as it is defined in Pfam, only spans four TM regions. Phobius predicts DUF1171 members to have either seven TM helices with an extracellular N terminus (with or without signal peptide) or eight TM helices with cytosolic N terminus, making a GPCR topology likely. Novel human GPCR predictions included one DUF1171 member (Q7Z2Z9) and three unannotated proteins (Q9H6H6, Q8N4V6, Q9P2C4). The latter two proteins appear to be splice variants of the same gene.
The largest novel family was DUF40 (PF01838), that contained 44 predicted GPCRs, all in C. elegans. The Pfam domain spans four or five of the seven TM helices found by GPCRHMM; the remaining TM helices are N-terminal to the Pfam domain. Out of the 40 full-length seed sequences of DUF40, GPCRHMM detects 27. Among these sequences, a conserved [DE]R-motif is present in the interface between the third helix and the second cytosolic loop, which is the location of the DRY motif in rhodopsin-like GPCRs. Not all members of DUF40 are detected by GPCRHMM; most of the undetected members contain extra TM helices, and seven proteins have a second DUF40 domain. The DUF286 (PF03383) domain is found in seven of the novel predictions, none of which has any functional annotation. A conserved arginine is present in the predicted second cytosolic loop, which could be related to the DRY motif. Sequences with the DUF621 (PF04789) and DUF1182 (PF06681) domains are also predicted by GPCRHMM, but none of the sequences have GPCR annotation. The DUF621 domain is cut short in Pfam, where it only spans about three TM helices.
In summary, GPCRHMM classified 55 proteins as GPCR that had no previous annotation. Another 65 predictions lack functional annotation but belong to Pfam DUFs. Of these, DUF1171 is unique in being found in all five proteomes, while the other DUF-matching novel predictions are worm-specific. These often span fewer than seven TM helices, and have in some cases either been duplicated or combined with other domains or sequences containing transmembrane segments. We observed great diversity in length and the number of predicted TM segments for many of the DUF domain members; hence, many were not predicted by GPCRHMM. This variation is either caused by poor gene predictions, or a result of rapid evolution of chemosensory receptors, generating a collection of truncated or aberrant pseudogenes.
Finally, we classified some of the predictions as suspected false positives, if they had a consistent and proven annotation (mostly transporters) or regions of low sequence complexity which can sometimes fool HMMs of the GPCRHMM type. Low complexity sequences were particularly common in the F. rubripes proteome, and this increased the number of false positives considerably.
Discussion
We have described GPCRHMM, a hidden Markov model tailored to the superfamily of G protein-coupled receptors. The model is intended for identification of remote members of the GPCR superfamily that cannot be detected by ordinary methods. By a cross-validated benchmark we showed that for identification of novel families, GPCRHMM is superior to the set of Pfam profile HMMs. We also compared GPCRHMM to general-purpose topology predictors and found that GPCRHMM is considerably better at discriminating GPCR sequences from non-GPCR sequences.
The main model assumption in GPCRHMM is that GPCRs have high-level features that are more conserved than the primary sequence and make them distinguishable from other proteins. We have described conserved patterns in loop lengths, but also clear differences in amino acid composition between TM helices, extracellular, and cytosolic loops. These features were incorporated into GPCRHMM by maximum likelihood parameter estimation methods.
We applied GPCRHMM to five proteomes and detected a large number of sequences that have no other annotation. The results from C. elegans are particularly interesting, and include a large number of sequences with no annotation, which are prime candidates for being previously undetected GPCRs.
Strikingly, GPCRHMM gives strong negative predictions to sequences of the arthropod odorant receptor family (corresponding to Pfam domain 7tm_6). Since this family was excluded from model training we were cautious about the biological relevance of these predictions. Could we have biased GPCRHMM by excluding the 7tm_6 family to the extent that it could not recognize the family? To get an indication of whether this was the case, we scored the 7tm_6 sequences by each of the 11 HMMs from the cross-validation benchmark. These HMMs are trained on all but one sequence family. We compared the 7tm_6 scores to the scores of the sequences belonging to the excluded family, and the 7tm_6 sequences overall scored much lower. Only nine of the 311 sequences in the training set scored below the top-scoring 7tm_6 sequence. This lends further support to the notion that 7tm_6 proteins are very different from known GPCRs. Since forcing a GPCR TM topology onto 7tm_6 proteins requires violating the positive inside rule, there is a lot indicating that the 7tm_6 proteins are not members of the GPCR superfamily.
The arthropod odorant receptors are related to the arthropod gustatory receptor family (Clyne et al. 2000) (Trehalose_recp in Pfam) and a small family in C. elegans (Robertson et al. 2003). GPCRHMM also predicts these families to be non-GPCRs. A speculation would be that the three families form a superfamily of environment-sensing receptors that have little in common with odorant receptors in mammals, and probably have a different membrane topology. The experimental work to determine the TM topology and possible G protein coupling remains to be done.
GPCRHMM has strengths and limitations compared to sequence similarity methods. The obvious strength is its potential to find receptor proteins with little or no sequence similarity to other GPCRs but with the conserved topology features. A possible limitation is that GPCRHMM by design cannot detect fragment sequences. In that sense, GPCRHMM and similarity-based methods complement each other in protein family discovery. For example, we suggest in this article that a number of Pfam DUFs are domains within GPCRs. Currently, available similarity-based tools cannot say that these proteins are GPCRs. GPCRHMM, on the other hand, cannot cluster sequences to construct proper families.
While it is not yet fully known what the seven-TM helix conformation offers that is necessary for GPCR function, the conserved topology provides a means for detecting novel families that have diverged in primary sequence. The HMM technology is highly suited for such topology-based detection, as it allows modeling distributions of sequence length and amino acids. GPCRHMM should be valuable for researchers interested in whether a sequence is a GPCR or not, as well as for determining the location of TM helices of known GPCRs.
GPCRHMM is freely available through the Web server http://GPCRHMM.cgb.ki.se/.
Materials and methods
Train and test data
We used the Pfam (Bateman et al. 2004) classification to obtain a diverse set of training sequences. From Pfam we extracted either the seed or full sequence sets of 13 families that are known or putative GPCRs according to GPCRDB (Horn et al. 2001). The seed sets are curated and contain a representative sample of trusted member sequences, but the full sets include more sequences. At this initial stage we decided to use the seed set if it contained at least 20 sequences, or else we used the full set. We also extracted the sequences of the bacteriorhodopsin family and the protein kinase family, both known not to be G protein-coupled.
All families were analyzed by “all-against-all” sequence searches using the HMMER 2.3.2 package (Eddy 1998). Using the search tool of this package (hmmsearch, with an E-value cutoff high enough to report all scores), the calibrated profile HMM of each family was used to score the full-length sequences of all other families. For each family pair, this generated two lists of E-values. We used the best of the two median E-values as a measure of similarity between the families. Using the median match rather than the best match compensates for the fact that some families contain many more sequences than others, and due to the larger sample have a bigger chance of generating extreme matches.
Having a similarity (or distance) measure between all families, we constructed a UPGMA tree using the Statistica package. Two families (7tm_6 and Mlo) emerged as dissimilar to any of the other families and were excluded from the training set. The remaining 11 families were used for model training after reduction of sequence redundancy. As input to the redundancy reduction we used the Pfam seed set of sequences if it held at least 50 sequences, or else we used the full sets. The redundancy reduction was carried out for each family individually based on sequence identity as reported by Blastp. We reduced the majority of the 11 family sets to a maximum sequence identity of 60%. Exceptions were made for the two closely related C. elegans chemoreceptor families (7tm_4/PF01461 and 7tm_5/PF01604), which were reduced to 30% sequence identity, and the secretin and metabotropic glutamate families (7tm_2/PF00002 and 7tm_3/PF00003), for which a level of 40% was used. The latter is motivated by a wish to keep the rhodopsin family (7tm_1) the largest in the dataset as it is the most commonly occurring receptor family. The final training set contained 311 sequences (Table 1).
Each amino acid in the training set was labeled to indicate whether it is cytosolic, extracellular, TM, or part of a signal peptide. It is known that UniProt records as well as prediction methods often misplace the exact location of TM helices. To circumvent this problem we aligned all training sequences to their respective Pfam HMM, using hmmalign. We gave each sequence position a label according to the UniProt annotation if available or else based on a prediction by Phobius (Käll et al. 2004). Phobius was executed in constrained mode, by fixing the C terminus to the cytosol. Each alignment column was then labeled based on a majority decision, and finally, each sequence position was relabeled by the consensus label of the column to which it was aligned. We verified that the HMM covered all TM regions at least partly, and that overhanging parts of TM segments were aligned sufficiently well by hmmalign. The thus labeled sequences were input to the length analysis and to the model training procedure.
Three datasets were used for assessing the performance of GPCRHMM: (1) the GPCRDB (release 8.0) sequence set, downloaded from http://www.gpcr.org. Bacteriorhodopsin (which are proton pumps), 7tm_6, and Mlo sequences were removed, and the sequences belonging to the Pfam taste receptor family (TAS2R) were added (these sequences are incorporated into later versions of the GPCRDB). The dataset was filtered to produce a nonredundant positive dataset. (2) A nonredundant negative dataset comprising 731 soluble and 340 TM sequences with known topology. The dataset was derived from the set used for training Phobius (Käll et al. 2004), by removing all proteins having a 7TM topology. (3) A negative dataset of all the 510 E. coli sequences predicted by Phobius to have at least five TM regions. Prokaryotic proteomes do not contain GPCRs so this set is entirely negative.
Minimum error rate calculation
For GPCRHMM, 11 models were trained from sequences in all but one of the families. Sequences of the remaining family were then “buried” in the negative dataset composed of a mix of soluble and TM proteins (no GPCRs), and searched for using the model trained on the other 10 families. The procedure was repeated for all families. Similarly, the Pfam HMM searches were carried out using all but one of the families, which was likewise buried in the negative test set.
The accuracies of the methods were compared using the cutoff score (Scutoff) that gives the lowest number of false positives (FP) and false negatives (FN), called Minimum Error Rate (MER). An equation for MER is MER = minScutoff{FP(Scutoff) + FN(Scutoff)}
Calculating MER for GPCRHMM was done by sorting the scores for all sequences (positives and negatives) in a list and setting a cutoff to minimize the number of misclassifications. This gave 11 MER values, one for each family, and these were summed up to produce the total MER value of GPCRHMM. The Pfam HMM searches gave 10 scores for each sequence (one per HMM). We only considered the best of these scores (lowest E-value) to produce a sorted list of positives and negatives. The total MER was then calculated in the same way as for GPCRHMM.
Model
GPCRHMM is built up by a consecutive series of compartments, representing mainly the seven TM helices, the three cytosolic and the three extracellular loops, as shown in Figure 4. There are also compartments for the C-terminal region and for the N-terminal region, the latter containing compartments for modeling an optional signal peptide.
Although each compartment consists of many states, this does not add much to the number of parameters and complexity of the model. All states within a compartment have the same amino acid probabilities, and only two transition probabilities are used in the same compartment. The number of states and their connectivity produce an implicit length distribution. An alternative approach to model length distribution is using an explicit length model (a duration model) (Rabiner 1989; Burge and Karlin 1997). This can give a better fit to highly complicated distributions. However, because the database annotation of loop lengths is far from perfect we wanted to avoid closely fitting the data, and instead favored the more neutral implicit length model. In terms of parameter numbers and model complexity, both approaches are roughly equal.
We used two different layouts to model loop regions with either limited maximum length (Fig. 4B) or with unlimited maximum length (Fig. 4C). For loops with limited maximum length, two parallel linear chains of N states, one chain emitting amino acids and one silent, were connected such that it is possible to pass from each state to the next emitting or silent state. All transition probabilities to an emitting state are set to p and all transition probabilities to a silent state are set to 1 − p. The sequence length generated by this pattern has a binomial distribution.
For loops with unlimited maximum length, the pattern is shown in Figure 4C. It consists of a linear chain of N emitting states that all have self-transitions. Again, transition probabilities are set equal throughout the structure such that there is a probability p of staying in the current state and a probability 1 − p of continuing to the next state. The emitted sequence can be no shorter than N amino acids but there is no upper limit. The lengths of sequences generated by this pattern have a negative binomial distribution (Durbin et al. 1998).
Data on the first cytosolic loop (see Fig. 2) shows a narrow length interval that motivated modeling it by a binomial length distribution ranging from 0–16 amino acids (Fig. 4). A fixed length region of four amino acids was added, which gives a length interval of 4–20 amino acids that agrees with the observed loop length data. A similar architecture was used to model the second cytosolic loop with a loop length interval of 11–26 amino acids, which covers all but three cases. We regarded those as potentially false outliers and prioritized a specific model that can capture essentially all observed values.
The remaining four loops had less defined length (see Fig. 2) and we modeled them using the model with unlimited maximum length. A fixed-length region of seven states followed by three self-transition states model the first extracellular loop. This gives a minimum length of 10 amino acids and no maximum length. The second extracellular loop is modeled by a similar structure but with five states with self-transition, which gives a minimum loop length of 12 amino acids. Also, the third cytosolic loop is modeled by the same overall architecture: a fixed-length region of five amino acids followed by four states with self-transitions. The final (third) extracellular loop is biased towards very short lengths (Fig. 2), which was best modeled by a model of only two states with self-transitions. The N-terminal region of the mature protein is modeled by three states with self-transitions. Our analysis of amino acid composition suggested that the C-terminal region should be modeled by two separate compartments. We split it into a fixed length region of 15 states followed by one state with self-transition.
TM helices were modeled by a core compartment allowing from 7 to 25 amino acids (see Fig. 4D), flanked on each side by fixed-length five amino acid long “helical end” compartments. This is the same as the TM architecture used in Phobius (Käll et al. 2004).
We analyzed the similarities between amino acid distributions of all topological regions (Fig. 3), and this suggested a way to reduce the number of model parameters by linking the emission probabilities of different compartments. In the final model, the emission probabilities were set to be equal for the following regions: the extracellular loops; the TM helix core of the fourth, fifth, and sixth TM helix; the extracellular side of the fourth and sixth TM helix; the cytosolic side of the first, fourth, and fifth TM helix.
We found an increased number of positive residues near the ends of the second cytosolic loop compared to the other cytosolic loops (data not shown), and hence, we chose to divide this loop into three different compartments. Two flanking regions of five amino acids were given a different amino acid distribution than the remaining variable length region.
A signal peptide was present in 25% of the training set proteins. Signal peptides are composed of a hydrophobic region flanked by more hydrophilic regions followed by a cleavage site motif. Topology predictors often fail to discriminate the hydrophobic region of signal peptides from membrane spanning regions (Krogh et al. 2001), but with a combined signal peptide and TM model the number of false predictions can be reduced substantially (Käll et al. 2004). We therefore modeled signal peptides explicitly in GPCRHMM by a specific model compartment consisting of three consecutive regions (n, h, and c) followed by a cleavage site model. The n-region is 1–10 amino acids long, the h-region 6–20 amino acids long, and the c-region 1–12 amino acids long. The −3, −1 motif of a signal peptide’s cleavage site (Perlman and Halvorson 1983; von Heijne 1983), was modeled by four states with individual amino acid distributions. A transition branch in the Begin state allows sequences to transit directly from the Begin state to the N-terminal model compartment or “choose” to pass through the signal peptide model.
HMM parameter estimation
The parameters of GPCRHMM were estimated in a procedure similar to Phobius parameter estimation. The procedure includes five steps. First, the transition probabilities of the TM helix states and the signal peptide model were estimated from data by maximum likelihood. The TM helix lengths were fitted to a gamma probability distribution, while each of the signal peptide submodel lengths (n, h, and c) were fitted to a normal distribution. Since distributions are continuous while transitions are discrete, the fittings were obtained by integrating the transition probabilities over all states. Signal peptide transition probabilities were only used as an initial estimation, while the TM transition probabilities were left unchanged in subsequent steps.
In a second step the remaining model parameters were estimated by a noise injected Baum-Welch procedure (Krogh et al. 1994). Prior to the second step, three amino acids on either side of all TM/loop borders and internal signal peptide borders were “unlabeled.” Unlabeling introduces flexibility for the training procedure to correct for mislabeled amino acids that may occur in the training set. The model from the second step was used to relabel the training data.
The third step reestimated the TM and signal peptide length model parameters as in step one, but this time the relabeled data was used. In a fourth step all other parameters were reestimated from the relabeled data using a standard Baum-Welch procedure. This procedure sets model parameters to maximize the probability of each sequence. A fifth step updated the parameters using conditional maximum likelihood (Krogh 1994), by which the parameters are set to maximize the correct labeling rather than the probability of the sequences.
HMM evaluation
The Forward algorithm (Rabiner 1989) is the primary scoring function employed by GPCRHMM. This is calculated by summing the probabilities of all paths through the model. The reported score is a log-odds score: the logarithm of the ratio between the model score and the null model score. The null model is a one-state model with self-transition and with standard neutral background emission probabilities. A GPCRHMM score is thus indicating the fit of a sequence to GPCRHMM relative to a random model. The model architecture implies a lower bound on sequence length, and we introduced a prefiltering step that classes all proteins shorter than 200 amino acids as non-GPCRs.
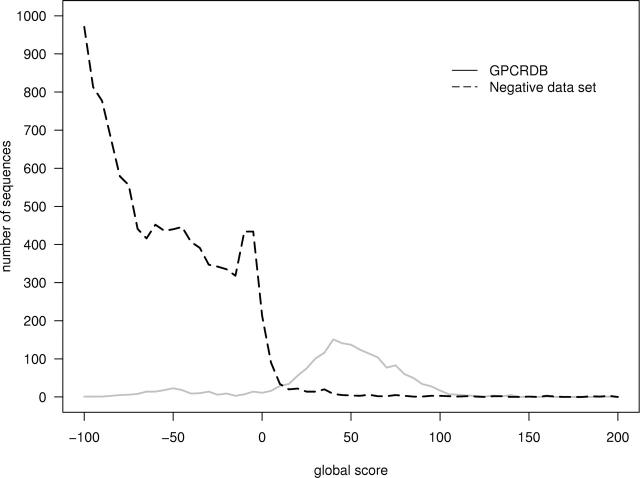
To get an idea of how well our method separates true from false we scored a nonredundant version of GPCRDB (dataset 1) and a reduced set of Swiss-Prot (release 42, 2004) where all sequences of >20% sequence identity to any sequence in GPCRDB had been removed (Fig. 5). We noticed that some soluble proteins were “fooling” the model; in particular, long cysteine-rich proteins. Our analysis is that these proteins fit the terminal loop regions well and accumulate enough score to compensate for poor resemblance to the core of GPCRHMM. To improve the scoring we therefore introduced a local score. The part of the query sequence modeled by GPCRHMM to span the first to seventh TM helix according to the prediction of the one-best algorithm (Schwartz and Chow 1990) was rescored by a core version of GPCRHMM lacking the signal peptide model and the N- and C-terminal models. In other words, the local score is testing for fit to the model only over the membrane spanning part of the protein. The local score removes a large number of false positives, while keeping the majority of true positives (Fig. 6). We call the scoring procedures global and local. The global score alone produces good results, and the local score should be seen as a refinement. By default, only sequences with global score above 0 bits are carried further for local score filtering.
Figure 5.
Large-scale testing of GPCRHMM. Shown is a histogram of GPCRHMM scores for GPCRDB (redundant sequences removed) and a large negative dataset (the Swiss-Prot database minus all sequences with >20% sequence identity to any protein in GPCRDB). Some high-scoring false positives occur, and to address this a local scoring procedure was devised (see Fig. 6). The majority of low scoring GPCRDB sequences are fly odorant receptors (7tm_6).
Figure 6.
GPCRHMM’s discrimination can be improved by applying a “local score.” Global and local scores are plotted for the sequences in GPCRDB and a large negative dataset as in Figure 5. The sequences from Figure 5 with a global score above 0 were rescored using a devised local score (see Materials and Methods). This improves the separation between true and false hits. We noted that a number of the high scoring negative sequences were actually putative GPCRs not part of GPCRDB (e.g., serpentine receptors). GPCRHMM’s default cutoffs are global score >0 and local score >0.
Tree building
Figure 3 was generated using the UPGMA method (Durbin et al. 1998). As distance measure between two relative amino acid distributions p = {pi} and q = {qi},i ∈ A, where A is the set of all amino acids, we chose a measure based on relative entropy:
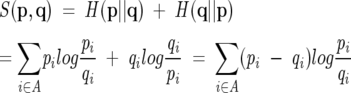 |
The measure was chosen because the relative entropy,
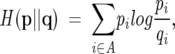 |
is not symmetrical in its original form. The measure is still not a true distance metric since it does not fulfill the triangle inequality, but should be sufficient for our purpose.
Algorithms and datasets
If nothing else is stated, the following algorithms, versions, and settings were used: Phobius, TMHMM 2.0, and HMMTOP 2.1 were run using default settings. 7TMHMM was obtained from the author and run using the forward score and a standard background model. QFC was also obtained from the author and run using the recommended cutoff level. HMMER 2.3.2 was used for the profile searches and always with default settings. GPCRHMM proteome predictions were based on an initial global score above −5, followed by a local score above 0. Pfam version 11.0 was used for retrieving profile HMMs. SEG 5.2.1, run with default settings, was used to detect low complexity sequences. Peptide sets from full genomes and their annotation were downloaded from Ensembl the following dates: Human (4 February 2004), Mouse (11 June 2004), F. rubripes (25 May 2004), and D. melanogaster (1 June 2004). The C. elegans peptide set was obtained from WormBase (wormpep123, 22 April 2004).
Acknowledgments
We thank Anders Krogh for the HMM software and helpful discussions, Steffen Möller for helpful discussions and for providing the 7TMHMM algorithm, and Gunnar Schulte for initial discussions. This work was supported by grants from Pfizer Inc. and the Swedish Knowledge Foundation.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051745906.
Supplemental material: see www.proteinscience.org
References
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32: D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockaert, J. and Pin, J.P. 1999. Molecular tinkering of G protein-coupled receptors: An evolutionary success. EMBO J. 18: 1723–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Clyne, P.J., Warr, C.G., Freeman, M.R., Lessing, D., Kim, J., and Carlson, J.R. 1999. A novel family of divergent seven-transmembrane proteins: Candidate odorant receptors in Drosophila. Neuron 22: 327–338. [DOI] [PubMed] [Google Scholar]
- Clyne, P.J., Warr, C.G., and Carlson, J.R. 2000. Candidate taste receptors in Drosophila. Science 287: 1830–1834. [DOI] [PubMed] [Google Scholar]
- Devoto, A., Hartmann, H.A., Piffanelli, P., Elliott, C., Simmons, C., Tar-amino, G., Goh, C.S., Cohen, F.E., Emerson, B.C., Schulze-Lefert, P., et al. 2003. Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J. Mol. Evol. 56: 77–88. [DOI] [PubMed] [Google Scholar]
- Durbin, R., Eddy, S.R., Krogh, A., and Mitchison, G.1998. Biological sequence analysis. Cambridge University Press, New York.
- Eddy, S.R. 1998. Profile hidden Markov models. Bioinformatics 14: 755–763. [DOI] [PubMed] [Google Scholar]
- FlyBase consortium. 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31: 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson, R., Lagerstrom, M.C., Lundin, L.G., and Schioth, H.B. 2003. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63: 1256–1272. [DOI] [PubMed] [Google Scholar]
- Gao, Q. and Chess, A. 1999. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 60: 31–39. [DOI] [PubMed] [Google Scholar]
- Gether, U. 2000. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr. Rev. 21: 90–113. [DOI] [PubMed] [Google Scholar]
- Hallem, E.A. and Carlson, J.R. 2004. The odor coding system of Drosophila. Trends Genet. 20: 453–459. [DOI] [PubMed] [Google Scholar]
- Hallem, E.A., Ho, M.G., and Carlson, J.R. 2004. The molecular basis of odor coding in the Drosophila antenna. Cell 117: 965–979. [DOI] [PubMed] [Google Scholar]
- Harris, T.W., Chen, N., Cunningham, F., Tello-Ruiz, M., Antoshechkin, I., Bastiani, C., Bieri, T., Blasiar, D., Bradnam, K., Chan, J., et al. 2004. WormBase: A multi-species resource for nematode biology and genomics. Nucleic Acids Res. 32: D411–D417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, F., Vriend, G., and Cohen, F.E. 2001. Collecting and harvesting biological data: The GPCRDB and NucleaRDB information systems. Nucleic Acids Res. 29: 346–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, Y., Ikeda, M., and Shimizu, T. 2004. Proteome-wide classification and identification of mammalian-type GPCRs by binary topology pattern. Comput. Biol. Chem. 28: 39–49. [DOI] [PubMed] [Google Scholar]
- Jones, A.M. 2002. G-protein-coupled signaling in Arabidopsis. Curr. Opin. Plant Biol. 5: 402–407. [DOI] [PubMed] [Google Scholar]
- Josefsson, L.G. and Rask, L. 1997. Cloning of a putative G-protein-coupled receptor from Arabidopsis thaliana. Eur. J. Biochem. 249: 415–420. [DOI] [PubMed] [Google Scholar]
- Käll, L., Krogh, A., and Sonnhammer, E.L. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338: 1027–1036. [DOI] [PubMed] [Google Scholar]
- Kim, J., Moriyama, E.N., Warr, C.G., Clyne, P.J., and Carlson, J.R. 2000. Identification of novel multi-transmembrane proteins from genomic databases using quasi-periodic structural properties. Bioinformatics 16:767–775. [DOI] [PubMed] [Google Scholar]
- Kim, M.C., Panstruga, R., Elliott, C., Muller, J., Devoto, A., Yoon, H.W., Park, H.C., Cho, M.J., and Schulze-Lefert, P. 2002. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416: 447–451. [DOI] [PubMed] [Google Scholar]
- Krogh, A.1994. Hidden Markov models for labeled sequences. In Proceedings of the 12th IAPR International Conference on Pattern Recognition, pp. 140–144. IEEE Computer Society Press, Los Alamitos, CA.
- Krogh, A., Brown, M., Mian, I.S., Sjoelander, K., and Haussler, D. 1994. Hidden Markov models in computational biology. Applications to protein modelling. J. Mol. Biol. 235: 1501–1531. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Mombaerts, P. 1999. Seven-transmembrane proteins as odorant and chemosensory receptors. Science 286: 707–711. [DOI] [PubMed] [Google Scholar]
- Möller, S.2001. “An environment for consistent sequence annotation and its application to transmembrane proteins.” Ph.D. thesis, University of Cambridge, Cambridge, UK.
- Möller, S., Vilo, J., and Croning, M.D. 2001. Prediction of the coupling specificity of G protein coupled receptors to their G proteins. Bioinformatics 17(Suppl 1): S174–S181. [DOI] [PubMed] [Google Scholar]
- Otaki, J.M. and Firestein, S. 2001. Length analyses of mammalian G-protein-coupled receptors. J. Theor. Biol. 211: 77–100. [DOI] [PubMed] [Google Scholar]
- Pandey, S. and Assmann, S.M. 2004. The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16: 1616–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman, D. and Halvorson, H.O. 1983. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J. Mol. Biol. 167: 391–409. [DOI] [PubMed] [Google Scholar]
- Rabiner, L.R. 1989. A tutorial on hidden markov models and selected applications in speech recognition. Proc. IEEE 77: 257–286. [Google Scholar]
- Remm, M. and Sonnhammer, E. 2000. Classification of transmembrane protein families in the Caenorhabditis elegans genome and identification of human orthologs. Genome Res. 10: 1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, H.M. 1998. Two large families of chemoreceptor genes in the nematodes Caenorhabditis elegans and Caenorhabditis briggsae reveal extensive gene duplication, diversification, movement, and intron loss. Genome Res. 8: 449–463. [DOI] [PubMed] [Google Scholar]
- ———. 2000. The large srh family of chemoreceptor genes in Caenorhabditis nematodes reveals processes of genome evolution involving large duplications and deletions and intron gains and losses. Genome Res. 10: 192–203. [DOI] [PubMed] [Google Scholar]
- Robertson, H.M., Warr, C.G., and Carlson, J.R. 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. 100(Suppl. 2): 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, R. and Chow, Y. 1990. The N-best algorithm: An efficient and exact procedure for finding the N most likely sentence hypotheses. Proc. Intl. Conf. on Acoust., Speech and Signal Proc. pp. 81–84.
- Spangenberg, C., Winterpacht, A., Zabel, B.U., and Lobbert, R.W. 1998. Cloning and characterization of a novel gene (TM7SF1) encoding a putative seven-pass transmembrane protein that is upregulated during kidney development. Genomics 48: 178–185. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Kadowaki, S., Haga, T., Takaesu, H., and Mitaku, S. 2002. Identification of G protein-coupled receptor genes from the human genome sequence. FEBS Lett. 520: 97–101. [DOI] [PubMed] [Google Scholar]
- Tsukada, S., Iwai, M., Nishiu, J., Itoh, M., Tomoike, H., Horiuchi, M., Nakamura, Y., and Tanaka, T. 2003. Inhibition of experimental intimal thickening in mice lacking a novel G-protein-coupled receptor. Circulation 107: 313–319. [DOI] [PubMed] [Google Scholar]
- Tusnady, G.E. and Simon, I. 1998. Principles governing amino acid composition of integral membrane proteins: Application to topology prediction. J. Mol. Biol. 283: 489–506. [DOI] [PubMed] [Google Scholar]
- von Heijne, G. 1983. Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem. 133: 17–21. [DOI] [PubMed] [Google Scholar]
- ———. 1986. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 5: 3021–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J.M., Friedman, C., Williams, E.M., Ross, J.A., Tonnes-Priddy, L., and Trask, B.J. 2002. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 11: 535–546. [DOI] [PubMed] [Google Scholar]
- Zhang, X. and Firestein, S. 2002. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 5: 124–133. [DOI] [PubMed] [Google Scholar]