Abstract
Recombination between coinfecting viruses had not been documented previously for a nonsegmented negative-strand RNA virus (mononegavirus). We investigated the potential of intermolecular recombination by respiratory syncytial virus (RSV) by coinfecting HEp-2 cells with two recombinant RSV (rRSV) mutants lacking either the G gene (ΔG/HEK) or the NS1 and NS2 genes (ΔNS1/2). These viruses replicate inefficiently and form pinpoint plaques in HEp-2 cells. Therefore, potential recombined viruses with a growth and/or plaque formation advantage should easily be identified and differentiated from the two parental viruses. Further identification of potential recombinants was aided by the inclusion of point mutation markers in the F and L genes of ΔG/HEK and the design of reverse transcription-PCR (RT-PCR) primers capable of detecting these markers. Independent coinfections and control single infections by these two rRSV mutants were performed. In one of six coinfections, an RSV variant was identified that produced plaques slightly larger than those of wild-type RSV in HEp-2 cells. RT-PCR and sequencing provided evidence that this variant was a recombined RSV (rec-RSV). The rec-RSV appeared to have been generated by a polymerase jump from the ΔG/HEK genome to that of ΔNS1/2 and back again in the vicinity of the SH-G-F genes. This apparently involved nonhomologous and homologous recombination events, respectively. The recombined genome was identical to that of the ΔG/HEK mutant except that all but the first 12 nucleotides of the SH gene were deleted and replaced by an insert consisting of the last 91 nucleotides of the G gene and its downstream intergenic region. This insert could have come only from the coinfecting ΔNS1/2 virus. This resulted in the formation of a short chimeric SH:G gene. Northern and Western blot analysis confirmed that the rec-RSV did not express the normal SH and G mRNAs and proteins but did express the aberrant SH:G mRNA. This provides an experimental demonstration of intermolecular recombination yielding a viable, helper-independent mononegavirus. However, the isolation of only a single rec-RSV under these optimized conditions supports the idea that RSV recombination is rare indeed.
RNA viruses maintain extensive genetic variability through which rapid evolution can occur. For example, this variability allows viruses to mitigate antiviral immune responses and to adapt to new hosts. RNA viruses generate this genetic diversity in a number of ways. The RNA-dependent RNA polymerase of RNA viruses is relatively error prone, by some estimates misincorporating once every 104 to 105 nucleotides (nt) (7). The resulting swarm of mutant genomes is referred to as a quasispecies and is thought to allow rapid adaptation of the virus to changes in its environment. Another method of maintaining genetic diversity is by nucleic acid exchange between genotypically different strains or variants during coinfection of the same host cell. The progeny genome formed as a result of this exchange contains elements of the two or more parental genomes. For segmented RNA viruses such as influenza virus and rotaviruses, the exchange of genetic material most commonly occurs by reassortment, which involves the exchange of entire gene segments, resulting in progeny viruses containing segments from the parental viruses (5, 12, 24). Of course, reassortment does not occur for nonsegmented genomes.
Another means of exchanging genetic information is by recombination, which involves the generation of chimeric molecules containing segments derived from more than one parental molecule. For RNA viruses, most (and possibly all) recombination events occur during RNA replication when the polymerase switches from one template (the donor) to another (the acceptor). This forms a chimeric progeny molecule containing elements derived from both parental templates (16, 26). Recombination is termed homologous if it involves donor and acceptor templates that share significant sequence relatedness, where base pairing likely guides the polymerase from one template to the next (11, 16). Homologous recombination can be “imprecise,” where the junction contains evidence of an insertion, deletion, or other mutation, or “precise,” where there is no evidence of such aberrations and the junction seems exact and clean. Recombination is termed nonhomologous if the donor and acceptor templates do not share significant sequence relatedness and hence base pairing apparently is not involved in the template switch.
Recombination is common for positive-sense RNA viruses such as picornaviruses (6), togaviruses (8), flaviviruses (9), retroviruses (13), and coronaviruses (11). Recombination also has been identified for influenza A virus (14) and Tula hantavirus (18), as examples of segmented negative-sense RNA viruses. However, recombination yielding viable progeny had not previously been documented experimentally for a nonsegmented negative-strand RNA virus (mononegavirus). Recombination leading to large deletions and the generation of defective interfering particles is very common for mononegaviruses, but these particles are not viable alone, likely do not represent an important reservoir for maintaining genetic diversity, and are not considered further here. In this study, we have examined the potential for intermolecular recombination during mixed infection in vitro by the mononegavirus human respiratory syncytial virus (RSV).
RSV is the prototype member of the genus Pneumovirus, of the family Paramyxovirus and the order Mononegavirales. The RSV strain A2 genome is a single negative-sense RNA of 15,222 nt (or 15,223 nt for the cDNA-derived version used here) that encodes 10 major subgenomic mRNAs and 11 viral proteins (Fig. 1). The minimum unit for RNA replication is composed of the viral genome encapsidated by the nucleocapsid N protein and associated with the phosphoprotein P and large polymerase subunit L, which together comprise the RSV replicase. Transcription requires in addition the M2-1 protein encoded by the upstream open reading frame of the M2 mRNA. The encapsidated genome serves as a template for transcription, in which the viral genes are copied into mRNAs, and for RNA replication, in which an exact positive-sense copy of the genome, or antigenome, is synthesized and encapsidated and serves in turn as the template for the synthesis of progeny genome.
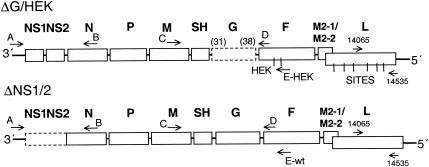
FIG. 1.
Gene maps of the two rRSV mutants, ΔG/HEK and ΔNS1/2, used for coinfection of HEp-2 cells. Genes are identified on top by the proteins which they encode: NS1 and NS2, nonstructural proteins 1 and 2; N, nucleocapsid protein; P, phosphoprotein; M, matrix protein; SH, small hydrophobic protein; G, attachment protein; F, fusion protein; M2-1, transcription antitermination factor; M2-2, RNA synthesis factor (M2-1 and M2-2 are encoded by two overlapping open reading frames in the same M2 mRNA); L, large polymerase protein. HEK and SITES are point mutations in the F and L genes, respectively, that are unique to the ΔG/HEK mutant (see Materials and Methods). Dashed boxes indicate the gene deletions of each rRSV mutant. The SH-F junction in ΔG/HEK contained the upstream 31 nt of the original SH-G junction fused to the downstream 38 nt of the original G-F junction (numbers in parentheses). In ΔNS1/2, the ATG of the N open reading frame was fused to the upstream nontranslated region of the NS1 gene. The placement of primers (A, B, C, D, E-HEK, E-wt, 14065, and 14535), used to differentiate the mutant constructs by RT-PCR (Table 1 and Materials and Methods), is indicated by arrows.
There were previously generated recombinant RSVs (rRSVs) that lack both of the nonstructural proteins NS1 and NS2 (ΔNS1/2) or lack the attachment protein G (ΔG/HEK) (22). (Note that the term “recombinant” in regard to rRSV refers to cDNA-derived virus, whereas the product of RNA recombination during mixed infection will be designated “recombined” RSV [rec-RSV].) Both of these rRSVs replicate similarly to wild-type (wt) rRSV (rA2) in Vero cells, whereas in HEp-2 cells they replicate much less efficiently and form small plaques. The growth restriction of these two viruses provided the possibility for recombination between the two genomes to generate chimeric progeny with improved growth, which would then be readily recovered and identified from the background of the two poorly growing parents. Indeed, this strategy provided identification of a chimeric rec-RSV generated by recombination between the two parents. To our knowledge, this study represents the first report of intermolecular recombination by a mononegavirus in an experimental setting.
MATERIALS AND METHODS
Construction of the ΔG/HEK and ΔNS1/2 RSV mutants.
The construction of ΔG/HEK has been described previously (22). Deletion of the G gene resulted in an SH-F intergenic region containing the 31 upstream nt from the SH-G intergenic region fused to the 38 downstream nt from the G-F intergenic region. In addition, this virus differs from its wt recombinant parent by the presence of six translationally silent restriction sites introduced into the L gene, collectively called the “restriction sites” (SITES) mutations. Specifically, these introduced sites are Bsu36I (nt position 9399), SnaBI (nt 11848), PmeI (nt 13342), RsrII (nt 14083), BstEII (nt 14318), and SnaBI (nt 14477) (25). There are also two amino acid changes, Lys-68-Glu and Gln-101-Pro, which were introduced in the F gene together with adjacent restriction site markers and are called the HEK mutations. These amino acid assignments match those found in a wt isolate of RSV that was used to derive a series of vaccine candidates and do not appreciably affect virus replication in vitro (25). To construct ΔNS1/2, the D51 plasmid containing the 5′ (leader complement) end of the antigenome through the SH gene end signal (21) was used as a template for PCR mutagenesis with Vent DNA polymerase (New England Biolabs) by the method of Byrappa et al. (3). This method involves the introduction of a mutation into an intact plasmid by PCR with two oligonucleotides that prime in opposite directions and produce a mutagenized linear plasmid, which is then self ligated and cloned. The 5′-phosphorylated primers used were as follows: forward primer, 5′-GCT CTT AGC AAA GTC AAG TTG AAT G-3′ (nt 1144 to 1168); reverse primer, 5′-CAT CTC TAA CCA AGG GAG TTA AAT TTA AGT GG-3′ (nt 101 to 70). The reverse primer contains the complement of the initiating methionine codon of NS1 (underlined) as well as the adjoining sequence in the NS1 untranslated region. The forward primer starts with the second codon of the N open reading frame, thus joining the NS1 untranslated region with the N open reading frame and resulting in deletion of both NS1 and NS2. The resulting PCR products were isolated by agarose gel electrophoresis, self ligated, and transformed into DH10B competent cells (Life Technologies). Structures were confirmed by restriction digestion, the structure of the AatII-AvrII fragment of D51ΔNS1/2 was confirmed by sequencing, and the fragment was then inserted into the same window of the full-length antigenome clone (D53) (4). The encoded rRSV lacking both NS1 and NS2 (ΔNS1/2) was recovered essentially as described previously (4, 20). The ΔG/HEK and ΔNS1/2 viruses were propagated in Vero cells, and viral titers were determined by plaque assay under methylcellulose on Vero cells with viral plaques being visualized by immunostaining with murine anti-F monoclonal antibodies followed by horseradish peroxidase-coupled anti-mouse immunoglobulin G antibodies and 4CN substrate (Kierkegaard & Perry Laboratories) as described previously (15).
Coinfection in HEp-2 cells.
Monolayers of HEp-2 cells at approximately 80% confluence in T-25 flasks were inoculated with either ΔNS1/2, ΔG/HEK, or both at a multiplicity of infection (MOI) of 2 PFU/cell for each virus. Six independent coinfections and duplicate single-virus infections were established. Virus was allowed to adsorb for 3 h at 37°C, after which the viral suspensions were replaced with 4 ml of Opti-MEM containing 4% fetal bovine serum (FBS). The 4-ml supernatant was harvested from each flask and replaced with fresh medium every 24 h for 7 days. The supernatants were clarified by centrifugation, snap-frozen on dry ice, and stored at −70°C for plaque analysis. Aliquots (600 μl) of each viral supernatant collected were analyzed by plaque assay as described above in both HEp-2 and Vero cells in six-well plates to assess plaque size and morphology. Viral RNA was extracted from a portion (400 μl) of each supernatant collected on day 4 and analyzed by reverse transcription-PCR (RT-PCR) with discriminatory primers (see below) to confirm the growth of the parental mutants in each coinfection and single infection.
Plaque purification and growth of rec-RSV.
Large wt plaques were observed in HEp-2 cells infected with supernatants collected from days 4 to 7 of coinfection 1. Individual viral plaques were isolated as follows: medium supernatants were adsorbed undiluted onto HEp-2 cell monolayers in six-well plates. The cells were incubated at 37°C for 2 h. The inocula were replaced with 3 ml of 0.8% agar (SeaKem) in modified L15 medium (Biowhittaker). The cells were incubated at 37°C for 6 days, until large plaques were seen in the cells infected with the coinfection 1 supernatant. The agar-medium was then overlaid with 1.5 ml of 0.8% agar-medium containing 5% neutral red and incubated for an additional 24 h. Seven large plaques were picked from coinfection 1 and transferred to 2 ml of Opti-MEM containing 1× SPG (0.218 M sucrose, 3.8 mM KH2PO4, 7.2 mM K2HPO4, 5.4 mM sodium glutamate). One milliliter was used to infect a fresh monolayer of HEp-2 cells, which was overlaid with agar-medium and treated in the same manner for plaque purification. The remaining 1 ml was snap-frozen and stored at −70°C. As a control, pinpoint plaques from coinfections 2 and 3 were picked and processed in parallel.
In this way, plaques were picked and passaged through HEp-2 cells three times. After the third round of purification, the virus from the agar-medium plug suspended in Opti-MEM-SPG was used to infect monolayers of HEp-2 cells to make small virus stocks. The rec-RSV isolates were designated rec-RSV 1-1 to 1-7. rec-RSV 1-1 and 1-2 were amplified further in Vero cells to prepare virus stocks that were used for the experiments shown in the figures.
Viral RNA extraction, RT-PCR, and sequencing.
Viral RNA was extracted from the supernatant with the QIAamp viral RNA minikit (Qiagen). The RNA was used as a template for reverse transcription with Superscript II and random hexamers (Invitrogen). PCR was performed using specific forward (positive-sense) and reverse (negative-sense) primers listed below and the Advantage cDNA polymerase mix (Clontech catalog no. 8417-1). The numbers in parentheses following each primer indicate its position in the complete 15,2223-nt sequence of wt rRSV.
The following set of primers was used to distinguish between the two parental viruses based on the presence or absence of the NS1 and NS2 genes, the G gene, and the HEK markers in the F gene (Fig. 1 and Table 1): A (forward), 5′ GAAAAAATGCGTACAACAAACTTGC 3′ (nt 5 to 29); B (reverse), 5′ TTATGGTGTCTTCTCTTCCTAACC 3′ (nt 1381 to 1358); C (forward), 5′ ACAAATTGGAAGCACACAGC 3′ (nt 3979 to 3998); D (reverse), 5′ TACCAACCAGTTCTCAGAGC 3′ (nt 5819 to 5800); E-wt (reverse), 5′ TTGTTGCTTGTGTGCTTTGC 3′ (nt 5971 to 5952); E-HEK (reverse), 5′ TTGTTGCTGGAGTACTTTGC 3′ (nt 5971 to 5952). Primer pairs A-B and C-D distinguished between the presence and absence of the NS1 and NS2 genes or the G gene, respectively, based on the size of the RT-PCR product (Fig. 1; Table 1). Primer pairs C-E-wt and C-E-HEK detected the presence or absence, respectively, of three nucleotide substitutions at positions 5963, 5966, and 5968 that result in the HEK point mutation Gln-101-Pro in F and the insertion of a nearby marker restriction site (Fig. 1; Table 1) (25). PCR was performed by 25 cycles of denaturation (88°C for 30 s), annealing (62°C for 30 s), and extension (68 or 72°C for 3 min), followed by a final 3-min extension at 68 or 72°C.
TABLE 1.
Primer pairs used to differentiate between the ΔG/HEK and ΔNS1/2 viruses based on product size and the presence or absence of HEK point mutations in the F gene
| Forward primera | Reverse primera | Virus | Product sizeb (bp) |
|---|---|---|---|
| A | B | wt rA2 | 1,327 |
| ΔG/HEK | 1,327 | ||
| ΔNS1/2 | 273 | ||
| C | D | wt rA2 | 1,801 |
| ΔG/HEK | 852 | ||
| ΔNS1/2 | 1,801 | ||
| C | E-wt | wt rA2 | 1,954 |
| ΔG/HEK | None | ||
| ΔNS1/2 | 1,954 | ||
| C | E-HEK | wt rA2/HEK | 1,963 |
| ΔG/HEK | 1,014 | ||
| ΔNS1/2 | None | ||
| 14065 | 14535 | All above | 521 |
See Materials and Methods for exact sequence positions and Fig. 1 for general location in the RSV gene map.
Primer pairs A-B and C-D differentiate between ΔG/HEK and ΔNS1/2 based on product size. Primer pairs C-E-wt and C-E-HEK prime only on wt F sequence (found in ΔNS1/2) and HEK F sequence (found in ΔG/HEK), respectively. Primer pair 14065-14535 primes on both wt L sequence (found in ΔNS1/2) and SITES L sequence (found in ΔG/HEK), which are then differentiated by nucleotide sequencing.
To determine the presence of the SITES mutations in the L gene, genomic RNA was subjected to RT-PCR with the random hexamer-derived cDNA described above and the following two primers: 14065 forward, 5′ GTATAGCATTCATAGGTGAAGGAGC 3′ (nt 14041 to 14065); 14535 reverse, 5′ ATTAAATACTGGGAATATATTCGCAGG 3′ (nt 14561 to 14535).
These two oligonucleotides prime on both the wt and SITES version of the L gene, yielding a 521-bp product that contains the positions of three of the six SITES mutations: this uncloned cDNA was then analyzed by sequencing with the Applied Biosystems Big Dye system (Perkin-Elmer). Additional primer pairs used to generate RT-PCR products for nucleotide sequencing are described in the Results.
In vitro growth analysis.
HEp-2 and Vero cell monolayers in six-well plates were infected in triplicate with either rA2, ΔG/HEK, ΔNS1/2, rec-RSV 1-1, or rec-RSV 1-2 at an MOI of 0.01. Live virus was diluted in Opti-MEM-2% FBS. After 3 h of adsorption at 37°C, the cells were washed three times with Opti-MEM and then 2 ml of Opti-MEM with 2% FBS was added. The entire 2-ml supernatant was removed and replaced with fresh medium every 24 h for 7 days. Supernatants were clarified by centrifugation, snap-frozen, and stored at −70°C for titration. Viral titers were determined by plaque assay in Vero cells with methylcellulose overlay and immunostaining (15).
Northern blot analysis.
Confluent Vero cells in six-well plates were infected with either rA2, ΔG/HEK, ΔNS1/2, rec-RSV 1-1, or rec-RSV 1-2 at an MOI of 3 or were left uninfected. Cells were harvested 24 h later, and half of each cell preparation was used for Northern blot analysis, while the other half was used for Western blot analysis (see below). Total RNA was extracted using Trizol-LS reagent (Invitrogen) and the RNeasy minikit (Qiagen). RNA was electrophoresed through a 1% agarose gel containing formaldehyde, transferred to a nitrocellulose membrane (Optitran, 0.2-μm pore size; Schleicher & Schuell), and hybridized with 32P-labeled double-stranded DNA (dsDNA) probes specific for G, SH, P, or N mRNA.
Western blot analysis.
Cell pellets were disrupted with 2× sample buffer (100 mM Tris-Cl [pH 6.8], 4% sodium dodecyl sulfate, 20% glycerol, 0.2% bromophenol blue, 200 mM dithiothreitol) and centrifuged through Qiashredders (Qiagen). Five microliters (approximately 7.5 × 105 cell equivalents) of each sample was electrophoresed through sodium dodecyl sulfate-4 to 20% polyacrylamide gels (Novex) and transferred to a nitrocellulose membrane. Membranes were incubated with rabbit polyclonal antisera to purified RSV or a peptide representing amino acids 186 to 201 of the RSV G protein or amino acids 53 to 64 of the RSV SH protein. Bound antibodies were visualized by incubation with horseradish peroxidase-coupled goat anti-rabbit immunoglobulin G antibodies and chemiluminescence (Amersham).
RESULTS
Strategy for identifying and isolating potential rec-RSV.
Intermolecular recombination during coinfection had not been documented previously for a mononegavirus, suggesting that it is at best a rare event. To facilitate the detection of possible rare recombinants in an experimental setting, we chose to “cross” two RSV gene deletion mutants, namely, ΔNS1/2, which lacks the NS1 and NS2 genes, and ΔG/HEK, which lacks the G gene. These viruses replicate well in Vero cells, whereas in HEp-2 cells they replicate inefficiently and form pinpoint plaques. The NS1 and NS2 genes are the first two genes in the gene order while the G gene is the seventh gene (Fig. 1): the rationale was that recombination in the intervening region might restore the full set of genes and produce a rec-RSV with improved replication efficiency that could be readily recovered and identified from the background of debilitated parental deletion viruses. Another consideration in this strategy was that the NS1, NS2, SH, and G genes, being nonessential for RSV replication in vitro, provided regions where nucleotide insertions or deletions that might arise due to imprecise recombination might be tolerated. In addition, the ΔG/HEK virus had been engineered to contain point mutations resulting in two amino acid substitutions in the F gene (called the HEK mutations), as well as translationally silent nucleotide substitutions in the L gene that resulted in the insertion of six restriction site markers (called the SITES mutations). The presence of these markers would discriminate between the two parental viruses with regard to the downstream F and L genes. Apart from these differences, the backbones of the ΔG/HEK and ΔNS1/2 viruses were identical. Thus, we infected six independent flasks of HEp-2 cells with the two viruses together, each at an input MOI of 2 PFU per cell, and harvested supernatants from infected cells every 24 h for 7 days. As controls, HEp-2 cells were infected at the same MOI with each of the parental viruses separately. We then subjected the supernatants to plaque analysis to screen for potential recombined viruses.
Plaque formation by mutant rRSVs.
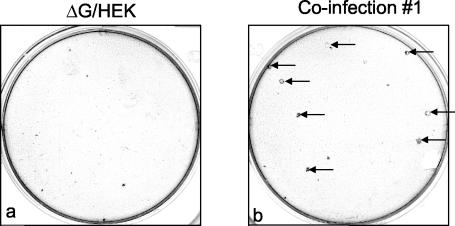
As expected, the ΔNS1/2 and ΔG/HEK single infections yielded viral supernatants that generated pinpoint plaques on HEp-2 cells and larger, more distinct plaques on Vero cells, as is characteristic of these deletion viruses (Fig. 2 and 3). In addition, extensive plaque analysis did not reveal any evidence of large-plaque variants in the ΔG/HEK and ΔNS1/2 virus stocks used for the infections (data not shown). Similarly, only pinpoint plaques were observed in HEp-2 cells infected by supernatants from five of the six coinfections (data not shown). However, analysis of supernatants collected on days 4 to 7 from coinfection 1 revealed, against the background of pinpoint plaques, a small number of large plaques that were somewhat greater in size than those of wt rRSV strain A2 (wt rA2) (Fig. 2b). When HEp-2 cells were exposed to 600 μl of viral supernatant collected on day 4 of coinfection 1, containing approximately 108,000 PFU, eight of these formed large plaques (Fig. 2b). Seven of these large plaques were picked, plaque purified three times in HEp-2 cells, and designated rec-RSV isolates 1-1 to 1-7. As a control, several small plaques also were picked and purified from the day 4 supernatants from coinfections 2 and 3: these retained their small-plaque phenotype throughout the plaque purification steps. Thus, the plaque morphology phenotypes of the parent mutant viruses and the rec-RSV were preserved.
FIG. 2.
Comparison of plaque size between ΔG/HEK and rec-RSV present in the harvest from coinfection 1, one of six coinfections. Plaque formation was detected by immunostaining as described in Materials and Methods after incubation under methylcellulose for 6 days. Note the pinpoint plaques formed in the ΔG/HEK infection (a) and the large-plaque variants observed in coinfection 1 (b; arrows) against the background of pinpoint plaques. Large plaques were not observed in the other coinfections or in any single infection involving ΔG/HEK, an example of which is shown, or ΔNS1/2 (data not shown).
FIG. 3.
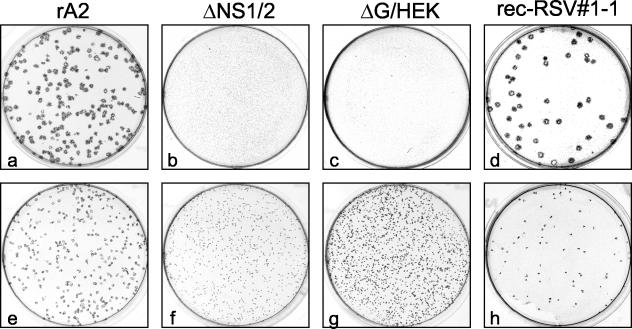
Photomicrographs of plaque formation in HEp-2 (a to d) and Vero (e to h) cells infected with rA2 (a and e), ΔNS1/2 (b and f), ΔG/HEK (c and g), or plaque-purified rec-RSV 1-1 (d and h) derived from one of the large plaques identified from the coinfection 1 progeny. Infected cells were incubated for 6 days under methylcellulose, and the plaques were visualized by immunostaining with a cocktail of anti-F monoclonal antibodies.
Following plaque purification, rec-RSV isolates 1-1 and 1-2 were used to infect both HEp-2 and Vero cell monolayers, and plaque formation 6 days postinfection was compared to that of wt rA2, ΔG/HEK, and ΔNS1/2 (Fig. 3). In HEp-2 cells, the plaques of rec-RSV were approximately 45% larger than those of wt rA2 and were easily differentiated from the pinpoint plaques of ΔG/HEK and ΔNS1/2 (Fig. 3a to d). In Vero cells, plaques formed by wt rA2 were slightly larger than those formed by the purified rec-RSV, and plaques formed by wt rA2 or rec-RSV were larger than those of ΔG/HEK and ΔNS1/2 (Fig. 3e to h). It should be noted that, in Fig. 3, the viruses that form larger plaques were infected at a lower MOI, and therefore, the differences in plaque number are not a reflection of the plaquing efficiency of each of the viruses. Only rec-RSV 1-1 is shown in Fig. 3: rec-RSV 1-2 yielded the same results (data not shown).
RT-PCR of plaque-purified viruses.
Primer pairs A-B and C-D (Fig. 1 and Table 1) were designed to span the NS1-NS2 genes and the SH-G genes, respectively, and thus differentiate between the ΔNS1/2 and ΔG/HEK parental mutants based on the size of the RT-PCR product. Primer pairs C-E-wt and C-E-HEK (Fig. 1 and Table 1) were designed to differentiate between viruses bearing the wt F gene and those bearing the F gene containing the HEK mutation encoding the Gln-101-Pro substitution.
RT-PCR of intracellular RNA from cells infected with wt rA2/HEK (a version of wt rA2 containing the HEK and SITES mutations but otherwise identical), wt rA2, and ΔG/HEK with primer pair A-B identified the presence of the NS1 and NS2 genes (1,327-bp product, Fig. 4a, lanes 1, 2, and 4) and the absence of these genes from ΔNS1/2 (273-bp product, Fig. 4a, lane 3). RT-PCR of wt rA2/HEK, wt rA2, and ΔNS1/2 with primer pair C-D yielded in each case an 1,801-bp product (Fig. 4b, lanes 1, 2, and 3, respectively), consistent with the expected presence of intact SH and G genes. In contrast, RT-PCR of ΔG/HEK with primer pair C-D yielded the expected 852-bp product (Fig. 4b, lane 4), consistent with the known deletion.
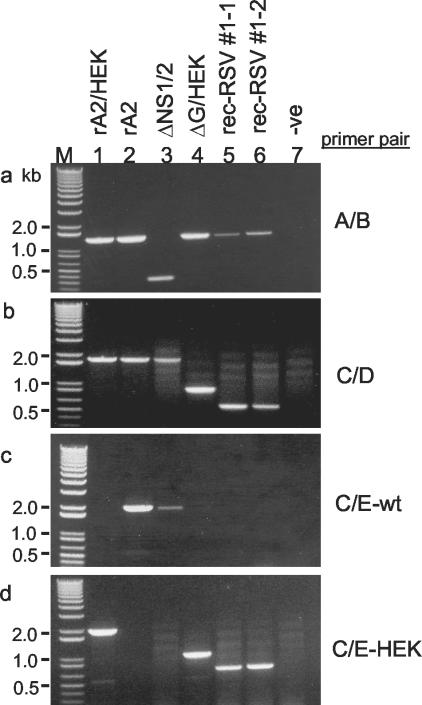
FIG. 4.
RT-PCR analysis of genomic RNA of parental viruses and rec-RSV from total intracellular infected-cell RNA. Primer pairs A-B (a), C-D (b), C-E-wt (c), and C-E-HEK (d) (Table 1 and Fig. 1) were used to identify gene deletions and the presence or absence of HEK point mutations in rA2/HEK (lane 1), wt rA2 (lane 2), ΔNS1/2 (lane 3), ΔG/HEK (lane 4), plaque-purified rec-RSV 1-1 (lane 5), and rec-RSV 1-2 (lane 6). Note that rA2/HEK and wt rA2 are two versions of wt rRSV that differ only by the presence of the HEK and SITES mutations in the former. M, DNA ladder; -ve (lane 7), a negative control lacking template. Primer pair A-B detects the presence or absence of NS1 and NS2 based on product size. Primer pair C-D detects the presence or absence of the G gene based on product size. Primer pairs C-E-wt and C-E-HEK also detect the presence of the G gene by product size and, in addition, are specific to wt and HEK sequence, respectively, in the F gene.
RT-PCR with primer pair C-E-wt yielded the expected 1,954-bp product in the case of wt rA2 and ΔNS1/2 (Fig. 4c, lanes 2 and 3) but not wt rA2/HEK or ΔG/HEK (Fig. 4c, lanes 1 and 4), confirming the specificity of this primer pair for the wt F gene. Conversely, primer pair C-E-HEK yielded the expected products for wt rA2/HEK and ΔG/HEK (Fig. 4d, lanes 1 and 4) and did not yield a product for wt rA2 or ΔNS1/2 (Fig. 4d, lanes 2 and 3), confirming its specificity for the F gene containing the Gln-101-Pro HEK mutation.
With their specificities confirmed, these primer sets were used to analyze the supernatants resulting from the six coinfections. RT-PCR analysis with primer pairs A-B (spanning NS1-NS2) and C-D (spanning SH-G) showed that both ΔNS1/2 and ΔG/HEK were present in all supernatants harvested on day 4, confirming that both parental viruses had replicated during the coinfections (data not shown). However, potential recombined viral genomes were not evident from analysis of the total supernatants.
Next, rec-RSV isolates 1-1 to 1-7 were analyzed by RT-PCR to determine their genotype. RT-PCR with primer pair A-B showed that the NS1 and NS2 genes were present in the seven isolates of rec-RSV as evidenced by the 1,327-bp product (Fig. 4a, lanes 5 and 6, and data not shown). Surprisingly, analysis of each of the seven isolates with primer pair C-D (spanning SH-G) yielded a product of ∼500 bp (Fig. 4b, lanes 5 and 6, and data not shown), which was smaller than either the 1,801-bp product derived from wt rA2 or the 852-bp product derived from ΔG/HEK. This suggested that the rec-RSV isolates had sustained an additional deletion in the SH-F region. RT-PCR with primer pairs C-E-wt and C-E-HEK showed that the seven rec-RSV isolates each contained the HEK point mutation of the ΔG/HEK parent, as evidenced by the 1,954-bp band present in Fig. 4d, lanes 5 and 6, and absent in Fig. 4c, lanes 5 and 6. Thus, these analyses indicated that the seven rec-RSV isolates were identical and contained the NS1, NS2, and F genes of the ΔG/HEK parent but contained a large deletion in the SH-G region represented by primer pair C-D.
Sequence analysis of selected regions of the rec-RSV isolates.
As described above, RT-PCR of the rec-RSV isolates with primer pair C-D (spanning SH-G) yielded an unexpectedly small product of ∼500 bp, suggesting that this region contained a large deletion. Thus, the ∼500-bp RT-PCR products from each of the seven rec-RSV isolates were sequenced; sequencing was performed directly on the uncloned material and yielded the same sequence for each isolate. This sequence showed that rec-RSV contained a large deletion in the SH gene, leaving only the first 12 nt of the gene including the gene start signal (Fig. 5a). In addition, immediately downstream of the truncated SH gene, rec-RSV contained an insertion of 91 nt of G, including the gene end signal. This in turn was followed by the complete 52-nt G-F intergenic region, of which only the downstream 38 nt were present in ΔG/HEK (Fig. 5a). In addition, the 12-nt segment from the SH gene and the 91-nt segment from the G gene were separated by a CA dinucleotide that could have been derived from either the SH or the G gene. This resulted in a 105-nt chimeric SH:G gene that contained an intact set of gene start and gene end transcription signals flanked by intact intergenic regions, thus encoding a short RNA; however, the RNA lacks a significant open reading frame. To confirm these results, an independent primer pair spanning the same general region (nt 4004 to 6094) was used to generate an RT-PCR product for sequencing, which produced the same results as described above.
FIG. 5.
Structure of rec-RSV. (a) Gene maps of wt rA2 and rec-RSV. The sequence of the chimeric SH:G gene resulting from the intracellular recombination event is shown in the box: italicized nucleotides (total of 12) were derived from the SH gene, nucleotides in regular text (total of 91) were derived from the G gene, and the intervening CA dinucleotide in boldface is the recombination junction, is present in both genes, and could be derived from either. (b) Alignment of the sequences on either side of the recombination junction in the SH:G gene. The top sequence is that of the parental ΔG/HEK virus, beginning at nt 4222 and showing the upstream end of the SH gene with the CA junctional dinucleotide in boldface; the bottom sequence is that of the parental ΔNS1/2 virus, beginning at nt 5495 and showing the downstream end of the G gene with the CA junctional dinucleotide in boldface; and the middle sequence is that of rec-RSV. This shows that the CA dinucleotide is one of the few scattered points of sequence identity in this region between the two parental templates. (c) Gene maps of the parental ΔG/HEK and ΔNS1/2 viruses showing the deduced path taken by the polymerase in jumping from the ΔG/HEK genome to that of ΔNS1/2 and back again (see the text). The upstream recombination junction was identified by the sequence analysis in panel a; the position of the downstream junction is unknown but, given the presence of a wt-like G-F junction and HEK markers in rec-RSV, presumably occurred at some point within the downstream 38 nt of the intergenic region preceding the F gene or within the F gene upstream of the first HEK marker (dotted lines) and occurred without any sequence insertion or deletion.
The chimeric SH:G gene apparently represented a recombination junction, with the upstream sequence derived from ΔG/HEK and the downstream sequence derived from ΔNS1/2. It was of interest to determine the extent of sequence relatedness between the two templates in the region of recombination. This is shown in Fig. 5b, in which the top line is the sequence of ΔG/HEK in the vicinity of the upstream end of the SH gene, the third line is that of ΔNS1/2 in the vicinity of the downstream end of the G gene, and the middle line is that of rec-RSV. This comparison shows that, apart from the shared CA dinucleotide, there is no significant sequence identity between the two sequence segments, indicating that a nonhomologous recombination event was involved.
The presence of the NS1 and NS2 genes and the upstream end of the SH gene in rec-RSV suggested that all of the sequence upstream of the junction was derived from the ΔG/HEK parent, while the downstream end of G and the adjoining intergenic sequence were derived from ΔNS1/2. However, as described above, RT-PCR with the C-E-wt and C-E-HEK primer pairs showed that the rec-RSV isolates also contained the HEK mutation in the F gene (Fig. 4c and d). This suggested that this region of the rec-RSV isolates also was derived from ΔG/HEK rather than ΔNS1/2. This would mean that a second recombination event had occurred, with its junction located somewhere between the G-F intergenic region and the second HEK mutation. To investigate this, RT-PCR was used to amplify nt 4384 to 4894 of the rec-RSV isolates, which were then sequenced. The sequence showed that the F gene contained both of the HEK markers of the ΔG/HEK parent and did not otherwise contain any nucleotide changes. Confirmation of the presence of the HEK markers supported the idea that a second recombination had occurred. This event must have occurred upstream of the HEK markers, within either the downstream 38 nt of the G-F intergenic region or the upstream 198 nt of the F gene. The lack of nucleotide changes indicated that this recombination event was homologous and precise.
The presence of the SITES markers in the L gene of the ΔG/HEK parent made it possible to determine the origin of the L gene present in the rec-RSV isolates. The seven isolates were subjected to RT-PCR to amplify part of the L gene that, in ΔG/HEK, contains three of the six SITES mutations. The primers were specific for sequence not involved in the SITES mutations (Materials and Methods) and thus would anneal to either wt or SITES L sequence. The resulting RT-PCR products were sequenced, showing that each of the seven isolates was identical and contained three SITES mutations, RsrII (nt 14083), BstEII (nt 14318), and SnaBI (nt 14477), unique to the ΔG/HEK mutant parent. The proposed recombination pathway deduced from these results is shown in Fig. 5c.
Growth of rec-RSV in HEp-2 and Vero cells.
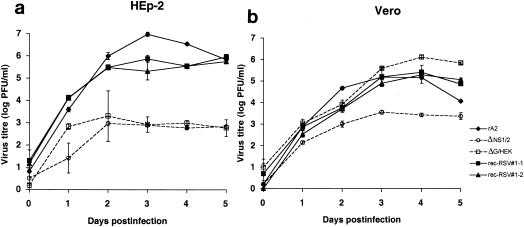
The growth kinetics of rec-RSV 1-1 and 1-2 in HEp-2 and Vero cells were compared to those of the parental and wt viruses. Triplicate cultures of both cell lines were infected with either rec-RSV 1-1 or 1-2 or with wt rA2, ΔG/HEK, or ΔNS1/2 at an MOI of 0.01. Viral supernatants were harvested daily for 5 days, and the resulting viral titers were determined (Fig. 6).
FIG. 6.
Growth kinetics of plaque-purified rec-RSV isolates 1-1 and 1-2 compared to wt rA2, ΔNS1/2, and ΔG/HEK. Triplicate cultures of HEp-2 (a) or Vero (b) cells were infected at an MOI of 0.01 PFU/cell. Supernatants were harvested daily and analyzed by plaque assay on Vero cells. Plaques were visualized by immunostaining with RSV F-specific antibodies. Mean titers (log10 PFU per milliliter) are shown with twice the standard error on either side of the mean.
In both HEp-2 and Vero cells, the two rec-RSVs displayed the same growth patterns. In HEp-2 cells, the titer of wt rA2 was significantly higher than that of rec-RSV from days 2 to 4, but by day 5 the two viruses were similar (Fig. 6a). In contrast, both the parental ΔNS1/2 and ΔG/HEK viruses grew more slowly and to approximately 1,000-fold-lower final titers than did wt rA2 and the rec-RSVs. In Vero cells, the growth kinetics of rA2 and the rec-RSVs were very similar (Fig. 6b). The kinetics of ΔG/HEK production in Vero cells was similar to that of the rec-RSVs and rA2 up to day 4, but thereafter the titer appeared to be significantly higher than that for the other viruses (Fig. 6b). This likely reflects the extensive cytopathology and reduction of viable cells observed for rA2 and rec-RSV infection by days 4 to 5 rather than a growth advantage for ΔG/HEK. ΔNS1/2 grew less efficiently than the other viruses in Vero cells, reaching final titers ∼100-fold less than those of rA2 (Fig. 6b).
Viral RNA expression by rec-RSV.
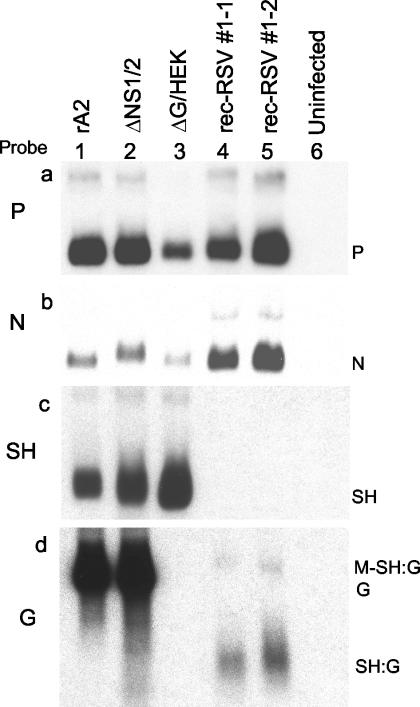
Total cellular RNA was extracted from Vero cells that had been infected by wt rA2, ΔG/HEK, ΔNS1/2, rec-RSV 1-1, or rec-RSV 1-2 or had been left uninfected. Northern blot hybridization was performed using dsDNA probes specific for the RSV N, P, SH, and G genes. All viruses including the rec-RSV expressed P and N mRNA, as expected (Fig. 7a and b). Monocistronic SH mRNA was expressed by wt rA2, ΔNS1/2, and ΔG/HEK, as expected, but not by the rec-RSV (Fig. 7c, lanes 1, 2, and 3 versus lanes 4 and 5). Full-length G mRNA was expressed by wt rA2 and ΔNS1/2 but not by either ΔG/HEK, as expected, or rec-RSV (Fig. 7d, lanes 1 and 2 versus lanes 3, 4, and 5). A small mRNA of a size appropriate to represent the chimeric SH:G gene was detected for rec-RSV by using the G-specific probe (Fig. 7d, lanes 4 and 5). The failure of the SH probe to detect this mRNA was expected, since it contains only 14 nt related to the SH gene (Fig. 5c). In addition, the RNA from rec-RSV contained a small amount of an mRNA that was marginally larger than full-length G mRNA and was of the appropriate size to be a readthrough of the M and novel SH:G genes (Fig. 7d, lanes 4 and 5).
FIG. 7.
Northern blot hybridization of total intracellular RNA isolated 24 h postinfection from Vero cells that were infected with either wt rA2 (lane 1), ΔNS1/2 (lane 2), ΔG/HEK (lane 3), plaque-purified rec-RSV 1-1 (lane 4), or plaque-purified rec-RSV 1-2 (lane 5) or were mock infected (lane 6). The RNAs were subjected to electrophoresis on formaldehyde gels, transferred to nitrocellulose, and hybridized to 32P-labeled dsDNA probes for the P (a), N (b), SH (c), or G (d) gene. The positions of monocistronic P, N, SH, and G and the putative SH:G mRNA are indicated: the faint upper band in lanes 4 and 5 is of the appropriate size to be a readthrough of the M and SH:G genes.
Protein synthesis by rec-RSV.
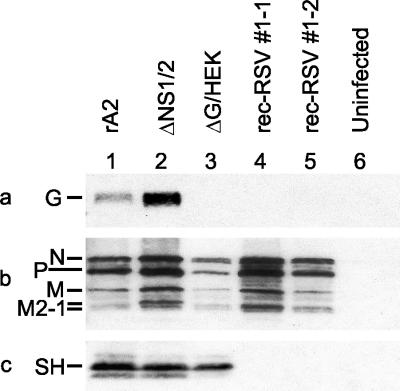
Further confirmation of the absence of functional SH and G genes from rec-RSV was provided by Western blot analysis. A portion of the cell pellets used to extract total RNA for Northern blot analysis was used for Western blot analysis. Expression of viral proteins was determined by Western blot analysis of total cellular proteins from the infected cells by using polyclonal antisera directed against either purified RSV (Fig. 8b) or peptides derived from the SH protein (Fig. 8c) or the G protein (Fig. 8a). Cells infected by any of the viruses expressed the N, P, M, and M2-1 proteins, as expected (Fig. 8b). Both wt rA2 and ΔNS1/2 expressed both G and SH (Fig. 8a and c, lanes 1 and 2). As expected, the ΔG/HEK virus-infected cells did not contain G (Fig. 8a, lane 3) but retained SH expression (Fig. 8c, lane 3). However, neither of the rec-RSV isolates directed the expression of G or SH (Fig. 8a and c, lanes 4 and 5), consistent with the absence of either functional gene in the recombined viruses.
FIG. 8.
Western blot analysis of cell-associated proteins from Vero cells that were infected with either wt rA2 (lane 1), ΔNS1/2 (lane 2), ΔG/HEK (lane 3), plaque-purified rec-RSV 1-1 (lane 4), or plaque-purified rec-RSV 1-2 (lane 5) or were mock infected (lane 6). Western blot analysis was performed using antisera against a peptide representing amino acids 186 to 201 of the G protein (a), purified RSV (b), or a peptide representing amino acids 53 to 64 of the SH protein (c). The relevant cropped portions of blots are shown.
DISCUSSION
Although recombination between the genomes of positive-sense RNA viruses is commonplace, the ability of mononegaviruses to undergo genetic recombination had not been previously described. Therefore, we designed experimental conditions to facilitate the detection of possible recombination events. To do this, we performed coinfection with two mutant RSVs that replicate inefficiently and form small plaques in HEp-2 cells due to deletion of the NS1 and NS2 genes (ΔNS1/2) or the G gene (ΔG/HEK). The NS1/2 and G gene deletions are separated in the RSV genome by more than 3,500 nt, providing the possibility that recombination might occur within this region and yield a virus with a complete set of genes that would grow efficiently and be readily isolated from the background of debilitated parental mutant viruses. Furthermore, the fact that the NS1, NS2, SH, and G genes are not essential for RSV replication in vitro meant that imprecise recombination that might occur would be tolerated within these genes. The fact that each of two parents replicated poorly in HEp-2 cells facilitated the identification of a virus that replicated efficiently since it had the potential to outgrow either parent and be detectable in their presence. Indeed, this strategy resulted in the identification of a rec-RSV. Unexpectedly, however, while this virus did exhibit a growth advantage over its parents, it was not due to the reacquisition of a deleted gene through recombination. Also, the recovered rec-RSV appeared to be the product of two recombination events, rather than one.
One of six coinfections with ΔG/HEK and ΔNS1/2 yielded rec-RSV with a growth advantage and the ability to form large plaques compared to the two parental deletion viruses. Seven isolates recovered from this one coinfection appeared to be identical, suggesting that all of the rec-RSVs from this experiment arose from a single recombined genome. The structure of the rec-RSV was investigated by identifying genetic markers, specifically the presence or absence of the NS1 and NS2 or G genes as well as the HEK and SITES markers in the F and L genes, respectively, of the ΔG/HEK parent. This, together with nucleotide sequence analysis, provided evidence that most of the genome of the rec-RSV was derived from the ΔG/HEK parent and that two recombination events were involved.
One recombination junction was identified by the finding that the first 12 nt of the SH gene of rec-RSV were fused to the last 91 nt of the G gene, the latter of which could have come only from the ΔNS1/2 parent. These two segments were separated by a CA dinucleotide that was represented in both the SH and G genes, which otherwise did not exhibit significant sequence relatedness, and thus might have been derived from either gene. It seems reasonable to suggest that, while base pairing evidently was not involved in aligning the two templates for recombination, this “homologous” dinucleotide probably helped guide the nascent strand to the new template. Thus, this recombination event fits the general category of “nonhomologous” (11), with the caveat that base pairing might have been required for reinitiation of polymerization. It also might be that the two templates were held in juxtaposition by base pairing elsewhere along the two strands, even if base pairing was not involved in the vicinity of the recombination.
The second recombination junction occurred within the last 38 nt of the G-F junction or the first 198 nt of the F gene. This was deduced from the finding that the rec-RSV contained (i) the complete 52-nt G-F intergenic region of wt RSV, which was present in its entirety in the ΔNS1/2 parent whereas only the downstream 38 nt were present in ΔG/HEK, and (ii) both HEK markers from ΔG/HEK. However, the exact junction point within this region could not be deduced because there were no sequence abnormalities. Thus, this recombination step appeared to be homologous and precise. Recombination of this type has not been described previously for a negative-strand RNA virus and would be difficult to detect without the use of multiple markers such as those used in the present study.
It seems likely that recombination took place during the synthesis of antigenomic RNA from genomic templates, since the latter are much more abundant in the infected cell, a factor that would increase the efficiency and probability of the event. If so, then the likely recombination pathway was that (i) synthesis began in the leader region of the ΔG/HEK genome and proceeded to the SH gene; (ii) the polymerase and nascent antigenome made a “nonhomologous,” imprecise jump from the upstream region of the SH gene of ΔG/HEK to the downstream region of the G gene of ΔNS1/2, where copying resumed across the end of the G gene, into the intergenic region and, possibly, the F gene; and (iii) the polymerase and nascent strand made a “homologous,” precise jump back to the ΔG/HEK strand and completed synthesis on that template (Fig. 5c).
For a recombination event to be considered as resulting in a recombined, viable virus, it must be sustained in subsequent generations and not lost through reversion to the principal parental genome (23). We have demonstrated that rec-RSV containing the SH:G chimeric gene and the other markers was maintained during rounds of plaque purification and amplification of the virus by serial passage in Vero cells. Northern and Western analysis showed that rec-RSV replicated efficiently despite the elimination of both G and SH proteins. Sequence information showed that there was no reversion to the ΔG/HEK genome. Therefore, the formation of a chimeric SH:G gene product is a stable recombination event and rec-RSV is a viable recombined virus.
Interestingly, the chimeric SH:G gene identified in rec-RSV is similar to that described by Karron et al. (10) for a cold-passaged, attenuated RSV subgroup B mutant called cp-52. The chimeric gene of both viruses has an SH gene start signal, a G gene end signal, and proportionally more of the G gene represented than the SH gene. The chimeric genes of the two viruses are of similar length, 91 bases for cp-52 and 105 bases for rec-RSV. The predicted chimeric mRNA is expressed in each case and in each case lacks a significant open reading frame. The recombination mechanism by which the deletion in the cp-52 genome occurred is not known, but the recombination probably also occurred during antigenome synthesis (for reasons of template abundance and probability, as discussed above) and presumably involved nonhomologous jumping from the upstream end of the SH of one template to the downstream end of the G gene of either the same template or a different template. The involvement of the SH-G region in recombination in two studies raises the possibility that it might be a “hot spot” for recombination, although this is only speculation at the present time. The rec-RSV contained intact NS1 and NS2 genes and lacked functional SH and G genes, and its genome was 1,274 nt shorter than that of wt RSV and 165 or 450 nt shorter than that of ΔG/HEK or ΔNS1/2, respectively. The growth advantage of rec-RSV compared to the ΔNS1/2 parent is easy to understand, since the NS1 and NS2 proteins are involved in counteracting the host interferon-mediated antiviral state (17). The basis for the improved growth of rec-RSV compared to ΔG/HEK is less clear but presumably involves the deletion of the SH gene and the shorter length of the genome: both of these factors have been shown to yield increases in growth or plaque size for RSV (1, 2, 19). It was unexpected that rec-RSV grew similarly to wt rA2 in HEp-2 cells because rec-RSV lacks the G protein, whose absence had previously been associated with reduced growth fitness in HEp-2 cells (22). It may be that the additional loss of SH and the reduction in the length of the genome played a role. It also might be that the chimeric SH:G gene played a role; there is no basis for this idea except for the odd coincidence mentioned above that the efficiently growing cp-52 also contained a similar chimeric gene. Thus, the acquisition of wt-like growth fitness by rec-RSV remains unexplained but is not a central issue here.
The recombination of two rRSV mutants to form a new progeny genome was detected in only one of the six coinfections established in this study and appeared to involve only a single recombined genome, despite the fact that the experimental conditions had been optimized to facilitate the generation and identification of recombined variants. This suggests that the formation of viable recombined RSV is rare in cell culture and therefore likely to be even more uncommon in nature, where the presence of large numbers of coinfected cells is less likely. This is offered with the caveat that our conditions would have detected recombinants that had a growth advantage.
Our expectation had been that, under optimal conditions, recombination would be detected more frequently, since large-plaque formation by ΔG/HEK should theoretically require only the reacquisition of the G gene by a single crossover within the ∼3,500 nt between the end of NS2 and the start of G. Since we were able to observe only one recombination event in six independent coinfections, it appears that the frequency of recombination, whether homologous or nonhomologous, is very low. However, the recent finding that recombination of influenza virus (14) RNA segments appeared to occur frequently indicates that this should be examined carefully for mononegaviruses. We are further refining our present system to examine RSV recombination more precisely.
Although the present study suggests that the rate of RSV recombination is very low, this does not mean that recombination has played no role in mononegavirus evolution. For example, the genomes of existing mononegaviruses share a common set of genes, N-P-M-(G/H/HN)-L, but also variably contain additional interspersed genes such as NS1, NS2, SH, and M2. Assuming that present-day mononegaviruses arose from a common ancestor, recombination might have played a role in the insertion or deletion of genes, although other mechanisms also are possible. A second example involves the differences in gene order between two pneumoviruses, namely, RSV (NS1-NS2-N-P-M-SH-G-F-M2-L) and human metapneumovirus (N-P-M-F-M2-SH-G-L). These differences include the absence of the NS1 and NS2 genes in human metapneumovirus and a difference in the order of the F-M2 gene pair relative to the SH-G gene pair. This latter difference illustrates a likely rearrangement in gene order, presumably mediated by recombination, involved in the divergence of these two pneumoviruses.
A third example comes from sequence analysis of RSV isolates, in which a segment of the SH gene of several isolates of one genetic lineage appeared to be derived from a second lineage (27). This suggested that the polymerase had jumped over, copied part of the SH gene, and jumped back. Here it is of interest that, once again, the SH gene was involved. These precedents not withstanding, the apparent low frequency of recombination for RSV suggests that it does not make an important contribution to the swarm of mutants that is characteristic of a population of mononegavirus and may not make a significant contribution to the maintenance and evolution of genetically diverse strains that are characteristic of RSV.
With regard to the development of RSV vaccine candidates containing defined, circumscribed attenuating mutations, even if recombination were frequent, it should not pose a concern for vaccine safety, since a cross between an attenuated strain and a wt strain would yield intermediately attenuated progeny that should not pose a threat in a world already full of wt RSV. Also, human and animal RSVs do not appear to have highly pathogenic variants or biotypes that might otherwise provide the possibility of the emergence of new virulent strains through recombination. Given the apparent low rate of recombination observed here, it is clear that recombination is not a concern for vaccine stability and safety.
Acknowledgments
We thank Kim-Chi Tran and Chris Hanson for technical assistance in cell culture and sequencing.
In the interests of full disclosure, we acknowledge that our laboratory receives support from Wyeth for the development of live-attenuated vaccines against RSV, although this particular project was not supported by this program.
REFERENCES
- 1.Bukreyev, A., S. S. Whitehead, B. R. Murphy, and P. L. Collins. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J. Virol. 71:8973-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukreyev, A., B. R. Murphy, and P. L. Collins. 2000. Respiratory syncytial virus can tolerate an intergenic sequence of at least 160 nucleotides with little effect on transcription or replication in vitro and in vivo. J. Virol. 74:11017-11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrappa, S., D. K. Gavin, and K. C. Gupta. 1995. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 5:404-407. [DOI] [PubMed] [Google Scholar]
- 4.Collins, P. L., M. G. Hill., E. Camargo, H. Grosfeld, R. M. Chanock, and B. R. Murphy. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl. Acad. Sci. USA 92:11563-11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condit, R. C. 2001. Principles of virology, p. 19-52. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 6.Cooper, P. D., A. Steiner-Pryor, P. D. Scotti, and D. Delong. 1974. On the nature of poliovirus genetic recombination. J. Gen. Virol. 23:41-49. [DOI] [PubMed] [Google Scholar]
- 7.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96:13910-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn, C. S., S. Lustig, E. G. Strauss, and J. H. Strauss. 1988. Western equine encephalitis virus is a recombinant virus. Proc. Natl. Acad. Sci. USA 85:5997-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes, E. C., M. Worobey, and A. Rambaut. 1999. Phylogenetic evidence for recombination in dengue virus. Mol. Biol. Evol. 16:405-409. [DOI] [PubMed] [Google Scholar]
- 10.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 94:13961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai, M. M. C. 1992. RNA recombination in animal and plant viruses. Microbiol. Rev. 56:61-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCullers, J. A., G. C. Wang, S. He, and R. G. Webster. 1999. Reassortment and insertion-deletion are strategies for the evolution of influenza B viruses in nature. J. Virol. 73:7343-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkelsen, J. G., and F. S. Pedersen. 2000. Genetic reassortment and patch repair by recombination in retroviruses. J. Biomed. Sci. 7:77-99. [DOI] [PubMed] [Google Scholar]
- 14.Mitnaul, L. J., M. N. Matrosovich, M. R. Castrucci, A. B. Tuzikov, N. V. Bovin, D. Kobasa, and Y. Kawaoka. 2000. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J. Virol. 74:6015-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy, B. R., A. V. Sotnikov, L. A. Lawrence, S. M. Banks, and G. A. Prince. 1990. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3-6 months after immunization. Vaccine 8:497-502. [DOI] [PubMed] [Google Scholar]
- 16.Nagy, P. D., and A. E. Simon. 1997. New insights into the mechanism of RNA recombination. Virology 235:1-9. [DOI] [PubMed] [Google Scholar]
- 17.Schlender, J., B. Bosser, U. Buchholz, and K.-K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sibold, C., H. Meisel, D. H. Kruger, M. Labuda, J. Lysy, O. Kozuch, M. Pejcoch, A. Vaheri, and A. Plyusnin. 1999. Recombination in Tula hantavirus evolution: analysis of genetic lineages from Slovakia. J. Virol. 73:667-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Techaarpornkul, S., N. Barretto, and M. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75:6825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teng, M. N., and P. L. Collins. 1998. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J. Virol. 72:5707-5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teng, M. N., and P. L. Collins. 1999. Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein. J. Virol. 73:466-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng, M. N., S. S. Whitehead, and P. L. Collins. 2001. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 289:283-296. [DOI] [PubMed] [Google Scholar]
- 23.Tolou, H. J. G., P. Couissinier-Paris, J.-P. Durand, V. Mercier, J.-J. de Pina, P. de Micco, F. Billoir, R. N. Charrel, and X. de Lamballerie. 2001. Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J. Gen. Virol. 82:1283-1290. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe, M., T. Nakagomi, Y. Koshimura, and O. Nakagomi. 2001. Direct evidence for genome segment reassortment between concurrently-circulating human rotavirus strains. Arch. Virol. 146:557-570. [DOI] [PubMed] [Google Scholar]
- 25.Whitehead, S. S., K. Juhasz, C.-Y. Firestone, P. L. Collins, and B. R. Murphy. 1998. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J. Virol. 72:4467-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80:2535-2543. [DOI] [PubMed] [Google Scholar]
- 27.Zheng, H. Q., G. A. Storch, C. Zang, T. C. T. Peret, C. S. Park, and L. J. Anderson. 1999. Genetic variability in envelope-associated protein genes of closely related group A strains of respiratory syncytial virus. Virus Res. 59:89-99. [DOI] [PubMed] [Google Scholar]