Abstract
Thermal unfolding monitored by spectroscopy or calorimetry is widely used to determine protein stability. Equilibrium thermodynamic analysis of such unfolding is often hampered by its irreversibility, which usually results from aggregation of thermally denatured protein. In addition, heat-induced protein misfolding and aggregation often lead to formation of amyloid-like structures. We propose a convenient method to monitor in real time protein aggregation during thermal folding/ unfolding transition by recording turbidity or 90° light scattering data in circular dichroism (CD) spectroscopic experiments. Since the measurements of turbidity and 90° light scattering can be done simultaneously with far- or near-UV CD data collection, they require no additional time or sample and can be directly correlated with the protein conformational changes monitored by CD. The results can provide useful insights into the origins of irreversible conformational changes and test the linkage between protein unfolding or misfolding and aggregation in various macromolecular systems, including globular proteins and protein–lipid complexes described in this study, as well as a wide range of amyloid-forming proteins and peptides.
Keywords: circular dichroism spectroscopy, irreversible protein unfolding, turbidity, light scattering, asparaginase-2, high-density lipoprotein, amyloid, protein structure/folding, conformational changes, stability and mutagenesis, enzymes, thermodynamics, hydrodynamics, aggregation
Thermal or chemical unfolding is generally used to determine thermodynamic protein stability, which is the Gibbs free energy difference between the folded and the unfolded states, ΔG = GU − GF. In thermal unfolding experiments, protein solution is heated at a constant rate, and changes in the protein conformation or their heat effects are monitored by spectroscopy or differential scanning calorimetry (DSC), respectively. The results, including the melting temperature (Tm), enthalpy (ΔH(Tm)), and heat capacity increment (ΔCp) of the unfolding, are used to determine protein stability function (ΔG(T)) (Privalov 1979; Pace et al. 1989). The key assumption behind this approach is that the protein unfolding is a thermodynamically reversible (that is, an equilibrium) transition. Although this assumption is usually valid for chemical denaturation, thermal denaturation (which is particularly widely used in protein stability studies) is often irreversible. The general root cause of this irreversibility is aggregation of the heat-unfolded polypeptide. Moreover, in many proteins, heat-induced misfolding leads to formation of β-sheet-rich fibrillar aggregates that resemble natural amyloid (Kusumoto et al. 1998; Gursky and Aleshkov 2000; Fandrich et al. 2003; Yang et al. 2003 and references therein). If aggregation occurs after completion of the unfolding transition, it does not necessarily preclude equilibrium thermodynamic analysis of the unfolding. However, if protein aggregation and unfolding are concomitant, the transition is inherently irreversible and cannot be analyzed by equilibrium thermodynamics. To discriminate between these possibilities and to monitor in real time by the same technique the heat-induced protein unfolding or misfolding and aggregation, we propose to record turbidity or 90° light scattering data in circular dichroism (CD) experiments and illustrate the usage of these methods for two different macromolecular systems.
CD spectroscopy is the method of choice for quantitative analysis of protein secondary structure and its unfolding (Kallenbach et al. 1996; Venyaminov and Yang 1996). The proposed application is based on the ability of the CD spectrometer to record not only differential absorption between the right- and the left-circularly-polarized light (which is proportional to CD) but also regular absorption or, more precisely, turbidity. This is accomplished by registering dynode voltage V, which is high voltage applied to the photomultiplier of the UV detector to compensate for the reduction in the light intensity that may result from light absorption and/ or scattering.
Light absorption follows Beer’s law, I/Io = log (clɛ), where Io is the intensity of the incident light, I is the light intensity after passing through the sample, c is the concentration of absorbing centers, l is sample path length, and ɛ is the extinction coefficient that depends on the nature of the absorbing centers and the wavelength of light, λ. For biological macromolecules, ɛ does not significantly change with temperature; hence clɛ≅const in CD experiments in which c = const and λ= const, and temperature is varied. Examples are (1) melting experiments in which the sample is heated at a constant rate and CD signal is recorded at λ= const as a function of temperature, Θλ(T), or (2) kinetic temperature-jump experiments in which the sample temperature is rapidly changed to a new constant value, and CD signal is recorded at λ= const as a function of time, Θλ(t). Since clɛ = const in such experiments, light absorption remains invariant; this is illustrated by constant dynode voltage, Vλ(T) = const, observed in reversible thermal unfolding of nonaggregating proteins (Gursky et al. 2002). Therefore, heat-induced changes in the dynode voltage may result only from the variations in the light scattering due to changes in the particle size and/or refractive index. The general cause for such changes is aggregation of the thermally unfolded or misfolded protein, which may be aided by chemical modifications, such as oxidation, that are accelerated at high temperatures.
An alternative way to monitor protein aggregation during thermal unfolding is to record 90° light scattering by using fluorescence attachment (which is available in AVIV or Jasco CD instruments). Light scattering, which depends on the particle size and refractive index, increases sharply as the particle size approaches the wavelength of light (λ ~ 200 nm in UV). Thus, formation of large protein aggregates will cause an increase both in the light scattering and in the dynode voltage. In our experience, changes in the particle size by a few nanometers (which is a typical size of protein aggregates) are readily detectable by 90° light scattering in CD experiments. Also, our analyses of the same system by light scattering (Benjwal et al. 2005) and turbidity (Gursky et al. 2002) suggest that the former method is more sensitive and yields better signal-to-noise ratio in the melting data.
Below, we describe the usage of dynode voltage and 90° light scattering in CD experiments to monitor irreversible heat-induced unfolding and concomitant increase in the particle size in two different systems: (1) asparaginase-2 from Escherichia coli (EcA2, Mw = 138 kDa), a homotetrameric enzyme of known structure that is used in leukemia treatment (Wriston and Yellin 1973; Swain et al. 1993; Derst et al. 1994), and (2) complex of human apolipoprotein C-1 (apoC-1, Mw = 6 kD) and dimyriostoil phosphatidylcholine (DMPC) that models nascent high-density lipoproteins (Gursky et al. 2002). Our results suggest that monitoring heat-induced changes in turbidity or light scattering together with CD provides a useful tool for the analysis of irreversible unfolding in diverse macromolecular systems, such as globular proteins and protein–lipid complexes described below. Furthermore, since heat-induced misfolding and formation of amyloid-like aggregates have been proposed to be a general property of proteins with diverse structures (Yang et al. 2003), our approach should be particularly useful for dissecting β-sheet folding from aggregation in these proteins.
Results and Discussion
Irreversible thermal unfolding and aggregation of EcA2 monitored by CD and turbidity
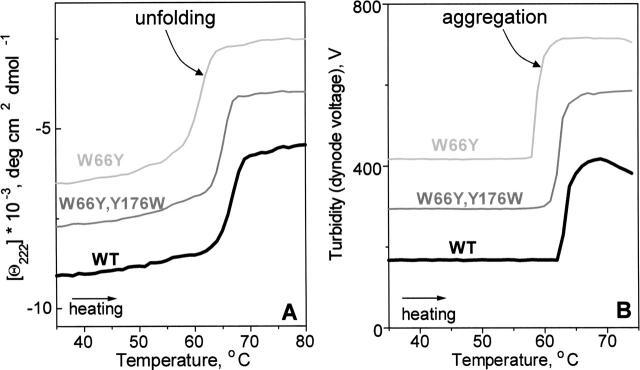
Figure 1A shows far-UV CD melting data recorded of EcA2 solution of 0.1 mg/mL protein concentration placed in a 2-mm cell. The sample was heated at a constant rate of 80 K/h, and the CD signal at 222 nm, which is proportional to the α-helical protein content, was recorded as a function of temperature, Θ222(T). Wild type EcA2, which is ~25% α-helical at room temperature, largely unfolds between 63° and 70°C. The unfolding is partially irreversible, as indicated by incomplete recovery of the CD and enzymatic activity upon heating and cooling from 25° to 80°C, and by the absence of the heat capacity peak in repetitive DSC scans (Verma 2005). Similarly, partially irreversible thermal unfolding of EcA2 mutants such as W66Y or (W66Y, Y16W) is observed by CD (Fig. 1A, thin lines) and by DSC (Verma 2005). To assess the cause of this irreversibility, turbidity (dynode voltage) was recorded upon heating protein solutions of 3.4 mg/mL concentrations placed in 5-mm cells. The V(T) data in Figure 1B were recorded together with the near-UV CD at 285 nm (data not shown); similar turbidity curves were observed at other wavelengths. The V(T) data of WT and mutant EcA2 show a large irreversible heat-induced increase by >300 V that is indicative of aggregation. Since a much smaller increase by only a few V can be readily detected by dynode voltage (Gursky et al. 2002), the protein concentration in these experiments can be reduced to low enough values (~0.1 mg/mL) to encompass the range used in far-UV CD experiments. The turbidity changes in EcA2 clearly correlate with the protein unfolding monitored by far-UV CD (Fig. 1A,B). For example, the onset of the unfolding in WT EcA2 indicated by CD and the onset of aggregation indicated by turbidity are observed at the same temperature of ~63°C (Fig. 1A,B, thick lines). Similar coincidence between the onset of unfolding and aggregation is detected in mutant proteins (Fig. 1A,B, thin lines). We conclude that the heat-induced α-helical unfolding in EcA2 and its mutants is concomitant with aggregation, which leads to intrinsic irreversibility and precludes thermodynamic analysis of this unfolding.
Figure 1.
Thermal unfolding and aggregation of wild type and mutant EcA2 monitored by CD and turbidity. (A) CD melting curves, Θ222(T), recorded at 222 nm report on α-helical unfolding. (B) Turbidity (dynode voltage) melting curves, V(T), recorded at 285 nm report on the heat-induced increase in the particle size due to protein aggregation. A reduction in V(T) of the WT observed upon heating >70°C reflects precipitation of the heat-unfolded protein. The data in A and B are shifted along the Y-axis to avoid overlap. Sample conditions are described in the text; heating rate is 80 K/h.
Thermal denaturation and fusion of lipoproteins monitored by CD and 90° light scattering
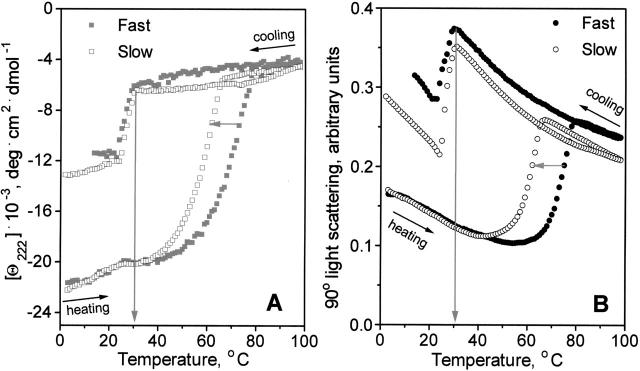
Another system in which thermodynamically irreversible heat unfolding leads to an increase in the particle size is lipoproteins. These are macromolecular complexes containing several protein and several hundred lipid molecules. ApoC-1:DMPC complexes used in this work are phospholipid bilayer disks that are 5.6-nm thick and 17 nm in diameter (Mehta et al. 2003) with amphipathic protein α-helices wrapped around the disk perimeter screening the lipid acyl chains from the aqueous milieu (Segrest et al. 1999; Lund-Katz et al. 2003). Earlier we showed that the heat-induced protein dissociation and unfolding from lipoproteins lead to particle fusion that compensates for the decrease in their polar surface (Gursky et al. 2002). Thermodynamic irreversibility of this transition is indicated by the hysteresis and scan rate effects on the far-UV CD melting data (Fig. 2A). The data in Figure 2A were recorded from DMPC complexes with L34P apoC-1; this apoC-1 mutant is largely unfolded in solution but is 65% helical in complex with the lipid (Mehta et al. 2003). Lipoprotein solution containing 0.02 mg/mL protein and 0.08 mg/mL lipid was placed in 5 mm cell, and CD data, Θ222(T), were recorded at 222 nm during sample heating and cooling from 2° to 98°C at a constant rate of 80 K/h (fast) or 11 K/h (slow). Simultaneously, 90° light scattering was recorded at 222 nm (Fig. 2B). Heat-induced increase in the light scattering shown in Figure 2B reflects particle fusion that was also observed by electron microscopy and nondenaturing gel electrophoresis (Gursky et al. 2002; Jayaraman et al. 2005); the particle diameter in this transition increases from 17 to ≥22 nm (Mehta et al. 2003). The heating portions of the data in Figure 2 clearly show that the protein unfolding (monitored by CD) and lipoprotein fusion (monitored by light scattering) occur in the same temperature range and are similarly affected by the changes in the scan rate. This is indicated by a −8° shift in the CD and light scattering heating curves observed upon slowing the heating rate from 80 to 11 K/h (horizontal arrows). Excellent agreement is also observed in the CD and light cooling curves that reflect partial protein refolding and concomitant lipoprotein reconstitution upon cooling <30°C (vertical arrows). We conclude that, in apoC-1:DMPC complexes, protein unfolding/dissociation and lipo-protein fusion into vesicles induced by heating, as well as protein refolding and lipoprotein reconstitution induced by cooling, are closely coupled. Protein unfolding in this system is thermodynamically irreversible due to the concomitant lipoprotein fusion that involves high kinetic barriers (Gursky et al. 2002); therefore, this unfolding is amenable to kinetic but not to thermodynamic analysis.
Figure 2.
Thermal denaturation and fusion of L34P apoC-1:DMPC complexes monitored by far-UV CD and 90° light scattering. (A) Heating and cooling CD data, Θ222(T), which were recorded upon heating at a rate of 80 K/h (▪) and 11 K/h (□), report on protein unfolding and dissociation from the lipoprotein. (B) Light scattering data, which were recorded at 222 nm simultaneously with the CD data in A, report on the increase in the particle size due to fusion. Horizontal arrows indicate shifts in the heating curves upon reduction in the scan rate from 80 (•) to 11 (○) K/h; vertical arrows indicate the onset of the protein refolding (A) and lipoprotein reconstitution (B) upon cooling. Protein and lipid concentrations are 20 μg/mL and 80 μg/mL, respectively.
Summary
Our results demonstrate that monitoring heat-induced changes in turbidity or light scattering in CD experiments can provide valuable insights into the origins of irreversibility in protein unfolding, such as protein aggregation or lipoprotein fusion described in this work. The proposed approach should be particularly useful for correlating heat-induced microscopic and macroscopic changes, such as β-sheet folding and formation of fibrillar aggregates, in a variety of amyloidogenic proteins, and thereby help to dissect protein misfolding and aggregation during amyloid fiber formation. Since turbidity and light scattering can be recorded simultaneously with CD, the approach requires no additional time or sample and facilitates very accurate correlation of the protein conformational changes and aggregation. This helps to find out whether the protein aggregation occurs during or after the unfolding, and thereby determine whether the unfolding is amenable to equilibrium thermodynamic analysis.
Materials and methods
Sample preparation
Asparaginase-2 and its mutants were expressed and purified as described in Harms et al. (1991). The protein purity was at least 95% as assessed by SDS/PAGE. The yield varied between 5 and 20 mg of purified enzyme per liter of E. coli culture, depending on the protein stability. The proteins were desalted using PD-10 desalting column (Amersham Pharmacia). The final buffer composition was 20 mM Na phosphate (pH 8.0).
Apolipoprotein C-1 (L34P mutant) was commercially synthesized using solid state synthesis and was purified by HPLC to 97%+ purity at Quality Control Biochemicals as described in Mehta et al. (2003). Lipoproteins were reconstituted from apoC-1 and DMPC (Avanti Polar Lipids) by co-incubation at 24°C using 1:4 mg/mg protein:lipid ratio. The complexes were visualized by negative staining electron microscopy before and after thermal denaturation to determine the morphology of the particles (disks or vesicles) and their size as described in Mehta et al. (2003). The buffer conditions were 10 mM Na phosphate (pH 7.6).
Circular dichroism, turbidity, and 90° light scattering data collection
CD and turbidity (dynode voltage) data were recorded as described in Gursky et al. (2002) by a UV detector in an AVIV 215 spectropolarimeter equipped with thermoelectric temperature control. Light scattering was recorded at the same wavelength as CD by an additional UV detector that is positioned at 90° to the direct beam and is a part of the fluorescence attachment; the filters from this attachment were removed. This assembly facilitates measurements of the light intensity at 90° in relative units but not on the absolute scale; hence it allows for qualitatively monitoring relative changes in the particle size but not for quantitatively analyzing the particle size distribution. In our experiments, the sample was positioned in the beam, and the light intensity at 90° was initially adjusted to 0.15 using “autoset” option of the fluorescence photomultiplier. Light scattering at 90° and turbidity (dynode voltage) at 0° were monitored simultaneously with the CD signal at the same characteristic wavelength (222 or 285 nm) during sample heating and cooling with 1 K increment and 30–300 sec data accumulation time, which corresponds to scan rates of ~80–11 K/h, respectively. The data were normalized to protein concentrations and expressed in units of molar residue ellipticity. Data analysis and display were done using ORIGIN software.
Acknowledgments
We thank Jack Aviv for continuous help and support of the CD instruments and for useful discussions. This work was supported by NIH grants GM 67260 and HL 26355, and by the German Research Foundation.
Abbreviations
CD, circular dichroism
DSC, differential scanning calorimetry
EcA2, asparaginase isoenzyme 2 from E. coli
apoC-1, apolipoprotein C-1
DMPC, dimyristoyl phosphatidylcholine
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051917406.
References
- Benjwal, S., Jayaraman, S., and Gursky, O. 2005. Electrostatic effects on the kinetic stability of model discoidal high-density lipoproteins. Biochemistry 44: 10218–10226. [DOI] [PubMed] [Google Scholar]
- Derst, C., Wehner, A., Specht, V., and Röhm, K.H. 1994. States and functions of tyrosine residues in Escherichia coli asparaginase II. Eur. J. Biochem. 224: 533–540. [DOI] [PubMed] [Google Scholar]
- Fandrich, M., Forge, V., Buder, K., Kittler, M., Dobson, C.M., and Diekmann, S. 2003. Myoglobin forms amyloid fibrils by association of unfolded polypeptide segments. Proc. Natl. Acad. Sci. 100: 15463– 15468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursky, O. and Aleshkov, S. 2000. Temperature-dependent β-sheet formation in β-amyloid Aβ(1–40) peptide in water: Uncoupling β-structure folding from aggregation. Biochim. Biophys. Acta 1476: 93–102. [DOI] [PubMed] [Google Scholar]
- Gursky, O., Ranjana, and Gantz, D.L. 2002. Complex of human apolipoprotein C-1 with phospholipid: Thermodynamic or kinetic stability? Biochemistry 41: 7373–7384. [DOI] [PubMed] [Google Scholar]
- Harms, E., Wehner, A., Jennings, M.P., Pugh, K.J., Beacham, I.R., and Röhm, K.H. 1991. Construction of expression systems for Escherichia coli asparaginase II and two-step purification of the recombinant enzyme from periplasmic extracts. Protein Exp. Purif. 2: 144–150. [DOI] [PubMed] [Google Scholar]
- Jayaraman, S., Gantz, D.L., and Gursky, O. 2005. Kinetic stabilization and fusion of apolipoprotein A-2:DMPC disks: Comparison with apoA-1 and apoC-1. Biophys. J. 88: 2907–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenbach, N.R., Lyu, P., and Zhou, H. 1996. CD spectroscopy and the helix-coil transition in peptides and polypeptides. In Circular dichroism and the conformational analysis of biomolecules, (ed. G. Fasman), pp. 201–259. Plenum Press, New York.
- Kusumoto, Y., Lomakin, A., Teplow, D.B., and Benedek, G.B. 1998. Temperature dependence of amyloid β-protein fibrillization. Proc. Natl. Acad. Sci. 95: 12277–12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund-Katz, S., Liu, L., Thuahnai, S.T., and Phillips, M.C. 2003. High density lipoprotein structure. Front. Biosci. 8: 1044–1054. [DOI] [PubMed] [Google Scholar]
- Mehta, R., Gantz, D.L., and Gursky, O. 2003. Effects of mutations on the reconstitution and kinetic stability of discoidal lipoproteins. Biochemistry 42: 4751–4758. [DOI] [PubMed] [Google Scholar]
- Pace, C.N., Shirley, B.A., and Thomson, J.A. 1989. Measuring conformational stability of a protein. In Protein structure: A practical approach (ed. T.E. Creighton). Oxford University Press, Oxford, UK.
- Privalov, P.L. 1979. Stability of proteins: Small globular proteins. Adv. Protein Chem. 33: 167–241. [DOI] [PubMed] [Google Scholar]
- Segrest, J.P., Jones, M.K., Klon, A.E., Sheldahl, C.J., Hellinger, M., De Loof, H., and Harvey, S.C. 1999. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274: 31755–31758. [DOI] [PubMed] [Google Scholar]
- Swain, A.L., Jaskolski, M., Housset, D., Rao, J.K., and Wlodawer, A. 1993. Crystal structure of Escherichia coli L-asparaginase, an enzyme used in cancer therapy. Proc. Natl. Acad. Sci. 90: 1474– 1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venyaminov, S.Y. and Yang, J.T. 1996. Determination of protein secondary structure. In Circular dichroism and the conformational analysis of biomolecules (ed. G. Fasman), pp. 69–107. Plenum Press, New York.
- Verma, S. 2005. “Systematic site-directed mutagenesis to characterize subunit interactions in E. coli asparaginase II, an enzyme used in leukemia treatment.” Ph.D. thesis, Philipps University Marburg, Marburg, Germany.
- Wriston, J.C., and Yellin, T.O. 1973. L-asparaginase: A review. Adv. Enzymol. 39: 185–248. [DOI] [PubMed] [Google Scholar]
- Yang, W.Y., Larios, E., and Gruebele, M. 2003. On the extended β-conformation propensity of polypeptides at high temperature. J. Am. Chem. Soc. 125: 16220–16227. [DOI] [PubMed] [Google Scholar]