Abstract
Recent studies indicate that Caenorhabditis elegans CED-4 interacts with and promotes the activation of the death protease CED-3, and that this activation is inhibited by CED-9. Here we show that a mammalian homolog of CED-4, Apaf-1, can associate with several death proteases, including caspase-4, caspase-8, caspase-9, and nematode CED-3 in mammalian cells. The interaction with caspase-9 was mediated by the N-terminal CED-4-like domain of Apaf-1. Expression of Apaf-1 enhanced the killing activity of caspase-9 that required the CED-4-like domain of Apaf-1. Furthermore, Apaf-1 promoted the processing and activation of caspase-9 in vivo. Bcl-XL, an antiapoptotic member of the Bcl-2 family, was shown to physically interact with Apaf-1 and caspase-9 in mammalian cells. The association of Apaf-1 with Bcl-XL was mediated through both its CED-4-like domain and the C-terminal domain containing WD-40 repeats. Expression of Bcl-XL inhibited the association of Apaf-1 with caspase-9 in mammalian cells. Significantly, recombinant Bcl-XL purified from Escherichia coli or insect cells inhibited Apaf-1-dependent processing of caspase-9. Furthermore, Bcl-XL failed to inhibit caspase-9 processing mediated by a constitutively active Apaf-1 mutant, suggesting that Bcl-XL regulates caspase-9 through Apaf-1. These experiments demonstrate that Bcl-XL associates with caspase-9 and Apaf-1, and show that Bcl-XL inhibits the maturation of caspase-9 mediated by Apaf-1, a process that is evolutionarily conserved from nematodes to humans.
Keywords: apoptosis, cell death
Programmed cell death or apoptosis, a morphologically distinguished form of cell death, is critical for development and tissue homeostasis in multicellular organisms (1, 2). The apoptotic mechanism is evolutionarily conserved and controlled by a genetic program (1–3). Genetic studies in the nematode Caenorhabditis elegans have identified three genes that play critical roles in the induction and execution of programmed cell death (3). Two nematode genes, ced-3 and ced-4, are required for the execution of the cell death program. The ced-3 product is homologous to the mammalian interleukin 1β-converting enzyme (4). The ced-9 gene functions upstream of ced-3 and ced-4 and protects cells that normally survive programmed cell death during worm development (5, 6). Biochemical analyses of CED-3, CED-4, and CED-9 have provided insight into the mechanism by which programmed cell death is regulated in the nematode. CED-4 interacts with CED-3 and CED-9 forming a multimeric protein complex (7–9). Furthermore, CED-4 promotes the activation of the death protease CED-3, and this activation is inhibited by CED-9 (10–12).
Several of the apoptosis regulatory genes identified in C. elegans have mammalian counterparts. A family of cysteine proteases (designated caspases) related to the C. elegans CED-3 appears to represent the effector arm of the apoptotic program (13). Each caspase contains conserved residues important for specific proteolytic activity cleaving after aspartic acid residues (13). Several caspases including caspase-4, -8, and -9 structurally resemble CED-3 in that they contain long prodomains and appear to act upstream in the caspase cascade (13, 14). Activation of downstream caspases through several stimuli leads to cleavage of target proteins and execution of the apoptotic program (13). Bcl-2 and Bcl-XL, two members of the Bcl-2 family, function as apoptosis inhibitors and are considered homologs of the nematode CED-9 (15). Several studies have shown that these apoptosis inhibitors regulate the activation of caspases (16, 17). However, the precise mechanism by which Bcl-2 and Bcl-XL control caspase activation and apoptosis remains controversial.
Recent studies have identified and partially characterized Apaf-1, a mammalian homolog of C. elegans CED-4 (18). The N-terminal region of Apaf-1 shares amino acid homology with CED-4 and the prodomains of CED-3 and several CED-3-like proteases with long prodomains. The region of homology shared between Apaf-1 and the prodomains of CED-3-like proteases has been termed caspase recruitment domain (19). The C-terminal region of Apaf-1 lacks homology with CED-4 and is composed of 12 WD repeats (18). In the presence of dATP and cytochrome c, a molecule that is released from mitochondria during apoptosis, Apaf-1 binds to caspase-9 and induces the activation of caspase-3, a downstream death protease (20).
In the current studies, we have sought to assess the interaction of the antiapoptotic Bcl-XL protein with Apaf-1 and CED-3-like caspases to determine if the regulation of the death mechanism is evolutionarily conserved in nematodes and mammals. The analyses indicate that Apaf-1 interacts with several caspases containing long prodomains including caspase-4, -8, -9, and CED-3. Furthermore, Bcl-XL physically associates with Apaf-1 and caspase-9 and inhibits Apaf-1-mediated maturation of caspase-9.
MATERIALS AND METHODS
Plasmid Construction.
An expression plasmid containing the Apaf-1 cDNA (18) was obtained from X. Wang (University of Texas Southwestern Medical Center, Dallas). Apaf-1, mutant N-Apaf-1, and C-Apaf-1 were amplified by PCR using the Apaf-1 cDNA as a template and cloned into the expression vector pcDNA3-Myc to produce C-terminal tagged Apaf-1 proteins. The human caspase-3, caspase-4, wild type, and mutant (C287S) caspase-9 cDNAs were cloned into the BamHI and XhoI sites of a pcDNA3-hemagglutinin (HA) or -Flag vectors to produce C-terminal tagged proteins. The plasmids pcDNA-3-Flag-Bcl-XL, pcDNA3-β-galactosidase, pcDNA3-Ced-3-HA (C360S), pcDNA3-caspase-8-HA and pcDNA3-caspase-8-AU1 mutant (C377S) were described previously (10, 21, 22). The pcDNA-p35 plasmid was a gift from V. Dixit (Genentech, South San Francisco, CA).
Transfection, Immunoprecipitation, and Western Blot Analysis.
Human embryonic kidney 293T cells (2–5 × 106) were transfected with 5 μg each of the indicated plasmid DNA by the calcium phosphate method. Protein immunoprecipitation and Western blot analysis with relevant antibodies were performed as described (7). Rabbit anti-caspase-9 antibody was a gift of X. Wang, and anti-poly(ADP-ribose) polymerase (PARP) antibody was purchased from Enzyme Systems Products (Livermore, CA). The proteins were detected by using an enhanced chemiluminescence system (Amersham).
Apoptosis Assay.
293T cells (1 × 105) were seeded in each well of 12-well plates. After 12 hr, cells were transiently transfected with 0.25 μg of the reporter pcDNA3-β-galactosidase plasmid plus various combinations of the following plasmids: 0.1 μg caspase-9, 0.4 μg each of Apaf-1-Myc (WT), N-Apaf-1 (N), C-Apaf-1 (C), untagged Apaf-1 (UT), or 1 μg pcDNA3-p35. pcDNA3 was used to adjust total plasmid DNA to an equal amount. Percentage of apoptotic cells was determined 18 hr after transfection in triplicate cultures as described (10).
Preparation of Recombinant Bcl-XL Protein.
The human Bcl-XL cDNA was excised from pSFFV-Bcl-XL (21) and cloned into the pET30a(+) plasmid (Novagen). Bcl-XL was purified from cultures of Escherichia coli BL21(DE3) expressing (His)6-Bcl-XL by Ni2+-resin column chromatography. The purity of the Bcl-XL preparation was at least 95% as determined by Coomassie blue staining. Recombinant Glu-Glu-tagged Bcl-XL was produced from a pAcoG-human-Bcl-XL construct in baculovirus-infected Sf9 cells (a gift of M. J. Fernandez Sarabia, Onyx Pharmaceuticals, Richmond, CA) and purified by affinity chromatography as described (23). The (His)6-tagged recombinant Bad protein was produced and purified from E. coli BL21(DE3) carrying pET30a(+)-Bad plasmid as described (24).
In Vitro Caspase-9 Assay.
S-100 extracts from 293T cells were prepared essentially as described by Liu et al. (25) and frozen at −80°C. Caspase-9 was translated in vitro from a pcDNA-3-caspase-9 plasmid in the presence of [35S]methionine (Amersham) by using a Promega TNT transcription/translation kit. Fifty or twenty micrograms of S-100 cellular extract was incubated with or without dATP (1 mM) or bovine cytochrome c (0.4 μg, Sigma) in the presence of 1 μl in vitro translated caspase-9 in a final volume of 25 μl. The mixtures were incubated for 30 min at 37°C and stopped by adding 5× SDS loading buffer and boiled for 5 min. To determine Apaf-1-dependence of the activation of caspase-9, 0.5 μl of normal rabbit serum or rabbit anti-Apaf-1 serum (a gift of X. Wang) was added to the S-100 extract. Recombinant Bcl-XL or Bad was preincubated with the S-100 extract for 30 min prior to addition of dATP and cytochrome c.
RESULTS
Apaf-1 Interacts with Multiple Caspases Containing Long Prodomains but not with Caspase-3.
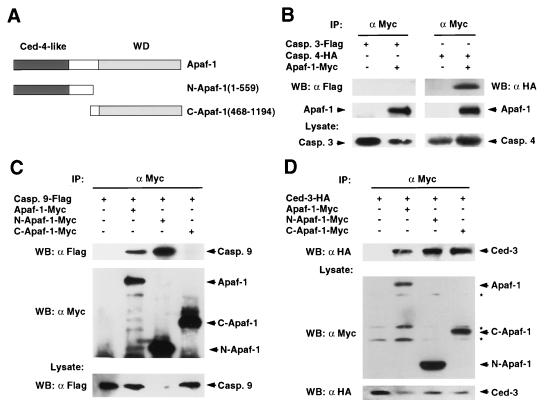
To determine if Apaf-1 interacts with caspases, we performed in vivo interaction analysis using full-length Apaf-1 or two deletion mutants of Apaf-1, N-Apaf-1 (residues 1–559), and C-Apaf-1 (residues 468-1194) comprising the CED-4-like and C-terminal regions of Apaf-1 respectively (Fig. 1A). We transiently cotransfected 293T cells with expression plasmids producing caspase-3, -4, -9, C. elegans CED-3, or control empty vector and Myc-tagged Apaf-1. Western blot analysis of protein complexes immunoprecipitated with anti-Myc antibody revealed that caspase-4, -9, and CED-3 were coimmunoprecipitated with Apaf-1 but caspase-3 was not (Fig. 1 B–D). The interaction of Apaf-1 with caspase-9 confirmed a recent study published while this manuscript was in preparation (20). Analysis of Apaf-1 mutants revealed that Apaf-1 interacted with caspase-9 through its N-terminal CED-4-like domain (Fig. 1C). However, both the CED-4-like and C-terminal regions of Apaf-1 interacted with the C. elegans CED-3 protease (Fig. 1D).
Figure 1.
Apaf-1 interacts with several caspases with long prodomains in mammalian cells. (A) Schematic drawing of the Apaf-1 constructs used in this study. (B–D) Interaction of Apaf-1 with caspase-4, caspase-9, and CED-3. 293T cells were cotransfected with the indicated tagged caspases and control vector, or tagged Apaf-1, N-terminal, or C-terminal Apaf-1 constructs (Apaf-1-Myc, N-Apaf-1-Myc, and C-Apaf-1-Myc, respectively). (Upper) Western blot analysis of immunoprecipitated protein complexes with indicated antibodies. (Lower) The expression of caspase-3 and -4 (B), caspase-9 (C), and CED-3 and Apaf-1 proteins (D). Reduced level of pro-caspase-9 in lane 3 of C is due to efficient N-Apaf-1-mediated processing of pro-caspase-9. IP: immunoprecipitation, WB: Western blot analysis. Asterisks indicate nonspecific bands.
Apaf-1 Interacts with the Death Effector Domain of Caspase-8.
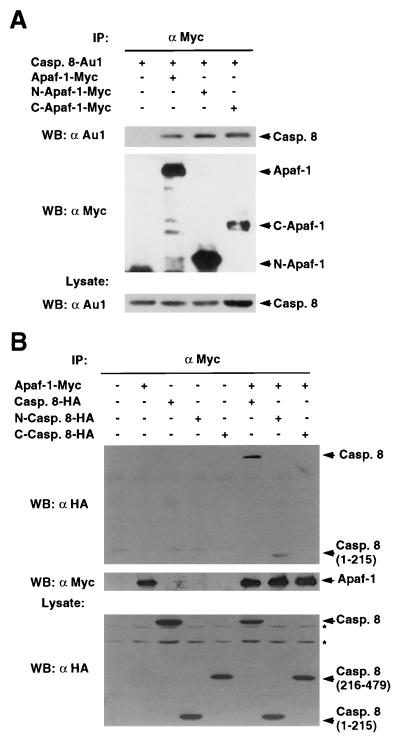
As caspase-8 contains a long prodomain with two death effector domains structurally related to the prodomain of CED-3 (19), we determined next if Apaf-1 also interacts with caspase-8. Western blot analysis of complexes immunoprecipitated with anti-Myc antibody revealed that caspase-8 was coimmunoprecipitated with Apaf-1 in 293T cells (Fig. 2A). Analysis of Apaf-1 mutants showed that caspase-8 associates with both the CED-4-like and C-terminal regions of Apaf-1 (Fig. 2A). To further dissect the Apaf-1-caspase-8 interaction, we engineered two deletion mutants to express either the N-terminal prodomain (residues 1–215) or the C-terminal catalytic domain (residues 216–479) of caspase-8. Immunoprecipitation analysis revealed that Apaf-1 interacts with the prodomain containing the death effector domains but not with the catalytic domain of caspase-8 (Fig. 2B).
Figure 2.
Apaf-1 interacts with the prodomain of caspase-8. (A) Interaction of Apaf-1 with caspase-8. (B) Apaf-1 interacts with the prodomain of caspase-8. Immunoprecipitation analysis was performed as described in Fig. 1. (Upper) Western blot analysis of immunoprecipitated protein complexes with indicated antibodies. (A and B, Lower) show the expression of caspase-8 (A) or Apaf-1 (B) in total lysates by Western blot analysis. IP: immunoprecipitation, WB: Western blot analysis. Asterisks indicate nonspecific bands.
Apaf-1 Promotes Both Activation of Caspase-9 and Caspase-9-Mediated Apoptosis in Mammalian Cells.
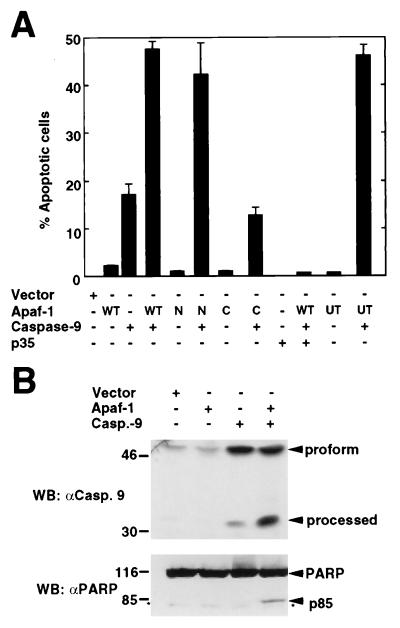
The interaction of Apaf-1 with caspase-9 prompted further experiments to assess whether Apaf-1 could regulate the activation of caspase-9. In these experiments, 293T cells were transiently transfected with constructs producing Myc-tagged full-length or truncated mutant Apaf-1 and caspase-9. Expression of wild-type Apaf-1 or the N-terminal mutant (CED-4-like) but not the C-terminal mutant of Apaf-1 enhanced the killing activity of caspase-9 (Fig. 3A). Apoptosis induced by Apaf-1 and caspase-9 was inhibited by baculovirus p35, an inhibitor of caspases (Fig. 3A). Untagged Apaf-1 also enhanced the killing activity of caspase-9, ruling out any artifact due to the Myc tag (Fig. 3A). We performed further analysis to determine whether the proteolytic processing of caspase-9, a step required for activation of caspases (13), was regulated by Apaf-1 in vivo. Expression of Apaf-1 enhanced the formation of a major cleavage product of caspase-9 (p35), that is formed during its proteolytic activation (Fig. 3B). To assess the activation of caspase-9, the same cellular lysates were immunoblotted with an antibody against PARP, a protein that is cleaved after caspase-9 activation (20, 25). Western blot analysis revealed that Apaf-1 promotes caspase-9 activation as determined by the detection of p85, a proteolytic fragment of PARP that results from its cleavage by caspases (Fig. 3B).
Figure 3.
Regulation of caspase-9 by full-length and N-terminal Apaf-1. (A) Apaf-1 enhances caspase-9-induced apoptosis. 293T cells were transiently transfected with a reporter pcDNA3-β-galactosidase plus the indicated plasmids. WT, full-length Apaf-1-Myc; N, N-terminal Apaf-1-Myc; C, C-terminal Apaf-1-Myc; UT, Untagged full-length Apaf-1. The results represent the percentage of blue cells that exhibit morphologic features of apoptosis and are given as the mean +/− SD of triplicate cultures. (B) Apaf-1 promotes caspase-9 activation in vivo. Lysates from cells transfected with the indicated plasmids were immunoblotted with anti-caspase-9 (Upper) or anti-PARP antibodies (Lower). Arrows indicate full-length and processed caspase-9 and PARP fragments. Asterisk indicates nonspecific band.
Bcl-XL Associates with Caspase-9 and Apaf-1 in Mammalian Cells.
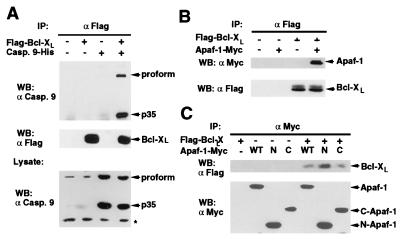
We examined next whether Bcl-XL could associate with caspase-9. The analysis showed that Bcl-XL and caspase-9 coimmunoprecipitated in vivo (Fig. 4A). As the nematode CED-9 protein, a homolog of mammalian Bcl-XL, interacts with CED-3 through CED-4 (8, 10), we determined if Bcl-XL could associate with Apaf-1. Western blot analysis of Bcl-XL complexes with anti-Myc antibody revealed that Apaf-1 coimmunoprecipitated with Bcl-XL (Fig. 4B). To verify these results, we performed reciprocal experiments in which wild-type Apaf-1 and Apaf-1 deletion mutants were immunoprecipitated with anti-Myc antibody. Western blot analysis with anti-Flag confirmed that Apaf-1 coimmunoprecipitated with Bcl-XL and revealed that Bcl-XL associates with both the CED-4-like domain and the C-terminal region that contains the WD repeats of Apaf-1 (Fig. 4C).
Figure 4.
Bcl-XL interacts with caspase-9 and Apaf-1. (A) Bcl-XL interacts with caspase-9. (Upper) Western blot analysis of immunoprecipitated Bcl-XL and coimmunoprecipitated pro-caspase-9 and processed form (p35). (Lower) The expression of caspase-9 in total lysate. (B and C) Bcl-XL interacts with Apaf-1. 293T cells were transfected with indicated plasmids and the lysates immunoprecipitated with anti-Flag (B) or anti-Myc (C) antibody. WT, full-length Apaf-1-Myc; N, N-terminal Apaf-1-Myc; C, C-terminal Apaf-1-Myc. Panels show Western blot analysis of coimmunoprecipitated Apaf-1 and Bcl-XL proteins. Asterisk indicates nonspecific band.
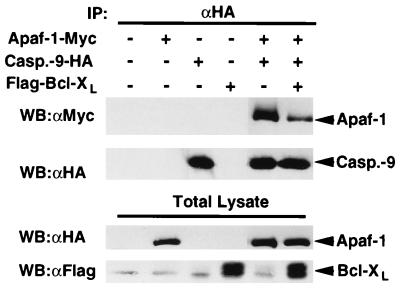
Bcl-XL Inhibits the Association of Apaf-1 with Caspase-9.
Because Bcl-XL associated with both Apaf-1 and caspase-9 in mammalian cells, we tested whether Bcl-XL could affect the Apaf-1-caspase-9 interaction. 293T cells were cotransfected with plasmids producing Apaf-1 and caspase-9 (C287S) in the presence or absence of Bcl-XL and caspase-9 complexes were immunoprecipitated with anti-HA antibody. Western blot analysis revealed that Bcl-XL inhibited the association of Apaf-1 with caspase-9 (Fig. 5). Western blot analysis of the same immunocomplexes and lysates showed comparable levels of caspase-9 and Apaf-1 (Fig. 5), indicating that the results were not due to differential expression of these proteins.
Figure 5.
Bcl-XL inhibits the association of Apaf-1 with caspase-9. 293T cells were transfected with indicated plasmids and the lysates immunoprecipitated with anti-HA antibody. (Upper) Western blot analysis of immunoprecipitated caspase-9 and coimmunoprecipitated Apaf-1. (Lower) Western blot analysis of total lysates with anti-Myc and anti-Flag antibody.
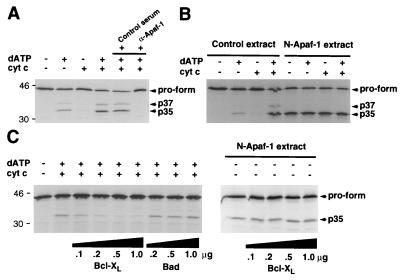
Purified Bcl-XL Inhibits Apaf-1-Mediated Maturation of Caspase-9.
To determine if Bcl-XL could regulate the activity of caspase-9, we performed biochemical experiments by using a cell-free system in which exogenously added dATP and cytochrome c induce the activation of caspase-9 (20). Processing of caspase-9 into p37, p35, and p10–12 forms in cellular extracts was dependent on dATP and cytochrome c as reported (20). However, we observed a partial cytochrome c dependence in our system presumably due to leakage of cytochrome c during extract preparation (Fig. 6A). The activation of caspase-9 was inhibited by rabbit anti-Apaf-1, but not control serum, confirming that activation of caspase-9 by cytosolic extracts was dependent on Apaf-1 (Fig. 6A). In contrast to caspase-9, we did not observe any enhancement of caspase-4 or caspase-8 processing using this in vitro system (data not shown). Significantly, an N-terminal mutant of Apaf-1 (residues 1–559) promoted the activation of caspase-9 independently of dATP and cytochrome c (Fig. 6B).
Figure 6.
Purified Bcl-XL inhibits Apaf-1-dependent maturation of caspase-9 in vitro. (A) Apaf-1-dependent processing of caspase-9 by Apaf-1 in vitro. In the last two lanes normal rabbit serum (control) or anti-Apaf-1 serum were added 30 min prior to the reaction. (B) N-terminal Apaf-1 mutant (residues 1–559) activates caspase-9 independently of cytochrome c and dATP. Extracts from 293T cells transfected with pcDNA3 (control extract) or pcDNA3-N-Apaf-1 (N-Apaf-1 extract) were used in the analysis. (C) Regulation of caspase-9 maturation by recombinant Bcl-XL. Cellular extracts were incubated with indicated amounts of recombinant Bcl-XL or Bad and then cytochrome c and dATP were added to the reaction. (Right) Extracts from cells expressing a N-terminal Apaf-1 mutant (residues 1–559) were incubated with indicated amounts of recombinant Bcl-XL. Proform and processed caspase-9 are indicated by arrows.
We purified Bcl-XL from E. coli to test the ability of Bcl-XL to regulate Apaf-1-dependent activation of caspase-9. Recombinant Bcl-XL purified from E. coli inhibited the maturation of caspase-9 in a dose-dependent manner (Fig. 6C). In control experiments, recombinant Bad purified from E. coli did not inhibit the processing of caspase-9 (Fig. 6C). In addition, recombinant Bcl-XL purified from SF9 insect cells also inhibited the maturation of caspase-9 in this cell-free system (data not shown). Because Bcl-XL associates with both caspase-9 and Apaf-1, we used the constitutively active Apaf-1 mutant (residues 1–559) to assess the possibility that caspase-9 activation is regulated directly by its interaction with Bcl-XL. Fig. 6C shows that Bcl-XL could not inhibit caspase-9 activation mediated by the N-Apaf-1 mutant. These results indicate that Bcl-XL requires wild-type Apaf-1 to inhibit caspase-9 activation. Therefore it is unlikely that Bcl-XL inhibits caspase-9 solely through a direct interaction with caspase-9.
DISCUSSION
In the present studies, we have shown that Apaf-1, a mammalian homolog of C. elegans CED-4 interacts with several caspases including caspase-4, -8, -9, and the nematode CED-3 protease in mammalian cells. These results indicate that the association of Apaf-1 with caspases is not restricted to caspase-9 (20). All of these interacting caspases contain long prodomains and are thought to function upstream in a caspase cascade (14). The prodomains of caspase-9 and CED-3 contain a caspase recruitment domain that is conserved in several apoptosis regulatory molecules including Apaf-1, Rip-associated ICHi/CED-3 homologous protein with a death domain (RAIDD), and cellular inhibitors of apoptosis (IAPs) (19, 20, 26). Caspase-8 is a proximal caspase that is involved in apoptosis mediated through the tumor necrosis factor family of death receptor pathways (27, 28). Caspase-8 contains two death effector domains in its prodomain (27, 28). We have shown here that caspase-8 interacts with Apaf-1 through its prodomain region that contains the death effector domains. In contrast, caspase-3, a death protease that functions downstream of caspase-9 and lacks a long prodomain (20), failed to interact with Apaf-1. These results indicate that Apaf-1 can interact with several other upstream caspases in addition to caspase-9. However, the relevance of the interaction between Apaf-1 and caspase-4 or caspase-8 is unclear. In contrast to caspase-9, Apaf-1 did not promote processing of these other caspases in our in vitro system. It is possible that Apaf-1 still plays a role in the activation of caspase-4 and/or caspase-8, but the regulation may require different factors that those required for caspase-9.
Previous studies have demonstrated that Bcl-XL can associate with caspase-1 and caspase-8 in mammalian cells (8). Here we have shown that Bcl-XL associates with caspase-9. We further demonstrate that Apaf-1 interacts with Bcl-XL. This result is consistent with previous observations with the C. elegans regulators CED-9 and CED-4 in which CED-4 associated with CED-9 as well as human Bcl-XL (7, 10). It is unclear, however, if the interaction of Bcl-XL with caspase-9 is direct or rather through endogenous Apaf-1 or another CED-4 homolog because caspase-9 and Apaf-1 can form a dimeric complex in mammalian cells (Fig. 1). Because Apaf-1 is expressed in 293T cells (Y.H. and G.N., unpublished observation), it is conceivable that Apaf-1 or another Apaf-1-like molecule could mediate the interaction of caspase-9 with Bcl-XL.
Several models have been proposed to explain the apoptosis inhibitory effect of Bcl-2 family members. It has been hypothesized that these proteins exert their antiapoptotic function by preventing the release of cytochrome c from the mitochondria to the cytosol (29, 30). In support of this model, Bcl-XL and Bcl-2, which reside in the outer mitochondria and other intracellular membranes, have been reported to prevent the release of cytochrome c, a molecule that binds to Apaf-1 and participates in the activation of caspase-9 (29–33). This mechanism could be mediated by the ability of Bcl-XL to maintain the integrity of the mitochondria membrane through the formation of ion channels or other mechanisms (33). However, several observations are not consistent with the hypothesis that antiapoptotic Bcl-2 family members function primarily to regulate cytochrome c release. First, in some systems cytochrome c release into the cytosol is not induced by apoptotic stimuli known to be inhibited by Bcl-XL (34). Second, Bcl-XL inhibits apoptosis in cells that are resistant to apoptosis induced by microinjection of cytochrome c, suggesting that the ability of Bcl-XL to inhibit cell death cannot be due solely to inhibition of cytochrome c release from mitochondria (35). Finally, both mutant Bcl-2 that targets to the endoplasmic reticulum, as well as the adenovirus Bcl-2-related E1B-19K protein that is largely excluded from mitochondria, can inhibit apoptosis (36, 37), suggesting that antiapoptotic Bcl-2 family members are active even if they do not reside in the mitochondria.
Our results suggest another model by which Bcl-XL could inhibit apoptotic cell death. This mechanism involves binding of Bcl-XL to Apaf-1 and caspase-9 and perhaps other death proteases such as caspase-4 and caspase-8. Recombinant Bcl-XL purified from E. coli or insect cells can inhibit the activation of caspase-9 that is dependent on Apaf-1. These results suggest that like its nematode homolog CED-9, Bcl-XL can regulate apoptosis by controlling the activation of caspase-9 through Apaf-1. Because recombinant Bcl-XL inhibited the maturation of caspase-9 mediated by Apaf-1, these results indicate that Bcl-XL can regulate caspase-9 activation independently of its association with intracellular membranes. In intact cells, the ability of Bcl-XL to inhibit caspase-9 activation could be enhanced or further regulated by its insertion into intracellular membranes. Our observation that Bcl-XL inhibited the interaction of Apaf-1 with caspase-9 suggests a mechanism by which Bcl-XL could inhibit caspase-9 activation. This could be the result of direct competition between Bcl-XL and caspase-9 for Apaf-1 and/or intracellular sequestration of Apaf-1 by Bcl-XL. These results do not rule out additional mechanisms of Bcl-XL function such as alteration of the interaction of Apaf-1 with cytochrome c and/or inhibition of conformational changes in the Apaf-1/caspase-9 complex by Bcl-XL (20). Further studies are clearly needed to fully understand the role of Apaf-1 in caspase activation and its regulation by Bcl-2 family members.
Acknowledgments
The authors thank L. del Peso for providing recombinant Bad protein; S. Chen, L. Ding, and T. Hlaing for technical help; and L. del Peso, D. Ekhterae, M. Gonzalez-Garcia, and T. Koseki for critical review of the manuscript. We also thank Drs. E. Alnemri, C. Distelhorst, V. Dixit, and particularly X. Wang for generously supplying reagents. This work was supported by National Institutes of Health Grants CA64556 and CA64421 and a grant from the U.S. Army Medical Research and Material Command. Y.H. was supported by a Postdoctoral Immunopathology Training Grant T32A107413-03 from the National Institutes of Health. M.A.B. was supported by a predoctoral fellowship from the U.S. Army Medical Research and Material Command. G.N. was supported by a Research Career Development Award K04 CA64421-01 from the National Institutes of Health.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: HA, hemagglutinin; PARP, poly(ADP-ribose) polymerase.
References
- 1.Thompson C B. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 2.White E. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Hengartner M O, Horvitz H R. Curr Opin Genet Dev. 1994;4:581–586. doi: 10.1016/0959-437x(94)90076-f. [DOI] [PubMed] [Google Scholar]
- 4.Yuan J, Shaham S, Ledoux S, Ellis H M, Horvitz H R. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 5.Hengartner M O, Ellis R E, Horvitz H R. Nature (London) 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- 6.Shaham S, Horvitz H R. Genes Dev. 1996;10:578–591. doi: 10.1101/gad.10.5.578. [DOI] [PubMed] [Google Scholar]
- 7.Wu D, Wallen H D, Núñez G. Science. 1997;275:1126–1129. doi: 10.1126/science.275.5303.1126. [DOI] [PubMed] [Google Scholar]
- 8.Chinnaiyan A M, O’Rourke K, Lane B R, Dixit V M. Science. 1997;275:1122–1126. doi: 10.1126/science.275.5303.1122. [DOI] [PubMed] [Google Scholar]
- 9.Spector M S, Desnoyers S, Hoeppner D J, Hengartner M O. Nature (London) 1997;385:653–656. doi: 10.1038/385653a0. [DOI] [PubMed] [Google Scholar]
- 10.Wu D, Wallen H D, Inohara N, Núñez G. J Biol Chem. 1997;272:21449–21454. doi: 10.1074/jbc.272.34.21449. [DOI] [PubMed] [Google Scholar]
- 11.Seshagiri S, Miller L K. Curr Biol. 1997;7:455–460. doi: 10.1016/s0960-9822(06)00216-8. [DOI] [PubMed] [Google Scholar]
- 12.Chinnaiyan A M, Chaudhary D, O’Rourke K, Koonin E V, Dixit V M. Nature (London) 1997;388:728–729. doi: 10.1038/41913. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, G. M. (1997) Biochem J. 326 (Pt. 1), 1–16. [DOI] [PMC free article] [PubMed]
- 14.Golstein P. Science. 1997;275:1081–1082. doi: 10.1126/science.275.5303.1081. [DOI] [PubMed] [Google Scholar]
- 15.Hengartner M O, Horvitz H R. Cell. 1994;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- 16.Chinnaiyan A, Orth K, O’Rourke K, Duan H, Poirier G G, Dixit V M. J Biol Chem. 1996;27:4573–4576. doi: 10.1074/jbc.271.9.4573. [DOI] [PubMed] [Google Scholar]
- 17.Armstrong R C, Aja T, Xiang J, Gaur S, Krebs J F, Hoang K, Bai X, Korsmeyer S J, Karanewsky D S, Fritz L C, et al. J Biol Chem. 1996;271:16850–16855. doi: 10.1074/jbc.271.28.16850. [DOI] [PubMed] [Google Scholar]
- 18.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann K, Bucher P, Tschopp J. Trends Biochem Sci. 1997;22:155–156. doi: 10.1016/s0968-0004(97)01043-8. [DOI] [PubMed] [Google Scholar]
- 20.Li P, Nijhawanl D, Budihardjol I, Srinivasula S, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 21.Boise L H, Gonzalez-Garcia M, Postema C E, Ding L, Lindsten T, Turka L A, Mao X, Núñez G, Thompson C B. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 22.Inohara N, Koseki T, Hu Y, Chen S, Núñez G. Proc Natl Acad Sci USA. 1997;94:10717–10722. doi: 10.1073/pnas.94.20.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubinfeld B, Crosier W J, Albert I, Conroy L, Clark R, McCormick F, Polakis P. Mol Cell Biol. 1992;12:4634–4642. doi: 10.1128/mcb.12.10.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Núñez G. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Kim C N, Yang J, Jemmerson R, Wang X. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 26.Duan H, Dixit V M. Nature (London) 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- 27.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 28.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, et al. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng T-I, Jones D P, Wang X. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 30.Kluck R M, Bossy-Wetzel E, Green D R, Newmeyer D D. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 31.Kharbanda S, Pandey P, Schofield L, Israels S, Roncinske R, Yoshida K, Bharti A, Yuan Z M, Saxena S, Weichselbaum R, et al. Proc Natl Acad Sci USA. 1997;94:6939–6942. doi: 10.1073/pnas.94.13.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim C N, Wang X, Huang Y, Ibrado A M, Liu L, Fang G, Bhalla K. Cancer Res. 1997;57:3115–3120. [PubMed] [Google Scholar]
- 33.Vander Heiden M G, Chandel N S, Williamson E K, Schumacker P T, Thompson C B. Cell. 1997;91:627–637. doi: 10.1016/s0092-8674(00)80450-x. [DOI] [PubMed] [Google Scholar]
- 34.Chauhan D, Pandey P, Ogata A, Teoh G, Krett N, Halgren R, Rosen S, Kufe D, Kharbanda S, Anderson K. J Biol Chem. 1997;272:29995–29997. doi: 10.1074/jbc.272.48.29995. [DOI] [PubMed] [Google Scholar]
- 35.Li F, Srinivasan A, Wang Y, Armstrong R C, Tomaselli K J, Fritz L C. J Biol Chem. 1997;272:30299–30305. doi: 10.1074/jbc.272.48.30299. [DOI] [PubMed] [Google Scholar]
- 36.Zhu W, Cowie A, Wasfy G W, Penn L Z, Leber B, Andrews D W. EMBO J. 1996;15:4130–4141. [PMC free article] [PubMed] [Google Scholar]
- 37.White E, Cipriani R. Proc Natl Acad Sci USA. 1989;86:9886–9890. doi: 10.1073/pnas.86.24.9886. [DOI] [PMC free article] [PubMed] [Google Scholar]