Abstract
Hepatitis C virus (HCV) is the only known positive-stranded RNA virus that causes persistent lifelong infections in humans. Accumulation of HCV RNA can be inhibited with alpha interferon (IFN-α) in vivo and in culture cells. We used cell-based assay systems to investigate the mechanisms responsible for the cytokine-induced inhibition of HCV replication. The results showed that IFN-α could suppress the accumulation of viral RNA by a noncytopathic pathway and could also induce apoptosis of virally infected cells in a concentration- and cell line-dependent fashion. Whereas the noncytopathic IFN-α response depended on a functional Jak-STAT signal transduction pathway, it did not appear to require double-stranded RNA-dependent pathways. Moreover, we found that functional proteasomes were required for establishment of the IFN-α response against HCV. Based on the results described in this study we propose a model for the mechanism by which IFN-α therapy suppresses HCV replication in chronic infections by both cytopathic and noncytopathic means.
Hepatitis C virus (HCV) is a positive-stranded RNA virus that can cause productive, lifelong infections. Hepatocytes are the primary target for HCV infections, but other cells can also harbor viral RNA (5, 36). Like other members of the Flaviviridae, HCV is an enveloped virus encoding a polyprotein that is proteolytically processed into 10 polypeptides (37). Three are structural proteins required for capsid formation (core) and assembly into enveloped virus particles (E1 and E2). Four virus-encoded proteins are enzymes, including cysteine and serine proteases (NS2 and NS3), an ATP-dependent helicase (NS3), and an RNA-directed RNA polymerase (NS5B). The functions of the remaining three polypeptides, p7, NS4B, and NS5A, in viral replication are not yet known.
An estimated 170 million people worldwide are persistently infected with HCV. Although the initial infection is asymptomatic, subsequent clinical manifestations of liver disease include fibrosis, cirrhosis, and hepatocellular carcinoma (1). Combination antiviral therapy with alpha interferon (IFN-α) and ribavirin, a purine nucleoside analogue, arrests disease progression and can lead to sustained recovery in 45 to 80% of treated patients (10). The parameters determining the success or failure of antiviral therapy are not understood, and their identification represents a major challenge in HCV biology. Interestingly, the response to IFN-α therapy can vary significantly depending on the viral genotype, ranging from 30 to 40% for genotype 1 to as high as 80% for genotypes 2 and 3. This suggests that viral determinants play an important role in regulating the cellular IFN response against HCV (24, 29).
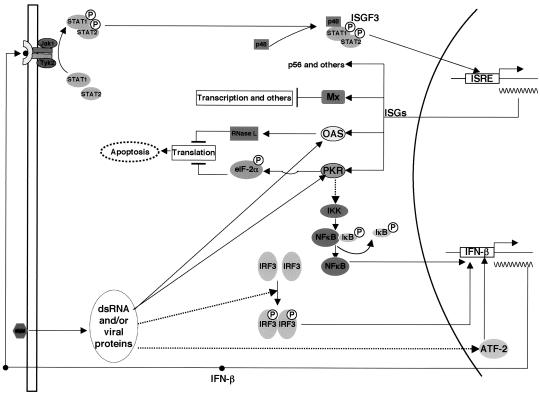
DNA microarray studies revealed that the antiviral response induced by IFNs alters the expression of hundreds of genes and, hence, is far more complex than previously anticipated (9). Little is known about the nature of the cellular proteins that specifically target viral components and, hence, are responsible for the inhibition of viral replication in the absence of cell death. In contrast, the major signal transduction pathways required for the innate immune response against many viruses have been elucidated. The first wave of IFN-induced genes depends on the phosphorylation of STAT1 and STAT2 and their interaction with IRF9 (p48) to form the transcription factor complex ISGF3 (see Fig. 4). In addition, viral double-stranded RNA (dsRNA) and other unknown viral factors are believed to play an important role in the establishment of an antiviral state. They can activate dsRNA-dependent enzymes such as protein kinase R (PKR) and 2′,5′-oligoadenylate synthase (OAS), as well as other still-elusive protein kinases (see Fig. 4) (42). IFN-α can induce a noncytopathic antiviral response or, alternatively, trigger apoptotic programs leading to the elimination of infected cells (46). For example, activated PKR can phosphorylate the alpha subunit of the eukaryotic initiation factor 2 (eIF-2α), leading to an arrest in translation and cell death (7). Similarly, activated OAS synthesizes 2′,5′-oligoadenylates and, in turn, activates RNase L which cause apoptosis through the degradation of rRNA (3). Alternatively, PKR can play a role in the noncytopathic response by activation of signal transduction pathways such as those of nuclear factor κB (NF-κB) and p38 mitogen-activated protein kinase (MAPK), which, in turn, can induce transcription of the IFN-β gene (19, 28).
FIG. 4.
Overview of the interactions between virus and the IFN system. Replication of viruses in cells produces dsRNA and viral proteins, which activate PKR and OAS/RNase L antiviral pathways and also signal to the promoter of the IFN-β gene by activating transcription factors IRF3, NF-κB, and ATF2. Secreted IFN binds to its receptor and activates receptor-associated Jak kinases, leading to the formation of the trimeric transcription factor ISGF3, which binds to the IFN-stimulated response element (ISRE) on promoters of IFN-stimulated genes (for reviews see references 41 and 44). Among the products of the several hundred genes induced by IFN, PKR, OAS/RNase L, and Mx are the best-characterized antiviral proteins, which inhibit different stages of viral replication and induce apoptosis of virally infected cells.
Nucleotide sequence analyses of HCV genomes isolated from Japanese patients revealed a correlation between the presence of mutations in a short segment of NS5A, termed the IFN sensitivity-determining region (ISDR), and resistance to antiviral therapy with IFN-α (11, 12). Subsequently, it was reported that the ISDR motif can bind to PKR (18). Importantly, the ISDR from IFN-resistant, but not from IFN-sensitive, HCV isolates appeared to be a substrate for PKR, suggesting that IFN treatment of chronically infected patients can lead to the selection of HCV variants with ISDRs that can bind and inactivate PKR (17, 45).
The development of a tissue culture system permissive for HCV replication in Huh7 cells has provided a new tool to investigate the IFN-α response against HCV (27). As was found in vivo, HCV RNA amplification in Huh7 cells was sensitive to the antiviral programs induced by IFN-α and IFN-γ (2, 16, 21). Surprisingly, in all cases examined so far, the 50% inhibitory concentrations (IC50) for IFN-α were very low, ranging from 0.5 to 3 IU/ml, and were not dependent on the nature of the ISDR (2, 15, 21). These observations stand in marked contrast with those made with other RNA viruses. The latter revealed that the IFN-α response in Huh7 cells was significantly reduced compared with those in other cell lines tested (22, 30). Hence, the results obtained with HCV replicons raised the possibility that a distinct antiviral pathway could be responsible for the IFN response against HCV in Huh7 cells.
To gain insight into the mechanisms responsible for the IFN response against HCV, we addressed the following questions. First, does HCV replication induce the dsRNA responses in infected cells? Second, is IFN-induced inhibition of HCV replication dependent on major known signal transduction pathways? Third, how does the IFN response lead to inhibition of viral replication? To investigate these questions, we took advantage of a novel HeLa-derived cell line permissive for replication of HCV subgenomes (50). We demonstrated that the IFN response against HCV could be both cytopathic and noncytopathic and that HCV replication per se could induce an antiviral state in cell lines expressing subgenomic replicons. Moreover, we obtained evidence for a role for proteasomes in inhibition of viral replication induced by IFN-α.
MATERIALS AND METHODS
Chemicals and reagents.
Recombinant IFN-α2b (intron A) was purchased from Schering-Plough. Cycloheximide, 2-aminopurine (2-AP), genistein, sodium salicylate, and wortmannin were obtained from Sigma. SB 203580, PD 98059, vanadate, PP2, rapamycin, and lactacystin were obtained from Calbiochem. Epoxomicin was obtained from Boston Biochem, and caspase inhibitor ZVAD-fluoromethyl ketone (ZVAD-FMK) was obtained from Enzyme Systems Products.
Cell culture.
Huh7 and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin G, streptomycin, nonessential amino acids, and l-glutamine. For the cell lines carrying HCV and Kunjin virus replicons, 500 μg of G418/ml was added to the medium. The GS4.1 cell line was derived from a subclone of FCA1 cells as described previously (21). SL1 is a HeLa cell line expressing HCV subgenomic replicon I377NS3-3′ (27, 50). The KUNCD20 cells represent a pool of approximately 200 colonies of G418-resistant HeLa cells obtained after transfection with the Kunjin virus replicon C20DXrepNeo RNA (23) (kindly provided by A. Khromykh, Sir Albert Sakzewski Virus Research Center, Brisbane, Australia).
Plasmids.
pCMV-E3L expressing the vaccinia virus E3L protein was obtained from Robert Schneider, New York University, New York. pln035 expressing virus-associated (VA) RNA was obtained from David Lazinski, Tufts University, Boston, Mass. pEF-HA-HPIV2 expressing the V protein of human parainfluenzavirus 2 (HPIV2) was obtained from Curt Horvath, Mount Sinai School of Medicine, New York, N.Y. To obtain cDNA clones of the gene encoding human Mx-1, Huh7 cells were treated with 100 IU of IFN-α/ml for 6 h and total cellular RNA was extracted with TRIzol reagent (Invitrogen) and first-strand cDNA was made with an oligo(dT)12-18 primer and Superscript II DNA polymerase (Invitrogen) by following the manufacturer's direction. For the amplification of Mx-A cDNA, the primers used were 5′-AGTATCGTGGTAGAGAGCTGC-3′ and 5′-TAATACGACTCACTATAGGGATGTGGCTGGAGATGC-3′. The purified PCR fragment was cloned into the pGEM-T Easy vector (Promega). The identity of the cloned fragment was verified by nucleotide sequence analysis.
RNA extraction and Northern blot hybridization.
Total cellular RNA was extracted with TRIzol reagent (Invitrogen) by following the manufacturer's direction. Five micrograms of total RNA was fractionated on a 1% agarose gel containing 2.2 M formaldehyde and transferred onto nylon membranes. Membranes were hybridized with riboprobes specific for plus-stranded HCV replicon RNA and Mx-A and β-actin mRNA in the conditions described previously (21).
Detection of eIF-2α phosphorylation by Western blotting.
For Western blot analysis of eIF-2α phosphorylation, cells were treated with 100 IU of IFN-α/ml for 12 h and then transfected with 2 μg of poly(I:C) per 60-mm-diameter plate by using Lipofectamine (Invitrogen). After 3 h of incubation, cells were lysed in high-salt radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 8.0], 250 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]). Proteins (40 μg) were separated on SDS-10% polyacrylamide gel and electrophoretically transferred to nitrocellulose membranes. Membranes were incubated with 50% methanol, washed extensively with water, and blocked with 3% casein in TNET buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.05% Tween 20). Membranes were incubated with rabbit polyclonal antibodies against eIF-2α (a gift from Robert Schneider, New York University) or phosphorylated eIF-2α (eIF-2α-P; Research Genetics, Inc.) diluted in blocking solution for 1 h and then washed extensively with TNET buffer. Membranes were then incubated with horseradish peroxidase-conjugated goat ant-rabbit and immunoglobulin G (IgG) (Amersham), respectively. The bound IgG was detected with Super-Signal chemiluminescence reagents (Pierce).
RPA.
For the analysis of IFN-β gene expression, cells were treated with 100 IU of IFN-α/ml for 12 h and then transfected with 2 μg of poly(I:C) per 60-mm plate by using Lipofectamine (Invitrogen). After 3 h of incubation, total cellular RNA was extracted with TRIzol reagent (Invitrogen), and IFN-β mRNA levels were determined by RNase protection assay (RPA) with the help of the RPAII kit purchased from Ambion. The probes complementary to IFN-β (GenBank accession no. M25460) and β-actin (GenBank accession no. BC013380) mRNAs spanned positions 272 to 650 and 1030 to 1250, respectively.
Annexin V-FITC staining.
SL1 cells were plated on coverslips in six-well plates 16 h prior to treatment. Cells were then mock treated or treated with 100 IU of IFN-α/ml in the absence or presence of 20 μM ZVAD-FMK. Coverslips were then put on slides and incubated with 100 μl of staining solution containing annexin V-fluorescein isothiocyanate (FITC) at room temperature for 10 to 15 min. After extensive washes with phosphate-buffered saline, slides were examined with a Nikon fluorescence microscope and photographed with a charge-coupled device camera.
Flow cytometry analysis.
To determine the fraction of apoptotic cells, we used the annexin V assay system (Roche Diagnostics GmbH). Cells were incubated with IFN-α (100 IU/ml) alone or together with 20 μM ZVAD-FMK for 24 h. The culture medium containing detached cells was collected, and the adherent cells were trypsinized and then combined with the detached cells. The cells were collected by centrifugation and washed once in phosphate-buffered saline. Pelleted cells were resuspended in binding buffer and were incubated with annexin V-FITC at room temperature for 10 to 15 min. The stained cells were then diluted with binding buffer and analyzed by flow cytometry (FACScan; Becton Dickinson).
RESULTS
IFN-α can induce noncytopathic and cytopathic antiviral responses against HCV replicons.
Recently we have been successful in establishing stable HeLa cell lines that express HCV subgenomes with an efficiency similar to that of Huh7 cells (Fig. 1) (50). Examination of the IFN-α response in HeLa-derived cell lines such as SL1 revealed a very similar dose-dependent reduction of virus replication. The IC50 of IFN-α in HeLa cell lines was generally in the range of 0.1 IU/ml, approximately 10-fold lower than the IFN-α IC50 in Huh7-derived cell lines. However, in marked contrast to observations made with Huh7 cell lines, treatment of SL1 and other HeLa-derived cell lines with more than 30 IU of IFN-α/ml induced cell death in a significant fraction of cells between 6 and 20 h posttreatment (Fig. 2A). Cell death was caused by apoptosis, as determined by annexin V staining, and could be prevented by the caspase inhibitor ZVAD-FMK. We determined the fraction of apoptotic cells before and after treatment with the cytokine. The results showed that IFN-α induced apoptosis in more than 30% of SL1 cells compared with 6 to 7% in untreated SL1 and parental HeLa cells (Fig. 2B). Several other HeLa-derived cell lines were examined to assure that the results obtained with SL1 cells reflected a general property of HeLa cells expressing HCV subgenomes. Moreover, to test more directly whether IFN-induced apoptosis was caused by viral replication, we used two methods to inhibit RNA replication in SL1 cells. The first was based on our observation that replication of HCV subgenomes is temperature sensitive and is inhibited at 39°C (J. A. Sohn and C. Seeger, unpublished observations). The second method relied on the availability of an inhibitor of the viral RNA polymerase. Consistent with a role for viral replication in the induction of apoptosis, cell death could be prevented when viral replication was inhibited by either incubation of the cells for 60 h at the elevated temperature or treatment with a viral polymerase inhibitor (Fig. 2B and result not shown).
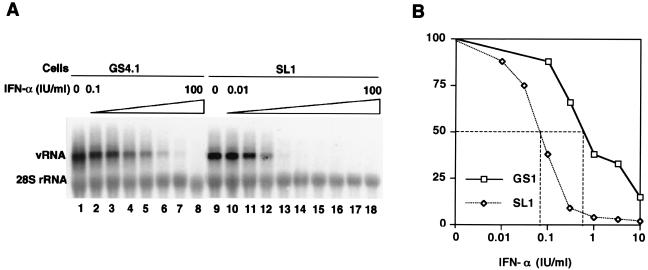
FIG. 1.
Antiviral activity of IFN-α in Huh 7 (GS4.1) and HeLa (SL1) cell lines containing HCV replicons. (A) Viral RNA (vRNA) levels present in GS4.1 and SL1 cells incubated with 0, 0.1, 0.3, 1, 3, 10, 30, and 100 IU of IFN-α/ml (lanes 1 to 9 and 12 to 18) and with 0.01 and 0.03 IU/ml (lanes 10 and 11) for 72 h were determined by Northern blot analysis. rRNA (28S rRNA) served as a control for the amount of RNA loaded per lane. (B) Amounts of HCV RNA were determined with a Fuji phosphorimager, and the values were plotted as the percentages of the values obtained with untreated cells in lanes 1 and 9.
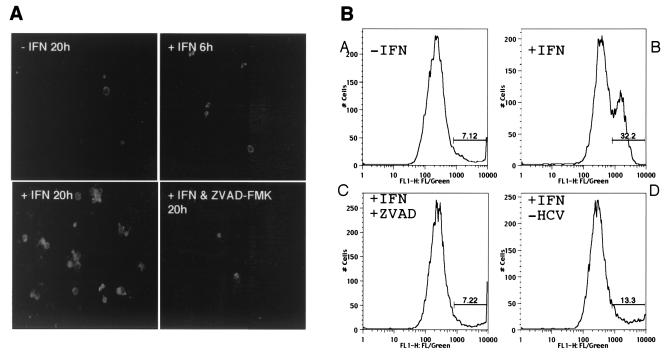
FIG. 2.
IFN-α induces apoptosis of SL1 cells. (A) Annexin V-FITC staining. SL1 cells grown on glass coverslips were left untreated (upper left) or treated with 100 IU of IFN-α/ml for 6 (upper right) or 20 h (lower left) or with 100 IU of IFN-α/ml and 20 μM caspase inhibitor ZVAD-FMK for 20 h (lower right). Cells were then processed for annexin V-FITC staining and viewed with a fluorescence microscope. (B) Flow cytometry analysis. SL1 cells were left untreated (A) or treated with 100 IU of IFN-α/ml (B) or with IFN-α and 20 μM ZVAD-FMK (C) for 24 h. To inhibit viral replication, SL1 cells were incubated at 39°C for 60 h and then treated with 100 IU of IFN-α/ml for 24 h (D). Cells were harvested and processed for annexin V-FITC staining and analyzed by flow cytometry. The percentages of FITC-positive cells are indicated.
These results raised the question of whether IFN-induced apoptosis reflected a general property of HeLa cells expressing viral replicons. To address this problem, we compared the IFN response against HCV in SL1 cells with that against the flavivirus Kunjin virus in HeLa cells. For this purpose we established a pool of HeLa cells, KUNCD20, expressing Kunjin virus subgenomic replicons lacking the structural genes, similar to the HCV subgenomic replicons (23). The Kunjin virus RNA levels in these cells were approximately fivefold higher than those observed with HCV in SL1 cells. In contrast to results with SL1 cells, treatment of KUNCD20 cells with different concentrations of IFN-α only slightly inhibited viral replication (Fig. 3). Importantly, we could not detect cell death in IFN-α-treated KUNCD20 cells either by light microscopy or annexin V staining (results not shown). These results indicated that IFN-induced apoptosis is a property of HCV-expressing HeLa cells rather than a general property of HeLa cells replicating viral RNA genomes.
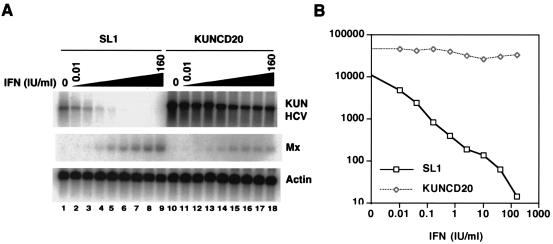
FIG. 3.
Comparison of IFN-α responses against HCV and flavivirus Kunjin virus replicons in HeLa cells. (A) SL1 and KUNCD20 cells were incubated with 0, 0.01, 0.04, 0.16, 0.625, 2.5, 10, 40, and 160 IU of IFN-α/ml (lanes 1 to 9 and 10 to 18, respectively) for 72 h, and viral RNA levels were determined by Northern blot analysis with a plus-strand-specific RNA probe for the neomycin phosphotransferase II gene. Mx-1 mRNA served as a control for IFN-α-induced gene expression. β-Actin mRNA served as a control for the amount of RNA loaded per lane. (B) The amounts of HCV and Kunjin virus replicon RNA (arbitrary units) were determined with a Fuji phosphorimager.
In summary, these results demonstrated that IFN-α could induce noncytopathic as well as cytopathic antiviral programs in cells expressing HCV replicons in a concentration- and cell type-dependent fashion. Moreover, the results showed that this antiviral program was specific for HCV replicons. Importantly, the results suggested that HCV replication could induce an innate cellular response that, in combination with IFN-α, could lead to apoptosis.
Does IFN-α inhibit HCV replication through the Jak-STAT signal transduction pathway?
Information about the signal transduction pathways responsible for execution of the IFN response has generally been obtained with cells treated with high concentrations (100 to 1,000 IU/ml) of the cytokine and with fibroblasts and epithelial cells, most of which cannot, to date, support HCV replication. Moreover, a recent study by Schlaak, et al. (40) revealed that the IFN response could vary in a cell type-dependent manner. In addition, we found that slight changes in cell culture conditions had major effects on HCV replication. Therefore, the observation that replication of HCV in both Huh7 and HeLa cells could be inhibited with low concentrations of the cytokine warranted a more careful study of the pathways involved in the antiviral program against HCV.
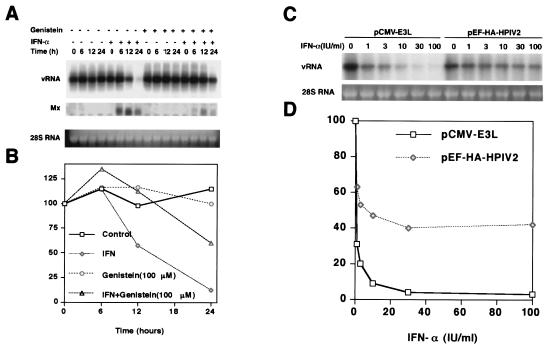
To investigate the nature of the IFN-α response against the HCV replicon, we took advantage of drugs that were known to inhibit specific components of selected signal transduction pathways that play a role in the antiviral response induced by IFN-α (Table 1). The current model for the signal transduction pathway induced by IFN-α predicts that the IFN receptor-associated tyrosine kinases Jak1 and Tyk2 are activated and, in turn, phosphorylate the transcription factors STAT1 and STAT2, which are required for the induction of the cellular antiviral program (Fig. 4) (41, 44). Incubation of GS4.1 cells with the tyrosine kinase inhibitor genistein suppressed the induction of the IFN-induced Mx-1 gene (Fig. 5A and B). Similarly, genistein antagonized the IFN-α response against the HCV replicon. An increase in the concentration of the drug from 100 to 300 μM led to a complete inhibition of the IFN response against HCV (results not shown). Consistent with this result, we found that the V protein of HPIV2 blocked the IFN response. The V protein of HPIV2 induces the degradation of STAT2 and, hence, inhibits the IFN-induced activation of gene expression (33) (Fig. 5C and D). IFN-α treatment of cells expressing HPIV2 led to a twofold reduction of viral RNA levels. When adjusted for the observed transfection efficiency, i.e., 40 to 45% of the cells express the V protein, the reduction corresponded to a complete inhibition of the IFN-α response. Finally, the decline of viral RNA levels was reduced in GS4.1 cells with IFN-α and cycloheximide, indicating that de novo protein synthesis was required for an antiviral response against HCV replication (Table 1).
TABLE 1.
Effects of inhibitors on the activity of IFN-α against the HCV replicona
| Drug, protein, or RNA | Concn | Primary target(s) | Effect |
|---|---|---|---|
| 2-AP | 10 mM | PKR and other kinases | − |
| Genistein | 300 μM | Tyrosine kinases | + |
| Cycloheximide | 10 μg/ml | Translation | + |
| Sodium salicylate | 5 mM | IKK | − |
| SB 203580 | 20 μM | p38 MAPK | − |
| PD 98059 | 50 μM | MEK kinase | − |
| Vanadate | 50 μM | Protein phosphatase | − |
| Wortmannin | 100 nM | PI3 kinase | − |
| PP2 | 50 μM | src kinase | − |
| Rapamycin | 200 nM | mTOR, translation | − |
| Lactacystin | 5 μM | 26S proteasome | + |
| Epoxomicin | 1 μM | 26S proteasome | + |
| V protein of HPIV2 | STAT2 | + | |
| E3L protein | PKR and OAS | − | |
| VA RNA | PKR | − |
GS4.1 cells were incubated with the indicated compounds for 2 h and then in the presence of 100 IU of IFN-α/ml for an additional 24 h. Viral RNA levels were determined by Northern blot analysis. Assays for V protein, E3L, and VA RNA are described in the legend to Fig. 5.
FIG. 5.
Inhibition of the IFN-α response by genistein and the V protein of HPIV2. (A) GS4.1 cells were incubated with 100 μg of genistein/ml for 2 h and then with 100 IU/ml IFN-α for an additional 24 h. Viral RNA levels were determined by Northern blot analysis. The cells were harvested at the indicated time points, and Mx-A mRNA and viral RNA levels were determined by Northern blot analysis. Ribosomal 28S RNA was used as a control for the amount of RNA loaded on each lane. (B) The amounts of viral RNA were measured with a phosphorimager and plotted as percentages of the values obtained with untreated cells. (C) GS4.1 cells were transfected with pCMV-E3L and pEF-HA-HPIV2 and treated with IFN-α at the indicated concentrations for 3 days. HCV RNA was subjected to Northern blot analysis. (D) Viral RNA levels were determined with a Fuji phosphorimager and plotted as the percentages of the values obtained with untreated cells.
IFN-α can activate, in addition to the STAT pathway, MAPKs, including extracellular signal-regulated kinase, p38 MAPK, and phosphatidylinositol 3 (PI3)-kinase-Akt pathways (8, 20, 35). However, in contrast to genistein, SB 203580, sodium salicylate, and wortmannin, known inhibitors of p38 MAPK, NF-κB, and PI-3 kinase, respectively, did not inhibit the IFN response at detectable levels, suggesting that the two major ancillary signaling pathways activated by IFN-α were not directly involved in inhibiting HCV replication in Huh7 cells (Table 1; results not shown).
In summary, the results showed that inhibition of HCV replication with IFN-α depended on a functional Jak-STAT pathway (Fig. 4). Hence, the results demonstrated that the IFN response against HCV was genuine and did not reflect an unspecific effect of the cytokine.
Does HCV replication induce an antiviral state in infected cells?
A critical step in activation of innate immunity is the induction of an antiviral state by dsRNA or viral proteins (Fig. 4) (47, 48). As shown above, evidence for such a virus-induced activation was also obtained from IFN-treated HeLa cells expressing HCV replicons. To investigate the nature of this HCV-induced activation, we determined the phosphorylation levels of eIF-2α and expression of IFN-β. eIF-2α is a substrate of PKR, which is activated by dsRNA that can accumulate as a consequence of viral RNA replication (43, 49). IFN-β gene transcription is activated through the coordinate actions of three families of transcription factors NF-κB, IRF3, and ATF2, all of which are activated by dsRNA and/or certain viral proteins (Fig. 4) (34, 48).
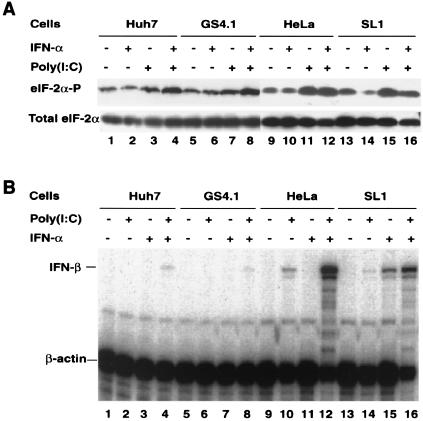
First we determined the levels of eIF-2α-P in Huh7, GS4.1, HeLa, and SL1 cells in the presence and absence of dsRNA and IFN-α. The results showed that eIF-2α-P levels were not significantly elevated in cells expressing HCV replicons (GS4.1 and SL1) compared with those in their parental cells (Huh7 and HeLa) (Fig. 6A). Similarly, incubation of cells with IFN-α did not induce eIF-2α-P levels. In contrast, transfection of cells with poly(I:C), mimicking dsRNA, augmented eIF-2α-P levels, particularly in HeLa and SL1 cells. Similar results were obtained when cells were primed with IFN-α prior to transfection with poly(I:C). These results were confirmed with several other cell lines derived from Huh7 and HeLa cells.
FIG. 6.
dsRNA response in parental Huh7 and HeLa cells and HCV replicon-containing GS4.1 and SL1 cells. (A) Phosphorylation of eIF-2α. Huh7, GS4.1, HeLa, and SL1 cells were left untreated (lanes 1, 5, 9, and 13) or treated with 100 IU of IFN-α/ml for 12 h (lanes 2, 4, 6, 8, 10, 12, 14, and 16) and then transfected with poly(I:C) and incubated for 3 h (lanes 3, 4, 7, 8, 11, 12, 15, and 16). eIF-2α-P and total eIF-2α were determined by Western blots analysis with a monoclonal antibody specific for eIF-2α-P and an antibody specific for total eIF-2α protein. (B) Induction of IFN-β mRNA by dsRNA and IFN-α. Parental Huh7 and HeLa cells and HCV replicon-containing GS4.1 and SL1 cells were left untreated (lanes 1, 5, 9, and 13) or treated with 100 IU of IFN-α/ml (lanes 3, 4, 7, 8, 11, 12, 15, and 16) for 12 h and then transfected with poly(I:C) (lanes 2, 4, 6, 8, 10, 12, 14, and 16) for 3 h. An RNase protection assay was performed with probes specific for IFN-β and β-actin mRNAs.
Second, we determined the levels of IFN-β mRNA in the four cell lines under the same conditions described above. In agreement with the above results, viral replication alone was not sufficient to activate IFN-β gene expression in both Huh7- and HeLa-derived cell lines (Fig. 6B). In Huh7 cells and GS4.1 and other Huh7-derived cells expressing HCV replicons, only a weak induction of IFN-β was observed when cells were primed with IFN-α and then transfected with poly(I:C). In contrast, IFN-β transcription was induced in HeLa and SL1 cells by poly(I:C) alone and particularly in combination with IFN-α. Remarkably, expression of IFN-β could be induced by IFN-α alone in SL1 cells but not in HeLa cells. Similar results were obtained with the HeLa-derived SL2 cell line (results not shown).
In summary, the results showed that, while both Huh7 and HeLa cells were competent to activate dsRNA-dependent signal transduction pathways, HCV replication alone was not sufficient to induce a detectable dsRNA response in these cells. This result could indicate that dsRNA either does not accumulate during HCV replication or cannot access PKR and other dsRNA binding proteins. Importantly, the results showed that, despite the apparent lack of biologically active dsRNA, viral replication could activate certain cellular signal transduction pathways that could cooperate with IFN-α to activate the transcription of the IFN-β gene.
Our results presented above favored a model predicting that IFN-α inhibited HCV replication by a mechanism that was independent of dsRNA-activated antiviral pathways. To test this model more carefully, we measured the IFN response in GS4.1 cells expressing the vaccinia virus E3L protein. E3L is known to sequester dsRNA and prevent PKR and OAS/RNase L activation (4, 38) (Fig. 4). Indeed, expression of E3L had no measurable effect on the IFN response against HCV (Fig. 5C and D). Experiments relying on simultaneous detection of E3L and the HCV protein NS5A in the same cell by immunofluorescence confirmed that IFN-α could inhibit HCV replication in cells expressing the E3L protein (results not shown). Finally, we found that the PKR inhibitors 2-AP and adenovirus VA RNA did not block the IFN response against HCV in Huh7 cells (Table 1).
What are the pathways that play a role in the IFN-α response against HCV?
A major question concerns the mechanism by which IFN-α induces the noncytopathic inhibition of HCV replication. DNA microarray analyses of IFN-α-treated GS4.1 cells and other Huh7-derived cell lines revealed the induction of several classes of genes belonging to known signal transduction and protein degradation pathways (J. Hayashi and C. Seeger, unpublished results) (6). In particular, several genes encoding proteasome subunits and ubiquitin-like proteins were among the genes most highly induced by IFN-α. Notably, kinetic studies of HCV replication in Huh7 cells indicated that replication complexes have a relatively short half-life, which is further reduced by IFN treatment (J.-T. Guo and C. Seeger, unpublished observations). Therefore, we surmised that the proteasome could play a role in inhibition of HCV replication in IFN-treated cells.
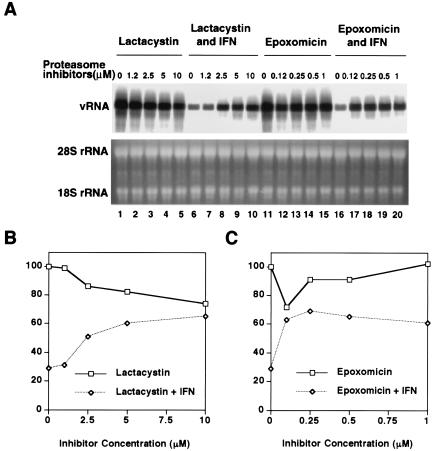
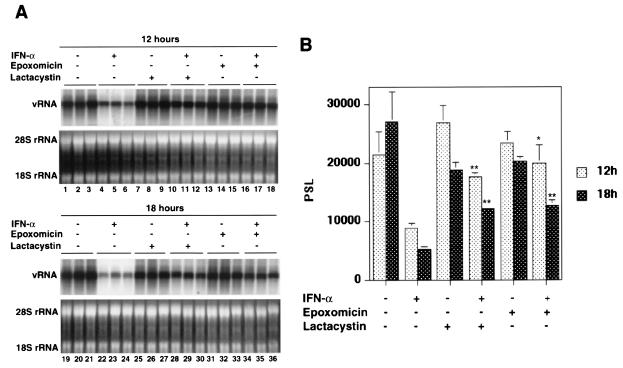
To test this hypothesis, we determined the outcome of combination treatment with IFN-α and the proteasome inhibitors lactacystin and epoxomicin for HCV RNA replication in GS4.1 cells. The cells were pretreated with different concentrations of the inhibitors for 1 h before IFN-α was added for an additional 6 h of combination treatment. Then the cells were incubated for 12 h before RNA was isolated and subjected to Northern blot analysis (Fig. 7A). The relatively short incubation period was necessary because of the known toxicity of proteasome inhibitors after longer incubation times. The results showed that HCV RNA levels dropped 70% within 18 h of IFN-α treatment, whereas in the presence of epoxomicin or lactacystin the reduction was only 30% (Fig. 7B). Lower doses of epoxomicin than of lactacystin were effective, which is consistent with the high specific activity of epoxomicin against the chymotrypsin-like activity of proteasomes (13, 31). Treatment with higher doses of lactacystin alone led to a slight reduction of HCV RNA levels. These results were confirmed with a second set of experiments. The cells were pretreated with 5 μM lactacystin and 1 μM epoxomicin, respectively, for 1 h before IFN-α was added for an additional 6 h of combination treatment. RNA was isolated from the treated cells either 6 or 12 h after incubation with IFN-α (Fig. 8). The results showed that, at both time points, viral RNA levels were significantly higher in cells that were exposed to the proteasome inhibitors than in cells that were treated with IFN-α alone.
FIG. 7.
Dose-dependent inhibition of the IFN-α response against subgenomes by lactacystin and epoxomicin. (A) Cells were incubated with lactacystin and epoxomicin at the indicated concentrations for 7 h and then for an additional 12 h without the drugs. One hour after incubation with the proteasome inhibitors, IFN-α (100 IU/ml) was added for 6 h to a fraction of the cell culture plates (lanes 6 to 10 and 16 to 20). Viral RNA levels were determined by Northern blot analysis. rRNA was used as a control for the amount of RNA present in the samples. (B and C) The amount of viral RNA was measured with a phosphorimager, and values were plotted as percentages of the values obtained with untreated cells.
FIG. 8.
Proteasome inhibitors block the IFN-α response against HCV replicons. (A) GS4.1 cells were left untreated (lanes 1 to 3 and 19 to 21) or treated with 100 IU of IFN-α/ml for 6 h (lanes 4 to 6 and 22 to 24), with 5 μM lactacystin (lanes 7 to 9 and 25 to 27) or 1 μM epoxomicin (lanes 13 to 15 and 31 to 33) alone for 7 h, or with 5 μM lactacystin (lanes 10 to 12 and 28 to 30) or 1 μM epoxomicin (lanes 16 to 18 and 34 to 36) alone for 1 h and then in the presence of 100 IU of IFN-α/ml for an additional 6 h. Cells were harvested at 12 (lanes 1 to 18) and 18 (lanes 19 to 36) h after addition of the cytokine. Viral RNA levels were determined by Northern blot analysis. rRNA was used as a control for the amount of RNA present in the samples. (B) The amount of viral RNA was measured with a phosphorimager and the mean values and standard deviations from three samples were plotted. *, P < 0.05; **, P < 0.01. PSL, arbitrary units.
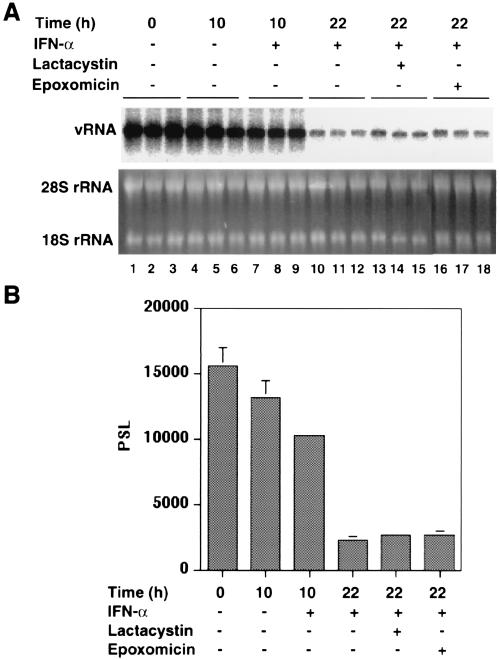
Finally, we sought to determine whether proteasome activity was required for induction of the IFN response or, more directly, for inhibition of HCV replication. To distinguish between these two possibilities, we first incubated GS4.1 cells with the cytokine for 10 h to induce the antiviral response. Then, the cells were incubated for 12 h in the presence of lactacystin or epoxomicin. Under these conditions, the IFN response against HCV remained effective and reduced RNA levels to similar extents independently of the presence of either of the two inhibitors (Fig. 9). Hence, these results indicated that the activity of proteasomes is required for the induction of the IFN response against HCV, but apparently not for direct inhibition of viral replication (see Discussion).
FIG. 9.
Proteasome inhibitors prevent establishment of an IFN-α response against HCV replicons. (A) GS4.1 cells were treated with IFN-α for 10 h, followed by treatment with the indicated proteasome inhibitors for 12 h. Cells were left untreated (lanes 1 to 3 and 4 to 6) or treated with 100 IU of IFN-α/ml for 10 h (lanes 7 to 18), followed by treatment with 10 μM lactacystin (lanes 13 to 15) or 1 μM epoxomicin (lanes 16 to 18) for 12 h. Cells were harvested at 0, 10, and 18 h after the cytokine treatment, as indicated. Viral RNA levels were determined by Northern blot analysis. rRNA was used as a control for the amount of RNA present in the samples. (B) The amount of viral RNA was measured with a phosphorimager and the mean values and standard deviations from three samples were plotted. PSL, arbitrary units.
DISCUSSION
In this study we have investigated the mechanism of the IFN-α response against subgenomic replicons of HCV in Huh7 and HeLa cells. The following conclusions can be drawn from our investigations. First, it can be concluded that IFN-α can inhibit HCV replication by both noncytopathic and cytopathic mechanisms. Our results demonstrating that SL1 cells treated with IFN-α (100 IU/ml) underwent programmed cell death raised the question of whether apoptosis contributes to the rapid decline of HCV RNA levels observed during the first 48 h of IFN therapy (32). The answer depends on whether HeLa or Huh7 cells mimic the scenario in HCV-infected hepatocytes in vivo. It is known from this and other studies that Huh7 cells exhibit an attenuated response to dsRNA and cannot induce an apoptotic program (results not shown) (22, 25, 30). In contrast, HeLa cells respond to dsRNA in a fashion similar to that in which primary chimpanzee hepatocytes respond. For example, treatment of chimpanzee primary hepatocyte cultures with poly(I:C) led to the induction of IFN-β, as shown in this report with HeLa cells (Fig. 6) (25). Therefore, it is possible that HeLa cells represent a more physiological model for hepatocytes in terms of IFN response than Huh7 cells. It was notable that only a fraction of SL1 cells died after treatment with IFN-α. One possibility is that apoptosis was induced in cells that replicated above-average levels of HCV RNA. In support of this possibility, we observed that reduction of viral levels by treatment with heat or HCV RNA polymerase inhibitors reduced the number of apoptotic cells after IFN-α treatment (Fig. 2 and results not shown). Based on our results we concluded that HeLa cells did not undergo apoptosis by default after IFN-α treatment. In fact, it appears that apoptosis is a hallmark of HCV-replicating HeLa cells, because HeLa cells replicating Kunjin virus RNA remained viable after IFN treatment. It will be interesting to determine whether Kunjin virus expresses a gene product that can interfere with apoptosis or whether Kujin virus replication does not induce an antiviral state, as shown for SL1 cells. Finally, to our knowledge, the observation reported here represents the first example of IFN-α-induced apoptosis of cells replicating an apparently noncytolytic RNA virus. Nevertheless, as mentioned above, we cannot yet exclude the possibility that the observed cell death is a property of HCV-expressing HeLa cells. To resolve this problem, it would be necessary to analyze liver biopsy samples from patients receiving IFN therapy.
Second, it can be concluded that the noncytopathic response can occur independently of dsRNA-dependent pathways. Although our results showed that dsRNA response pathways were at least partially functional in normal and HCV-replicating Huh7 cells and were intact in HeLa cells, as indicated by poly(I:C)-induced phosphorylation of eIF-2α and IFN-β gene transcription, viral replication per se did not induce such responses (Fig. 6). The expression of the vaccinia virus E3L protein or treatment of cells with the kinase inhibitor 2-AP had no measurable effect on the IFN response against the HCV replicon (Fig. 5 and Table 1). These observations were consistent with the notion that dsRNA-dependent antiviral pathways, such as PKR and RNase L pathways, were not involved in IFN-induced inhibition of HCV replication in Huh7 cells. Whether they play a role in HeLa cells is not yet known. Efforts to express E3L in SL1 cells were not successful due to the apparent toxicity of the protein, and treatment of HeLa cells with 2-AP for more than 12 h induced apoptosis (results not shown). Importantly, it is not yet known whether IFN-induced apoptosis in HCV-expressing cells is dependent on PKR or other dsRNA-dependent pathways (see below).
In summary, the results showed that, while both Huh7 and HeLa cells were competent to activate dsRNA-dependent signal transduction pathways, HCV replication alone was not sufficient to induce a detectable dsRNA response in these cells. This result could indicate that dsRNA either did not accumulate during HCV replication or was not accessible to PKR and other dsRNA binding proteins.
Third, it can be concluded that HCV replication can induce innate immune pathways. In SL1 and other HCV-expressing HeLa cell lines (results not shown), but not in normal HeLa cells, IFN-α induced the expression of IFN-β. We interpret this result to mean that HCV replication activated an unknown cellular factor, perhaps a viral activated kinase as proposed by Smith and colleagues (42), that, in turn, activated one or more transcription factors required for IFN-β transcriptional activation. Candidates include IRF3, NF-κB, and ATF-2 (Fig. 4). Because normal HeLa cells did not undergo apoptosis after IFN-α treatment, it can be concluded that HCV expression directs the IFN-α response into a cytopathic process. The mechanism responsible for this process remains to be elucidated. Interestingly, Foy et al. reported that in Huh7 cells activation of IRF3 is inhibited by the HCV protease, which indicates that IFN-β activation by poly(I:C) and IFN-α in SL1 cells could have occurred in an IRF3-independent fashion (14).
Fourth, it can be concluded that the antiviral activity of IFN-α against HCV depends, in part, on functional proteasomes. How IFN-induced antiviral programs inhibit viral replication noncytopathically is not yet understood. We demonstrated that, for HCV replication, proteasomes were required for this process. However, our hypothesis that proteasomes were directly involved in inhibition of HCV replication by increasing the turnover of replication complexes or viral proteins was not supported by our results. Instead we obtained evidence that induction of the IFN response was dependent on degradation of one or several proteins. Previously, it has been shown (26) that proteasome inhibitors attenuated the induction of certain IFN-stimulated genes. Because epoxomicin and lactacystin did not inhibit induction of Mx-A (results not shown), which is dependent on activation of the Jak-STAT pathway for the formation of ISGF3, proteasomes might be involved in the induction of the second-wave IFN-stimulated genes. Such a model is consistent with results published previously by Li and Hassel (26), who found that treatment of cells with proteasome inhibitors did not inhibit phosphorylation of STAT1 and binding of ISGF3 to DNA. As a consequence of our results, the number of IFN-induced genes that play a role in inhibition of HCV replication by IFN-α can be reduced to those that are repressed by epoxomicin. Their identity can be revealed with the help of DNA microarrays. Finally, similar to our observations, Robek and colleagues obtained results suggesting that the proteasome is involved in the IFN response against hepatitis B virus (39). However, as with HCV, the exact mechanism by which the proteasome exerts this antiviral activity is not yet known.
An important implication of our results for clinical IFN-α therapy and the pathogenesis of HCV infections is that besides the noncytopathic antiviral effects, IFN-α might also induce apoptosis of HCV-infected hepatocytes. At first glance, this possibility might be discounted because drug-induced cell death could lead immediately to the destruction of the infected liver. However, it is possible that only a fraction of hepatocytes express levels of HCV high enough to activate an apoptotic program in the presence of the high levels of IFN-α that are used for antiviral therapy. In this scenario cell death would occur unnoticed. An important consequence of such a scenario would be that cell killing could play a major role in the recovery from chronic HCV infections, similar to the situation in natural recovery from transient infections with woodchuck hepatitis virus, a model for hepatitis B virus infections (21a).
Acknowledgments
We thank Richard Katz, Glenn Rall, and Robert Silverman for helpful comments on the manuscript and acknowledge services provided by the FCCC tissue culture facility. We thank Curt Horvath, David Lazinski, Alexander Khromykh, Robert Schneider, Bob Silverman, and Jonathan Yewdell for providing reagents that were essential for this study. We acknowledge Ralf Bartenschlager and his colleagues for the communication of unpublished results.
This work was supported by grants from the National Institutes of Health and by an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. [DOI] [PubMed] [Google Scholar]
- 2.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1975. [DOI] [PubMed] [Google Scholar]
- 3.Castelli, J. C., B. A. Hassel, A. Maran, J. Paranjape, J. A. Hewitt, X. L. Li, Y. T. Hsu, R. H. Silverman, and R. J. Youle. 1998. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 5:313-320. [DOI] [PubMed] [Google Scholar]
- 4.Chang, H. W., J. C. Watson, and B. L. Jacobs. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 89:4825-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, T. T., K. C. Young, Y. J. Yang, H. Y. Lei, and H. L. Wu. 1996. Hepatitis C virus RNA in peripheral blood mononuclear cells: comparing acute and chronic hepatitis C virus infection. Hepatology 23:977-981. [DOI] [PubMed] [Google Scholar]
- 6.Cheney, I. W., V. C. Lai, W. Zhong, T. Brodhag, S. Dempsey, C. Lim, Z. Hong, J. Y. Lau, and R. C. Tam. 2002. Comparative analysis of anti-hepatitis C virus activity and gene expression mediated by alpha, beta, and gamma interferons. J. Virol. 76:11148-11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemens, M. J., and A. Elia. 1997. The double-stranded RNA-dependent protein kinase PKR: structure and function. J. Interferon Cytokine Res. 17:503-524. [DOI] [PubMed] [Google Scholar]
- 8.David, M., E. Petricoin III, C. Benjamin, R. Pine, M. J. Weber, and A. C. Larner. 1995. Requirement for MAP kinase (ERK2) activity in interferon alpha- and interferon beta-stimulated gene expression through STAT proteins. Science 269:1721-1723. [DOI] [PubMed] [Google Scholar]
- 9.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Bisceglie, A. M., and J. H. Hoofnagle. 2002. Optimal therapy of hepatitis C. Hepatology 36:S121-S127. [DOI] [PubMed] [Google Scholar]
- 11.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, N. Izumi, F. Marumo, and C. Sato. 1995. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J. Clin. Investig. 96:224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, Y. Ogura, N. Izumi, F. Marumo, and C. Sato. 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 334:77-81. [DOI] [PubMed] [Google Scholar]
- 13.Fenteany, G., and S. L. Schreiber. 1998. Lactacystin, proteasome function, and cell fate. J. Biol. Chem. 273:8545-8548. [DOI] [PubMed] [Google Scholar]
- 14.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 15.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 16.Frese, M., V. Schwarzle, K. Barth, N. Krieger, V. Lohmann, S. Mihm, O. Haller, and R. Bartenschlager. 2002. Interferon-gamma inhibits replication of subgenomic and genomic hepatitis C virus RNAs. Hepatology 35:694-703. [DOI] [PubMed] [Google Scholar]
- 17.Gale, M. J., Jr., M. J. Korth, and M. G. Katze. 1998. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin. Diagn. Virol. 10:157-162. [DOI] [PubMed] [Google Scholar]
- 18.Gale, M. J., Jr., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 19.Goh, K. C., M. J. deVeer, and B. R. Williams. 2000. The protein kinase PKR is required for p38 MAPK activation and the innate immune response to bacterial endotoxin. EMBO J. 19:4292-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goh, K. C., S. J. Haque, and B. R. Williams. 1999. p38 MAP kinase is required for STAT1 serine phosphorylation and transcriptional activation induced by interferons. EMBO J. 18:5601-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Guo, J. T., H. Zhou, C. Liu, C. Aldrich, J. Saputelli, T. Whitaker, M. I. Barrasa, W. S. Mason, and C. Seeger. 2000. Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infections. J. Virol. 74:1495-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keskinen, P., M. Nyqvist, T. Sareneva, J. Pirhonen, K. Melen, and I. Julkunen. 1999. Impaired antiviral response in human hepatoma cells. Virology 263:364-375. [DOI] [PubMed] [Google Scholar]
- 23.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinzie, J. L., P. H. Naylor, M. G. Nathani, R. R. Peleman, M. N. Ehrinpreis, M. Lybik, J. R. Turner, J. J. Janisse, M. Massanari, and M. G. Mutchnick. 2001. African Americans with genotype 1 treated with interferon for chronic hepatitis C have a lower end of treatment response than Caucasians. J. Viral Hepatitis 8:264-269. [DOI] [PubMed] [Google Scholar]
- 25.Lanford, R. E., B. Guerra, H. Lee, D. R. Averett, B. Pfeiffer, D. Chavez, L. Notvall, and C. Bigger. 2003. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(I)-poly(C), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J. Virol. 77:1092-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, X. L., and B. A. Hassel. 2001. Involvement of proteasomes in gene induction by interferon and double-stranded RNA. Cytokine 14:247-252. [DOI] [PubMed] [Google Scholar]
- 27.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 28.Maran, A., R. K. Maitra, A. Kumar, B. Dong, W. Xiao, G. Li, B. R. Williams, P. F. Torrence, and R. H. Silverman. 1994. Blockage of NF-κB signaling by selective ablation of an mRNA target by 2-5A antisense chimeras. Science 265:789-792. [DOI] [PubMed] [Google Scholar]
- 29.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 30.McNair, A. N., D. Cheng, J. Monjardino, H. C. Thomas, and I. M. Kerr. 1994. Hepatitis delta virus replication in vitro is not affected by interferon-alpha or -gamma despite intact cellular responses to interferon and dsRNA. J. Gen. Virol. 75:1371-1378. [DOI] [PubMed] [Google Scholar]
- 31.Meng, L., R. Mohan, B. H. Kwok, M. Elofsson, N. Sin, and C. M. Crews. 1999. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA 96:10403-10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 33.Parisien, J. P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters, K. L., H. L. Smith, G. R. Stark, and G. C. Sen. 2002. IRF-3-dependent, NFκB- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. USA 99:6322-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfeffer, L. M., J. E. Mullersman, S. R. Pfeffer, A. Murti, W. Shi, and C. H. Yang. 1997. STAT3 as an adapter to couple phosphatidylinositol 3-kinase to the IFNAR1 chain of the type I interferon receptor. Science 276:1418-1420. [DOI] [PubMed] [Google Scholar]
- 36.Radkowski, M., J. Wilkinson, M. Nowicki, D. Adair, H. Vargas, C. Ingui, J. Rakela, and T. Laskus. 2002. Search for hepatitis C virus negative-strand RNA sequences and analysis of viral sequences in the central nervous system: evidence of replication. J. Virol. 76:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 38.Rivas, C., J. Gil, Z. Melkova, M. Esteban, and M. Diaz-Guerra. 1998. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2-5A synthetase enzyme. Virology 243:406-414. [DOI] [PubMed] [Google Scholar]
- 39.Robek, M. D., S. F. Wieland, and F. V. Chisari. 2002. Inhibition of hepatitis B virus replication by interferon requires proteasome activity. J. Virol. 76:3570-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlaak, J. F., C. M. Hilkens, A. P. Costa-Pereira, B. Strobl, F. Aberger, A. M. Frischauf, and I. M. Kerr. 2002. Cell-type and donor-specific transcriptional responses to interferon-alpha. Use of customized gene arrays. J. Biol. Chem. 277:49428-49437. [DOI] [PubMed] [Google Scholar]
- 41.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 42.Smith, E. J., I. Marie, A. Prakash, A. Garcia-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava, S. P., K. U. Kumar, and R. J. Kaufman. 1998. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 273:2416-2423. [DOI] [PubMed] [Google Scholar]
- 44.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 45.Tan, S. L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284:1-12. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka, N., M. Sato, M. S. Lamphier, H. Nozawa, E. Oda, S. Noguchi, R. D. Schreiber, Y. Tsujimoto, and T. Taniguchi. 1998. Type I interferons are essential mediators of apoptotic death in virally infected cells. Genes Cells 3:29-37. [DOI] [PubMed] [Google Scholar]
- 47.Taniguchi, T., and A. Takaoka. 2002. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14:111-116. [DOI] [PubMed] [Google Scholar]
- 48.tenOever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76:3659-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams, B. R. 3 July 2001, posting date. Signal integration via PKR. Sci STKE 2001: RE2. [Online.] http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2001/89/re2. [DOI] [PubMed]
- 50.Zhu, Q., J.-T. Guo, and C. Seeger. 2003. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 77:9204-9210. [DOI] [PMC free article] [PubMed] [Google Scholar]