Abstract
The activation of most paramyxovirus fusion proteins (F proteins) requires not only cleavage of F0 to F1 and F2 but also coexpression of the homologous attachment protein, hemagglutinin-neuraminidase (HN) or hemagglutinin (H). The type specificity requirement for HN or H protein coexpression strongly suggests that an interaction between HN and F proteins is required for fusion, and studies of chimeric HN proteins have implicated the membrane-proximal ectodomain in this interaction. Using biotin-labeled peptides with sequences of the Newcastle disease virus (NDV) F protein heptad repeat 2 (HR2) domain, we detected a specific interaction with amino acids 124 to 152 from the NDV HN protein. Biotin-labeled HR2 peptides bound to glutathione S-transferase (GST) fusion proteins containing these HN protein sequences but not to GST or to GST containing HN protein sequences corresponding to amino acids 49 to 118. To verify the functional significance of the interaction, two point mutations in the HN protein gene, I133L and L140A, were made individually by site-specific mutagenesis to produce two mutant proteins. These mutations inhibited the fusion promotion activities of the proteins without significantly affecting their surface expression, attachment activities, or neuraminidase activities. Furthermore, these changes in the sequence of amino acids 124 to 152 in the GST-HN fusion protein that bound HR2 peptides affected the binding of the peptides. These results are consistent with the hypothesis that HN protein binds to the F protein HR2 domain, an interaction important for the fusion promotion activity of the HN protein.
Infection by the paramyxovirus Newcastle disease virus (NDV) requires two glycoproteins, the attachment protein and the fusion protein (F protein) (reviewed in reference 11). The attachment protein, hemagglutinin-neuraminidase (HN), binds to sialic acid-containing receptors, and the F protein directs membrane fusion between the viral and the cellular membranes. Unlike many viral F proteins, paramyxovirus F proteins do not require the acid pH of endosomes for the activation of fusion activity; thus, other mechanisms for F protein activation must be invoked (7). Because of this acid pH independence, infected cells expressing both the HN and the F proteins can fuse with adjacent cells to form multinuclear cells or syncytia, a process that is assumed to be similar to virus-cell fusion (reviewed in reference 11).
The NDV F protein is synthesized as a precursor, F0 (11), that must be proteolytically cleaved to activate F protein fusion activity. Cleavage at amino acid 117 produces disulfide-linked F2 and F1 polypeptides derived from the amino-terminal and carboxyl-terminal domains of F0, respectively (reviewed in reference 11). The F1 polypeptide has one and perhaps two fusion peptides (18, 19). Upon initiation of fusion, fusion peptides are thought to insert into target membranes, docking the protein to these membranes (reviewed in references 6, 7, and 19). Paramyxovirus F1 polypeptides have two heptad repeat (HR) regions, one (HR1) located just carboxyl terminal to the more amino-terminal fusion peptide and the other adjacent to the transmembrane domain (HR2) (reviewed in reference 11). Studies of peptides with sequences of these HR domains (9, 12, 22, 38, 40, 41), characterization of mutations within these domains (16, 23, 24, 27), and results of similar studies of human immunodeficiency virus and influenza virus (6) have led to the hypothesis that F proteins are synthesized and transported to cell surfaces in a metastable conformation in which the HR domains are not associated and the fusion peptides are masked (6, 19, 37). Upon fusion activation, F proteins are thought to undergo a series of conformational changes that result in the insertion of fusion peptides into target membranes and the interaction of the HR1 and HR2 domains to form a very stable complex (1). The formation of this complex is thought to pull target and attack membranes in close proximity, allowing subsequent fusion events (1, 42).
In most paramyxovirus systems, F protein cleavage is not sufficient to activate these conformational changes in the F protein; rather, coexpression of the attachment protein is also required (8, 11). The HN protein provides more than an attachment or membrane-docking function, since mutants with mutations in the HN protein can retain attachment activity but are defective in fusion promotion (21, 25, 26, 28). It has been proposed that the attachment of the HN protein to its receptor serves to activate the F protein (11), but the nature of this activation is poorly understood. The HN protein and the F protein must be from the same virus, with a few exceptions, indicating that virus-specific interactions between HN and F proteins (8) are required for fusion directed by the F protein. Indeed, an interaction has been shown in several systems (4, 13, 20, 29, 39). It seems likely that this interaction is somehow linked to F protein activation.
The domain of the HN protein that interacts with the F protein has not been directly identified, but studies of hybrid HN proteins by three different laboratories have all implicated the membrane-proximal ectodomain in virus-specific fusion promotion activity (5, 31, 36). With this approach, the specificity for the NDV F protein has been mapped to amino acids 55 to 141 of the NDV HN protein, a sequence adjacent to the HN protein transmembrane domain of this type 2 glycoprotein (5). Because the F protein HR2 domain is adjacent to the transmembrane domain and because the HR2 domain should remain uncomplexed with the HR1 domain prior to activation, we examined whether there was a specific interaction between peptides with sequences from the F protein HR2 domain and the HN protein membrane-proximal ectodomain. We report a specific interaction between the F protein HR2 domain and an HN protein domain from amino acids 124 to 152. That this region of the HN protein is important for fusion promotion is shown by the properties of mutant proteins with alterations in this domain.
MATERIALS AND METHODS
Cells and vectors.
Cos-7 cells, obtained from the American Type Culture Collection, were maintained in Dulbecco's modified Eagle's medium supplemented with nonessential amino acids, vitamins, penicillin-streptomycin, and 10% fetal calf serum. The NDV HN and F protein genes were expressed in Cos-7 cells by using pSVL (Pharmacia) as previously described (27).
Peptides.
Peptide HR2a-B corresponds to a 20-amino-acid segment (amino acids 477 to 496) of the HR2 domain of the F protein (Fig. 1A) (40). Peptide HR2b-B corresponds to a 40-amino-acid segment (amino acids 452 to 491) of the HR2 domain of the F protein (Fig. 1A). Both peptides were obtained from Tufts University School of Medicine Peptide Core Facility. Both peptides included three glycine residues at the amino terminus of the NDV sequence, and a biotin residue was covalently linked to the amino terminus of each peptide. The peptides were purified at the Tufts University School of Medicine Peptide Core Facility by high-pressure liquid chromatography.
FIG. 1.
HN and F protein peptide sequences. (A) Positions in the intact F protein of sequences represented by two synthetic peptides, HR2a and HR2b. The top line diagrams the membrane-proximal ectodomain of the F protein. The position of the transmembrane domain (TM) is indicated by the hatched box. The sequences included by peptide HR2a-B (amino acids 477 to 496) and by peptide HR2b-B (amino acids 452 to 491) are indicated. (B) Positions in the HN protein of sequences cloned in frame at the carboxyl terminus of the GST fusion protein. The top line represents the HN protein membrane-proximal ectodomain. The position of the TM is indicated by the hatched box. The HN protein sequences in different GST fusion proteins are shown below. Position A corresponds to amino acid 49, while position B corresponds to amino acid 152. The numbers in the B series of GST fusion proteins refer to amino acid at the beginning of the HN protein sequence in each GST fusion protein. The numbers in the A series of GST fusion proteins refer to the amino acid at the end of the HN protein sequence.
GST-HN fusion proteins.
Glutathione S-transferase (GST) fusion protein genes were constructed with pET-42a(+) (Novagen). HN protein sequences inserted at the carboxyl terminus of the coding region for GST were generated by PCRs with start primers containing a BamHI site and end primers containing a translation stop codon followed by an EcoRI site. Start primers were as follows: START A, GCATATGGATCCGAGGCTAGCACA; START 65, AGCATACCAACTGCGGGATCCAGGGCA; START 77, TCTGCACTCGGATCCAATCAG; START 97, TGAATCTCCGTTGGGATTCCTAAA; and START 124, AACAGCGGATCCGGGGCACCTGTT. End primers were as follows: End B, TTCTTGGAATTCAGATCAATAGAATGA; End 76, CCTATCTACGAATTCCTGCTAGGAACCGAG; End 93, GATTCGGTGAATTCCAATGCCTACGGAGAT; End 118, CCCACACCCGAATTCTCATGCAGCTCC; and End 131, GTTCTTTACCGAATTCCCCGATCTAATCTGG. START A is designed to link amino acid 49 in the HN protein sequence to the carboxyl terminus of GST. End B corresponds to amino acid 152 in the HN protein sequence. Other numbers in the start and end primers refer to amino acids in the HN protein sequence. PCR products were ligated to BamHI/EcoRI-cut pET-42a(+) DNA and transformed into HB101 competent cells (Gibco/BRL/Life Technologies). GST-HN fusion protein sequences were verified by DNA sequencing. The plasmids were then introduced into BL21(DE3) (Novagen) for the preparation of fusion proteins.
GST-HN fusion proteins were prepared according to protocols specified by Novagen. Briefly, a log-phase culture of BL21(DE3) was induced to express the GST protein by exposure to isopropyl-β-d-thiogalactopyranoside (IPTG) (final concentration, 0.4 mM) for 2 h. The bacteria were collected by centrifugation, resuspended in gel sample buffer (17% glycerol, 0.1 M Tris-HCl [pH 6.8], 0.06% bromophenol blue, 5% sodium docecyl sulfate, 1 M β-mercaptoethanol), and boiled.
Peptide binding to GST-HN fusion proteins.
Extracts of bacteria expressing GST fusion proteins were diluted in gel sample buffer and loaded onto 10% polyacrylamide gels. After electrophoresis, the gels were equilibrated in transfer buffer (25 mM Tris, 192 mM glycine, 5% methanol [pH 8.2]) and transferred to Immobilon P (Millipore Corp.) membranes by electrophoresis. The membranes were blocked in phosphate-buffered saline (PBS) containing 0.5% Tween 20 (PBS-Tween 20) and 10% nonfat dried milk for 2 h at room temperature or overnight at 4°C. The membranes were washed in PBS-Tween 20 and incubated with either HR2a-B peptide or HR2b-B peptide (7 μg/ml) diluted in PBS-Tween 20 containing 0.5% nonfat milk for 1 h at room temperature. The membranes were washed and then incubated with Neutravidin that was coupled to horseradish peroxidase (5 μg/ml) (Pierce Corp.) and that was diluted in PBS-Tween containing 0.5% nonfat milk for 1 h at room temperature. The membranes were washed extensively, and bound peptides were detected by using an enhanced chemiluminescence Western blotting detection reagent system (Amersham).
For experiments with undenatured, purified fusion proteins, GST-HN fusion proteins were purified from bacterial cell extracts by using glutathione affinity columns (Novagen).
Site-specific mutagenesis.
The HN protein gene mutants were generated by PCR with the appropriate oligomer for each mutation. Amino acid 133 was changed from an isoleucine to a leucine, and amino acid 140 was changed from a leucine to an alanine. The forward primer, SEB 21 (AACTGCTCCTCAGTGGATGTTGC), bound to the multiple cloning cassette in the vector. Reverse primers contained the mutation and an SpeI site: I133L, TCACTAGTATCATCCACAATAAGTTCTTTACCTATCCCCCCGAGGTAATCTGG; and L140A, TCACTAGTATCATCCACAATTGCTTCTTTACC. PCR products were digested with XhoI/SpeI, and the 1.25-kb product was gel purified. The HN protein gene was reassembled in pSVL by using the XhoI/SpeI PCR product and the SpeI/SpeI 4.9-kb fragment corresponding to the 3′ end of the HN protein gene.
The genes of the mutant DNAs were sequenced in their entirety to verify the proper orientation of the SpeI/SpeI fragment and to determine that the rest of the gene remained unchanged by the mutagenesis reaction. Mutants are named in the single-letter code, and the names indicate the amino acid in the wild type, the position of the change, and the amino acid in the mutant.
Transfections.
Transfections with Lipofectamine (Gibco/BRL/Life Technologies) were done essentially as recommended by the manufacturer. Cos-7 cells were plated at 3 × 105 per 35-mm plate. At 20 to 24 h later, the cells were transfected. For each 35-mm plate, a mixture of DNA (0.5 to 1.5 μg, depending upon the experiment) in 0.1 ml of OptiMem (Gibco/BRL/Life Technologies) and 10 μl of transfection reagent in 0.2 ml of OptiMem was incubated at room temperature for 40 min, diluted with 0.7 ml of OptiMem, and added to a plate that had been washed twice with 2 ml of OptiMem. The cells were incubated for 5 h, DNA-Lipofectamine was removed, and 2 ml of supplemented Dulbecco's modified Eagle's medium was added.
Flow cytometry.
Cells transfected with 0.75 μg of plasmid DNA/35-mm plate for 48 h were removed from the plates after trypsin (50 μg/ml) digestion, washed in PBS containing 1% bovine serum albumin and 0.02% azide (fluorescence-activated cell sorting [FACS] buffer), and incubated with anti-NDV antibody (1:2,500 dilution) for 1 h on ice. After being washed three times with FACS buffer, the cells were incubated for 1 h on ice with goat anti-rabbit immunoglobulin G coupled to Alexa dye 488 (Molecular Probes) (1:1,400 dilution). After being washed three times with FACS buffer, the cells were resuspended in PBS containing 2% paraformaldehye and subjected to flow cytometry (University of Massachusetts Medical School Flow Cytometry Facility). The background was cells transfected with vector alone and incubated with both primary and secondary antibodies.
Fusion assays. (i) Syncytium formation.
Cos-7 cells were cotransfected with wild-type or mutant HN protein genes (0.75 μg/plate) and the wild-type F protein gene (0.75 μg/plate). At 24 h posttransfection, the cells were removed from the plates with trypsin (Gibco/BRL/Life Technologies), mixed with twice the number of untransfected Cos-7 cells, and replated at 5 × 105 cells/35-mm plate. The number of nuclei in 40 fusion areas was counted to determine the average size of the syncytia at each time point as previously described (26). Values obtained after transfection with the vector alone were subtracted.
(ii) Content mixing.
Content mixing was measured by using modifications of a previously described protocol (14). Briefly, a plasmid encoding a tetracycline-responsive transcriptional activator, tTA (Clontech), was transfected (1 μg/35-mm plate) with pSVL-HN (0.75 μg/35-mm plate) and pSVL-F (0.75 μg/35-mm plate) DNAs. A separate population of cells was transfected 24 h later with a plasmid encoding the β-galactosidase protein under the control of the tetracycline-responsive transcriptional activator (Clontech) (1 μg/35-mm plate). After 20 h, the cells transfected with the plasmid encoding the β-galactosidase protein were removed from the plate with trypsin and placed on top of the HN protein- and F protein-expressing cells. At 47 h posttransfection of the HN protein- and F protein-expressing cells, when fusion was evident, the monolayers were lysed (Promega cell lysis buffer), and extracts were assayed for β-galactosidase activity. Activity due to background fusion typical of Cos-7 cells was measured after transfection of cells with comparable amounts of the vector alone. Values obtained were subtracted from values obtained with cells expressing wild-type or mutant HN proteins and wild-type F protein.
RESULTS
GST-HN fusion proteins with sequences from the membrane-proximal domain of the HN protein.
Sequences encoding various lengths of the membrane-proximal region of the NDV HN protein, from amino acids 49 to 152, were inserted in frame at the carboxyl terminus of the GST protein sequence as described in Materials and Methods (Fig. 1B). This region of the ectodomain of the HN protein was chosen for two reasons. First, mutational analyses of the NDV HN protein, from amino acids 88 to 110, have shown this region to be important for fusion promotion activity (28). Second, a hybrid HN protein with sequences from the NDV HN protein and the PIV3 HN protein has implicated this region in type-specific interactions with the NDV F protein (5). The GST fusion protein containing the sequence from the entire membrane-proximal domain was designated GST-AB (Fig. 1). This amino acid sequence contains a cysteine residue (amino acid 123) which could interfere with the formation or recovery of the fusion protein. Thus, a second fusion protein in which the cysteine codon was replaced with a tryptophan codon was also produced (GST-AB-W). Tryptophan was chosen as the replacement codon since some strains of NDV encode an HN protein with tryptophan at this position.
Other GST-HN fusion proteins were also produced with various sequences from this region of the HN protein (Fig. 1). GST-HN fusion proteins with overlapping deletions of the amino-terminal region of the AB sequence were made (GST-65B, GST-77B, GST-97B, and GST-124B) (numbers in each fusion protein refer to the amino acid at the beginning of the HN protein-specific sequence). In addition, GST-HN fusion proteins with overlapping deletions of the carboxyl-terminal region of the AB sequence were made (GST-A131, GST-A118, GST-A93, and GST-A76) (numbers in each fusion protein refer to the amino acid at the end of the HN protein-specific sequence).
Interactions of HR2 peptides with GST-HN fusion proteins.
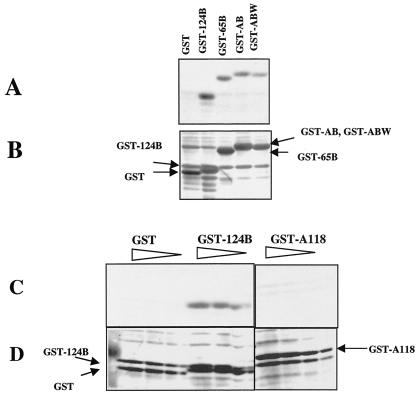
Peptides with sequences from the HR2 domain were chemically synthesized (Fig. 1A). Peptide HR2a-B contains residues from amino acids 477 to 496 of the F protein. Peptide HR2b-B contains residues from amino acids 452 to 491 of the F protein and includes more of the HR2 sequence. Peptide HR2a-B was previously shown to bind specifically to a peptide with a sequence from the HR1 domain of the NDV F protein but not to nonspecific peptides (41). To determine whether this peptide also had an affinity for the membrane-proximal domain of the HN protein, a blot containing the GST-AB fusion protein was incubated with peptide HR2a-B, and binding was detected by using biotin covalently coupled to the peptide. Figure 2A shows that peptide HR2a-B does bind to the GST-AB fusion protein (lane 4) but does not bind to GST without the HN protein sequence (lane 1). Figure 2A also shows that the peptide binds to GST-AB-W, although not as well as to GST-AB. In addition, the peptide binds to GST fusion proteins missing different amounts of the AB amino-terminal sequence. The GST-65B fusion protein is missing 16 amino acids present in GST-AB, but its binding is similar to that detected for GST-AB. GST-124B, which contains only amino acids 124 to 152, binds to the peptide even better than GST-AB. Figure 2B shows the same blot stained with Coomassie blue. The amounts of GST and GST fusion proteins in the blot are similar; thus, binding cannot be accounted for by different amounts of fusion proteins in the blot.
FIG. 2.
Binding of peptide HR2a-B to GST fusion proteins. (A and C) Binding of peptide HR2a-B to GST fusion proteins containing HN protein sequences. Binding was detected by using the biotin label on the peptide as described in Materials and Methods. (B and D) Total protein on blots shown in panels A and C, respectively, as detected by Coomassie blue staining. (A and B) Equivalent amounts of GST fusion proteins (indicated at the top of panel A and on the side of panel B), as determined by preliminary analysis of Coomassie blue-stained gels containing cell extracts, were loaded in the lanes. (C and D) Different relative amounts (5×, 3×, 2×, and 1×) of GST fusion proteins (indicated at the top of panel C and on the side of panel D) were loaded in the lanes.
In initial experiments, purified, nondenatured GST fusion proteins were spotted on Immobilon P, and the blots were incubated with peptide HR2a-B as previously described for the detection of an interaction of this peptide with an F protein HR1 peptide. The results obtained with this protocol were identical (data not shown) to those shown in Fig. 2, in which GST fusion proteins were denatured prior to transfer to Immobilon P. GST-124B bound HR2a-B, while GST-A118 did not.
The results shown in Fig. 2A suggest that the sequences in the GST-124B fusion protein are sufficient for the binding of peptide HR2a-B. To determine whether the peptide will also bind to sequences more amino terminal to the 124B sequence, the binding of peptide HR2a-B to GST-124B was compared to its binding to GST-A118 (Fig. 2C and D). The peptide did not bind to GST-A118.
The blot shown in Fig. 2C and D contained a fivefold range of concentrations of fusion proteins in order to determine whether binding was dependent upon protein concentrations. The results, quantitated by densitometry, are shown in Fig. 3A. Clearly, peptide binding to GST-124B increased with increasing amounts of the fusion protein, while no binding to either GST or GST-A118 was detected at any protein concentration. Similar results were obtained with a longer HR2 peptide, HR2b-B. No binding was detected to GST-A118, while the peptide bound to GST-124B (Fig. 3B). In addition, the degrees of binding of both HR2 peptides to GST-124B were virtually identical.
FIG. 3.
HR2 peptides bind only to HN protein sequences from amino acids 124 to 152. The binding of peptides HR2a-B and HR2b-B to GST fusion proteins containing only the amino-terminal (GST-A118) or only the carboxyl-terminal (GST-124B) sequences of the membrane-proximal domain of the HN protein was evaluated. The binding of each peptide to a fivefold or fourfold range of GST fusion protein concentrations in blots similar to those shown in Fig. 2C was quantitated by densitometry. (A) Results of binding of peptide HR2a-B to GST-124B and to GST-A118. (B) Results of binding of peptide HR2b-B to GST-124B and to GST-A118. Identical results were obtained in three separate experiments.
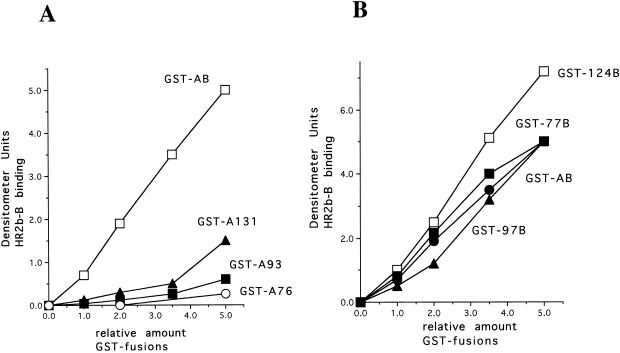
These results implicate amino acids 124 to 152 in HR2 binding. To verify this conclusion, GST fusion proteins with overlapping deletions of the carboxyl-terminal AB sequence were tested for interactions with peptides HR2a-B and HR2b-B. Blots similar to those shown in Fig. 2C were quantitated; the results obtained with peptide HR2b-B are shown in Fig. 4A. Loading of the GST fusion proteins was quantitated by Coomassie blue staining. Clearly, only GST-A131 showed any binding to the peptide. This fusion protein contains a short sequence in common with the GST-124B fusion protein, suggesting that it is this sequence or a sequence between amino acids 118 and 131 that is responsible for the slight binding detected. Identical results were obtained with peptide HR2a-B (data not shown).
FIG. 4.
Binding of HR2b-B to the HN protein membane-proximal sequence with overlapping deletions. Blots similar to those shown in Fig. 2C were incubated with HR2b-B, and binding was quantitated as described in the legends to Fig. 2 and 3. (A) Results of binding to the HN protein membrane-proximal sequence with overlapping deletions of the carboxyl-terminal region compared to binding to a GST fusion protein containing the entire membrane-proximal sequence (GST-AB). (B) Results of binding to the membrane-proximal sequence with overlapping deletions of the amino terminus compared to binding to a GST fusion protein containing the entire membrane-proximal sequence (GST-AB). Identical results were obtained in three separate experiments.
Similarly, the binding of peptide HR2b-B to GST fusion proteins with overlapping deletions of the amino-terminal region of the AB sequence was evaluated. Binding of the peptide to GST-77B and GST-97B was virtually the same as binding to GST-AB. Interestingly, the level of binding of the peptide to GST-124B was higher than that to the longer GST fusion proteins. Perhaps the conformation of the HN protein sequence in the longer GST fusion proteins in the blot was slightly different from that in the GST-124B fusion protein, decreasing the affinity of the peptide for the 124B sequence present in the longer GST fusion proteins.
GST fusion proteins with sequences past amino acid 152 did not show increased binding to either peptide (data not shown). GST fusion proteins containing sequences more amino terminal to amino acid 49 were not made because of potential difficulties with the hydrophobicity of this transmembrane domain sequence.
Fusion promotion activities of HN proteins with mutations in the 124B region.
The results described above implicate the HN protein domain from amino acids 124 to 152 in interactions with the F protein HR2 domain. However, no mutational analyses of this region of the HN protein have been reported; thus, it is difficult to assess the significance of these results to the fusion promotion activity of the intact protein. To determine whether this region of the HN protein is indeed involved in fusion promotion activity, mutant proteins with single amino acid changes at two different amino acids in the middle of this region were characterized for their fusion promotion activities. The isoleucine residue at amino acid 133 was changed to leucine and the leucine residue at amino acid 140 was changed to alanine to produce HN-I133L and HN-L140A. Since the only residues in the HR2 domain that influence fusion are those in the leucine zipper motif (16, 23), the I133 and L140 residues were chosen as likely candidates for interactions with this motif. The two mutant proteins were expressed at wild-type levels (data not shown) and were expressed on cell surfaces at levels comparable to that of the wild type (Table 1). The attachment activities and neuraminidase activities of the proteins are also shown in Table 1. The neuraminidase activities of the mutants were 41 and 57% that of the wild type, while the attachment activities, as measured by red blood cell binding, were 144 and 50% that of the wild type.
TABLE 1.
Properties of mutant proteins
| DNA | Mean ± SD % wild-typea
|
||
|---|---|---|---|
| Surface expressionb | Activity
|
||
| Attachmentc | Neuraminidased | ||
| HN wild type | 100 | 100 | 100 |
| HN-I133L | 127 ± 18 | 144 ± 21 | 41 ± 5 |
| HN-L140A | 118 ± 13 | 50 ± 4 | 57 ± 13 |
Values were from at least three separate experiments.
Determined by FACS analysis as described in Materials and Methods.
Defined as the binding of avian red blood cells to monolayers of cells transfected with wild-type or mutant HN cDNAs. Binding was quantitated by measuring the hemoglobin released after lysis of the attached red blood cells.
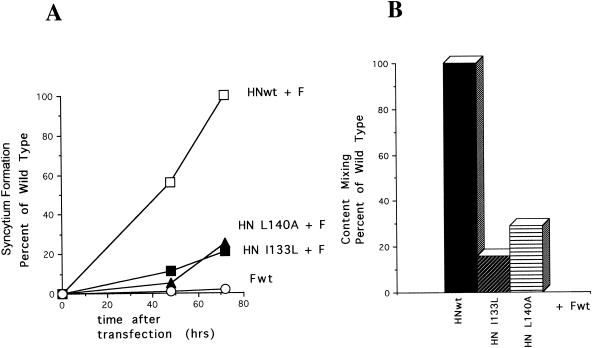
These mutations affected syncytium formation (Fig. 5A). Interestingly, these mutant proteins could mediate some syncytium formation, but the kinetics of formation of the syncytia were considerably slower than those of the wild type. The fusion promotion activities of these mutant proteins were also determined by content mixing (Fig. 5B). This measure of the fusion promotion activities of the mutant HN proteins also indicates a defect in these activities of the proteins. These combined results indicate that alterations in this region of the HN protein can affect the fusion promotion activities of the protein.
FIG. 5.
Fusion promotion activities of HN protein mutants. (A) Kinetics of syncytium formation directed by wild-type F protein (Fwt) in the presence of wild-type HN protein (HNwt), HN-I133L, and HN-L140A. Identical results were obtained in two separate experiments. (B) Fusion promotion activities of mutant HN proteins as measured by content mixing as described in Materials and Methods. The values obtained with wild-type HN protein were set at 100%. Identical results were obtained in three separate experiments.
HR2 peptide binding to mutant GST fusion proteins.
The fusion activities of the mutant HN proteins could be due to differences in binding to the HR2 domain of the F protein. To determine whether the amino acid mutations affect the binding of the GST-124B sequence to HR2 sequences, peptide HR2b-B was incubated with blots containing increasing amounts of the GST-124B fusion protein with changes at positions corresponding to amino acids 133 and 140. Binding to the resulting blots was quantitated as described in the legends to Fig. 2 and 3; the results are shown in Fig. 6. GST-124B with an I-to-A change at position 133 and GST-124B with an L-to-A change at position 140 both showed significantly less binding to the peptide than wild-type GST-124B. Thus, amino acid changes in the HN protein domain that interacts with HR2 sequences can affect binding to the HR2 peptide. Thus, the phenotype of intact mutant HN proteins could be accounted for by altered interactions with the HR2 domain of the intact F protein.
FIG. 6.
Binding of HR2 peptides to HN sequences with single amino acid changes. Different concentrations of GST, GST-124B, GST-124B-I133L, and GST-124B-L140A fusion proteins were blotted on Immobilon P as described in the legend to Fig. 2. Peptide HR2b-B was added to the blot, and binding was detected as described in the legend to Fig. 2 and Materials and Methods. The amount of binding was quantitated as described in the legends to Fig. 3 and 4. Results are the averages of three separate experiments.
DISCUSSION
Fusion mediated by most paramyxovirus F proteins requires not only proteolytic cleavage of F0 but also coexpression of the viral attachment protein (11). The attachment activity of the protein is necessary but not sufficient. Based on the original observation by Hu and Compans that the requirement for HN protein in fusion is virus specific (8), it has been proposed that there is a type-specific interaction between the paramyxovirus HN and F proteins necessary for fusion, an interaction that is likely related to F protein activation (10, 11).
Because of properties of hybrid HN proteins with sequences from two different HN proteins, it seemed likely that the HN protein membrane-proximal ectodomain is involved in the interaction with F protein (5, 31, 36). The F protein interacting domains are not clear. However, given the proximity of the HN domain implicated to the membrane, it seemed reasonable to hypothesize that a membrane-proximal domain of the F protein may be involved in HN protein interactions. The HR2 domain is a likely candidate since this domain is adjacent to the F protein TM domain. Furthermore, a complex of HR2 with the HN protein membrane-proximal domain would serve to prevent HR1-HR2 complex formation until fusion activation. We, therefore, asked if the F protein HR2 domain could complex with a sequence from the HN protein membrane-proximal domain.
Using synthetic peptides, we demonstrated an interaction between the F protein HR2 sequences and sequences derived from the membrane-proximal domain of the HN protein. This is the first report of a direct identification of an F protein domain that may be involved in HN protein interaction. There is one report characterizing domains in hybrid F proteins that may be involved in type-specific interactions with HN proteins (35). In this study, numerous domains were implicated including the HR2 domain. Another recent report provides indirect evidence for an interaction of a peptide with the Sendai virus F protein HR2 sequence to Sendai virus HN protein (33).
Using overlapping deletions of the HN protein sequence we found a specific interaction between the HR2 peptides and sequence from amino acids 124 to 152 of the HN protein sequence but little interaction with more amino-terminal sequences. That the sequence from amino acids 124 to 152 is involved in binding to an F sequence was not surprising since HN protein chimeras containing NDV HN protein membrane-proximal sequence must include amino acid 141 to confer NDV F protein specificity (5). It was surprising, however, that amino acids 49 to 118 did not bind the HR2 sequences. Mutation of this region, between amino acids 80 and 110, of the HN protein did eliminate the fusion promotion activity of the protein (28). Perhaps the domain from amino acids 80 to 110 of HN protein affects the conformation of the region from amino acids 124 to 152 or this region interacts with another F protein domain.
The region from amino acids 124 to 152 of the HN protein has not been previously characterized by mutation. Therefore, to determine directly if this domain in the intact HN protein has a role in fusion promotion, point mutations were made at amino acids 133 and 140. Indeed, both mutations affected fusion promotion activity without significantly affecting the surface expression. Furthermore, mutant I133L had attachment activity higher than that of the wild type, and so a lack of attachment activity cannot account for the phenotype of this mutant. Mutant L140A had approximately 50% the attachment activity of the wild type, a decrease that could not totally account for the decrease in fusion promotion. The neuraminidase activities of the mutant proteins were decreased by approximately 60 and 30%. It is unlikely that this decrease could directly account for the decrease in fusion promotion since neuraminidase activity may be significantly reduced without affecting fusion promotion (2, 25). These combined results suggest the region from amino acids 124 to 152 is involved in fusion promotion. Furthermore, these amino acid alterations affected the binding of the HR2 peptide to this HN protein sequence as measured by the binding of the HR2 peptide to GST fusion proteins. Thus, it is possible that altered interaction between the HR2 domain of the F protein and the HN protein could account for defective fusion promotion activities of the mutant HN proteins.
Our results do not rule out the possibility that other regions of both the HN and F proteins interact. Indeed, a hybrid HN protein containing only amino acids 55 to 141 of the NDV HN protein did not work with the NDV F protein (5). Adding the NDV TM domain or separately adding the globular head domain was necessary for type-specific fusion promotion. This result would suggest that while the membrane-proximal domain may be necessary it is not sufficient for a type-specific interaction necessary for fusion. Other reports also implicate more carboxyl-terminal sequences in the globular head domain of the HN protein in type-specific interactions (31, 36).
Two models have been proposed for the role of the HN protein-F protein interaction, models that differ in the role of attachment in inducing conformational changes in F protein required for the onset of fusion (10, 15, 26, 30). One model proposes that HN and F proteins interact only after HN protein receptor binding and this interaction initiates F protein conformational changes required for fusion (10, 17). An alternative model is that HN and F proteins form a metastable complex prior to HN protein attachment (29, 32, 34). Such a complex would prevent HR1-HR2 interactions that would form in F proteins expressed without HN protein. In this second model, HN protein attachment with a concomitant conformational change would release the F protein stimulating the cascade of F protein conformational changes required for fusion, changes that result in the formation of the six stranded coiled coil composed of HR1 and HR2 domains. Both models include the idea that the HN protein undergoes a conformational change upon attachment. Based on the reactivity of cell surface F protein to a peptide antibody specific for the HR1 domain, we have presented evidence consistent with the second model (15) as has Takimoto et al. (30). Our finding of an interaction between the HN protein membrane-proximal domain and the F protein HR2 domain is also consistent with this second model.
The crystal structure of the NDV HN protein has been solved by Crennell et al. (3). Remarkably, Crennell et al. reported two different conformational forms of the HN protein, one was crystallized at pH 4.5 with an empty catalytic site (Fig. 7A), while the other was crystallized at more neutral pH and contained a filled site (Fig. 7B). Crennell et al. have proposed that the two forms represent alternative conformations of the HN protein, one pre attachment and the other postattachment. They have suggested that the molecule undergoes a conformational transition upon attachment and cleavage of sialic acid, a transition that may be linked to fusion activation. While the membrane-proximal domain from amino acids 49 to 123 was missing from these structures due to the method of solubilization of material for crystallization, the domain from amino acids 124 to 152 was present in the structure and formed part of the globular head domain. Interestingly, the domain implicated here in binding to F protein HR2 (Fig. 7, dark blue residues) is in a very different orientation in the molecule in the two forms of HN protein. In the structure with an empty catalytic site, the domain lays along the side of the molecule (Fig. 7A and B) while in the structure with a filled site, the domain is located on the underside of the globular head domain (Fig. 7C and D). Movement from one form to the other may serve to alter the association of this domain from the F protein HR2 domain, activating the F protein.
FIG. 7.
Location of the 124B domain in the two conformations of the HN protein. Shown are space-filling representations of the two forms of NDV HN protein as determined by Crennell et al. (3). The representations were constructed by using the Protein Explorer Program (University of Massachusetts) with the coordinates reported by Crennell et al. (1e8t and 1e8v). The structures are colored according to the positions within the sequences, with dark blue representing the most amino-terminal sequences and red representing the most carboxyl-terminal sequences. Selected amino acids in the region from amino acids 124 to 152 are indicated. (A and C) Side views of the two forms of the HN protein dimer. (B and D) Underside (membrane-proximal side) of the dimer.
It must be noted, however, that the NDV HN protein used for determination of structure was from a strain of NDV that does not form disulfide-linked HN protein dimers. In other strains of NDV, including strain AV used here, a disulfide bond covalently links cysteine residues at position 123. As noted by Crennell et al. (3), in the structure of the attachment-ready form (Fig. 7A and B), the residues at position 123 in adjacent molecules are too far apart to form a disulfide linkage. This observation raises the possibility that the prebinding form of the HN protein from the AV strain is not in precisely the conformation shown in Fig. 7A and B. However, the domain may well be in a conformation different from that in the structure with a filled catalytic site.
Acknowledgments
This work was supported by grant AI 30572 from the National Institutes of Health.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 2.Corey, E. A., A. M. Mirza, E. Levandowsky, and R. M. Iorio. 2003. Fusion deficiency induced by mutations at the dimer interface in the Newcastle disease virus hemagglutinin-neuraminidase is due to a temperature-dependent defect in receptor binding. J. Virol. 77:6913-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068-1074. [DOI] [PubMed] [Google Scholar]
- 4.Deng, R., Z. Wang, P. Mahan, M. Marinello, A. Mirza, and R. M. Iorio. 1999. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with homologus F protein in the promotion of fusion. Virology 253:43-54. [DOI] [PubMed] [Google Scholar]
- 5.Deng, R., Z. Wang, A. Mirza, and R. Iorio. 1995. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209:457-469. [DOI] [PubMed] [Google Scholar]
- 6.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 8.Hu, X., R. Ray, and R. W. Compans. 1992. Functional interactions betweeen the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66:1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joshi, S. B., R. E. Dutch, and R. A. Lamb. 1998. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology 248:20-34. [DOI] [PubMed] [Google Scholar]
- 10.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis of changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 12.Lambert, D. M., S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erickson, G. Merutka, and S. R. Petteway. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. USA 93:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malvoisin, E., and T. F. Wild. 1993. Measles virus glycoproteins: studies on the structure and interaction of the haemagglutinin and fusion proteins. J. Gen. Virol. 74:2365-2372. [DOI] [PubMed] [Google Scholar]
- 14.McGinnes, L., T. Sergel, J. Reitter, and T. Morrison. 2001. Carbohydrate modifications of the NDV fusion protein heptad repeat domains influence maturation and fusion activity. Virology 283:332-342. [DOI] [PubMed] [Google Scholar]
- 15.McGinnes, L. W., K. Gravel, and T. G. Morrison. 2002. The Newcastle disease virus HN protein alters the conformation of the F protein at cell surfaces. J. Virol. 73:12622-12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGinnes, L. W., T. Sergel, H. Chen, L. Hamo, S. Schwertz, and T. G. Morrison. 2001. Mutational analysis of the membrane proximal heptad repeat of Newcastle disease virus fusion protein. Virology 289:343-352. [DOI] [PubMed] [Google Scholar]
- 17.Morrison, T. G., C. McQuain, and L. McGinnes. 1991. Complementation between avirulent Newcastle disease virus and a fusion protein expressed from a retrovirus vector: requirements for membrane fusion. J. Virol. 65:813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peisajovich, S. G., O. Samuel, and Y. Shai. 2000. Paramyxovirus F1 protein has two fusion peptides: implications for the mechanism of membrane fusion. J. Mol. Biol. 296:1353-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peisajovich, S. G., and Y. Shai. 2002. New insights into the mechanism of virus-induced membrane fusion. Trends Biochem. Sci. 27:183-190. [DOI] [PubMed] [Google Scholar]
- 20.Plemper, R. K., A. L. Hammond, D. Gerlier, A. K. Fielding, and R. Cattaneo. 2002. Strength of envelope protein interaction modulates cytopathicity of measles virus. J. Virol. 76:5051-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porotto, M., M. Murrell, O. Greengard, and A. Moscona. 2003. Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion. J. Virol. 77:3647-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rapaport, D., M. Ovadia, and Y. Shai. 1995. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 14:5524-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reitter, J., T. Sergel, and T. Morrison. 1995. Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J. Virol. 69:5995-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sergel, T., L. McGinnes, and T. Morrison. 2001. Mutations in the fusion peptide and adjacent heptad repeat inhibit folding or activity of the Newcastle disease virus fusion protein. J. Virol. 75:7934-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sergel, T., L. W. McGinnes, and T. G. Morrison. 1993. The fusion promotion activity of the NDV HN protein does not correlate with neuraminidase activity. Virology 196:831-834. [DOI] [PubMed] [Google Scholar]
- 26.Sergel, T., L. W. McGinnes, M. E. Peeples, and T. G. Morrison. 1993. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology 193:717-726. [DOI] [PubMed] [Google Scholar]
- 27.Sergel-Germano, T., C. McQuain, and T. Morrison. 1994. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J. Virol. 68:7654-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stone-Hulslander, J., and T. Morrison. 1999. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J. Virol. 73:3630-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stone-Hulslander, J., and T. G. Morrison. 1997. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 71:6287-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takimoto, T., G. L. Taylor, H. C. Connaris, S. J. Crennell, and A. Portner. 2002. Role of hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. J. Virol. 76:13028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70:6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka, Y., B. R. Heminway, and M. S. Galinski. 1996. Down-regulation of paramyxovirus hemagglutinin-neuraminidase glycoprotein surface expression by a mutant fusion protein containing a retention signal for the rough endoplasmic reticulum. J. Virol. 70:5005-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomasi, M., C. Pasti, C. Manfrinato, F. Dallocchio, and T. Bellini. 2003. Peptides derived from the heptad repeat region near the C-terminal of Sendai virus F protein bind the hemagglutinin-neuraminidase ectodomain. FEBS Lett. 536:56-60. [DOI] [PubMed] [Google Scholar]
- 34.Tong, S., and R. Compans. 1999. Alternative mechanisms of interaction between homotypic and heterotypic parainfluenza virus HN and F proteins. J. Gen. Virol. 80:107-115. [DOI] [PubMed] [Google Scholar]
- 35.Tsurudome, M., M. Ito, M. Nishio, M. Kawano, K. Okamoto, S. Kusagawa, H. Komada, and Y. Ito. 1998. Identification of regions on the fusion protein of human parainfluenza virus type 2 which are required for haemagglutinin-neuraminidase proteins to promote cell fusion. J. Gen. Virol. 79:279-289. [DOI] [PubMed] [Google Scholar]
- 36.Tsurudome, M., M. Kawano, T. Yuasa, N. Tabata, M. Nishio, H. Komada, and Y. Ito. 1995. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 213:190-203. [DOI] [PubMed] [Google Scholar]
- 37.Wiessenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp-41. Nature (London) 387:426-430. [DOI] [PubMed] [Google Scholar]
- 38.Yao, Q., and R. W. Compans. 1996. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology 223:103-112. [DOI] [PubMed] [Google Scholar]
- 39.Yao, Q., X. Hu, and R. Compans. 1997. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surfaces. J. Virol. 71:650-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young, J. K., R. P. Hicks, G. E. Wright, and T. G. Morrison. 1997. Analysis of a peptide inhibitor of paramyxovirus (NDV) fusion using biological assays, NMR, and molecular modeling. Virology 238:291-304. [DOI] [PubMed] [Google Scholar]
- 41.Young, J. K., D. Li, M. C. Abramowitz, and T. G. Morrison. 1999. Interaction of peptides with sequences from the Newcastle disease virus fusion protein heptad repeat regions. J. Virol. 73:5945-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]