Abstract
Soybean seed coat peroxidase (SBP) is a peroxidase with extraordinary stability and catalytic properties. It belongs to the family of class III plant peroxidases that can oxidize a wide variety of organic and inorganic substrates using hydrogen peroxide. Because the plant enzyme is a heterogeneous glycoprotein, SBP was produced recombinant in Escherichia coli for the present crystallographic study. The three-dimensional structure of SBP shows a bound tris(hydroxymethyl)aminomethane molecule (TRIS). This TRIS molecule has hydrogen bonds to active site residues corresponding to the residues that interact with the small phenolic substrate ferulic acid in the horseradish peroxidase C (HRPC):ferulic acid complex. TRIS is positioned in what has been described as a secondary substrate-binding site in HRPC, and the structure of the SBP:TRIS complex indicates that this secondary substrate-binding site could be of functional importance. SBP has one of the most solvent accessible δ-meso haem edge (the site of electron transfer from reducing substrates to the enzymatic intermediates compound I and II) so far described for a plant peroxidase and structural alignment suggests that the volume of Ile74 is a factor that influences the solvent accessibility of this important site. A contact between haem C8 vinyl and the sulphur atom of Met37 is observed in the SBP structure. This interaction might affect the stability of the haem group by stabilisation/delocalisation of the porphyrin π-cation of compound I.
Keywords: Soybean peroxidase, crystal structure, thermal stability, active site interactions, haem accessibility, recombinant peroxidase
Soybean peroxidase (SBP) is a 37 kD glycoprotein expressed in the seed coat of soybean ∼20 days after anthesis (Gillikin and Graham 1991). It belongs to the family of secretory plant peroxidases (class III) (Welinder et al. 1992), all oxidizing a broad range of organic and inorganic substrates at the expense of H2O2 (Dunford 1999):
(i) Resting state + H2O2 → Compound I + H2O
(ii) Compound I + AH → Compound II + Ȧ
(iii) Compound II + AH → Resting state + Ȧ + H2O
The most well-known member of this class of peroxidases is horseradish peroxidase isoenzyme C (HRPC) to which SBP shows 57% amino acid sequence identity. Some secretory plant peroxidases have been shown to correlate with lignification (Smith et al. 1994; Østergaard et al. 2000) and some have been associated with suberisation (Roberts and Kolattukudy 1989; Bernards et al. 1999; Quiroga et al. 2000).
The potential applications of SBP are considerable as a result of a high thermal stability (McEldoon and Dordick 1996) and a high reactivity and stability at low pH (McEldoon et al. 1995; Nissum et al. 2000). The observed inactivation temperature of SBP has been reported to be 90.5°C, while that of the classical HRPC is 81.5°C, and that of Coprinus cinereus peroxidase (a class II peroxidase from the fungus C. cinereus with a similar activity) is 65°C (McEldoon and Dordick 1996). In the study of McEldoon et al. (1995), it was observed that SBP undergo some irreversible inactivation upon heating, which seems to proceed through the loss of haem. It was shown further that SBP has a higher affinity for haem than HRPC. At low pH, with a pH optimum of 2.4, SBP can catalyze the oxidation of veratryl alcohol with similar second order rate constants for the compound I and II reduction, but without being inactivated by haem loss. Both k2 and k3 for SBP are, however, considerably lower than for lignin peroxidase (LIP) (Nissum et al. 2000; McEldoon et al. 1995). Under the same experimental conditions, HRPC is inactivated by haem loss (McEldoon et al. 1995; Munir and Dordrick 2000).
Crystal structures of several haem-containing peroxidases now are available including members of all three classes of plant peroxidases. They all seem to share the enzymatic mechanism for compound I formation (i), although with minor variations in the role of the catalytic distal arginine residue (Poulos and Kraut 1980). In the case of HRPC, the reaction of the intermediates with small substrates (ii, iii) has been described in detail (Dunford 1999), and an enzymatic mechanism has been proposed (Henriksen et al. 1999).
In this paper we describe the recombinant expression of SBP in Escherichia coli and in vitro folding and purification. The three-dimensional structure of SBP is described with emphasis on its enhanced resistance to denaturation and haem loss compared to HRPC. The structure of SBP described in this study is the sixth crystal structure of a secretory plant peroxidase (Schuller et al. 1996; Gajhede et al. 1997; Henriksen et al. 1998; Mirza et al. 2000; Østergaard et al. 2000). This means that information that makes it possible to identify the subtle structural differences resulting in different function of peroxidase isoenzymes now is available. Detailed studies also can shed light on why a plant like Arabidopsis thaliana express ∼100 different peroxidase isoenzymes (Østergaard et al. 1998).
Results and discussion
The peroxidase fold
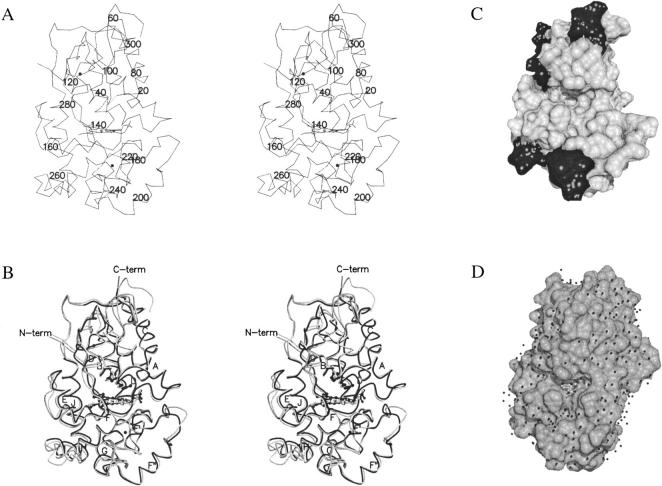
All SBP main and side chain atoms show coherent and significant 2Fo-Fc electron density, except for the two probably disordered C-terminal residues not included in the model. Superposition and structural alignment of class III plant peroxidase structures show that SBP is similar to the previously determined structures of peanut peroxidase (PNP) (Schuller et al. 1996), HRPC (Gajhede et al. 1997), A. thaliana peroxidase A2 (ATP A2) (Østergaard et al. 2000), and A. thaliana peroxidase N (ATP N) (Mirza et al. 2000) with r.m.s.d. of 0.91, 0.94, 0.77 and 0.97 Å, respectively. A stereo view of the SBP trace is seen in Figure 1A ▶, while a superposition of SBP and HRPC is shown in Figure 1B ▶. All 13 α-helices and the two β-strands previously found to be characteristic of class III peroxidases (Gajhede et al. 1997) also are present in SBP. A structural alignment of the class III peroxidases with the programs STAMP (Russell and Barton 1992) and ALSCRIPT (Barton 1993) shows that also when SBP is included in a structural alignment, the significant variation in backbone conformation of the secretory plant peroxidases is restricted to the seven variable regions defined by Mirza et al. (2000). The variable regions are located in the half of the molecule where also the substrate access channel is found, but not in its immediate proximity (Fig. 1C ▶).
Fig. 1.
(A) A stereo drawing of the Cα trace of SBP. The structure is viewed facing the substrate access channel. Residue numbers are included for every 20th amino acid. (B) Stereo drawing of SBP (light grey) with HRPC (black) superimposed. The part of the HRPC trace that corresponds to variable regions defined by a STAMP alignment (Russell and Barton 1992) of class III peroxidases is colored dark grey. Conserved secondary structural elements are labeled according to the labeling of secondary structural elements in peanut peroxidase. R38, H42 and haem are shown in ball and stick like the two structural Ca2 + that appear as black spheres. (C) The solvent accessible surface of SBP in the same orientation as A and B. The surface of variable regions is black. (D) The solvent-accessible surface of HRPC with ferulic acid bound in its substrate-binding site as defined by the HRPC-FA1 binding mode (Henriksen et al. 1999). This structure is rotated slightly compared to the orientations of A, B, and C to give a better look into the substrate binding site. The small black spheres are water molecules found in the HRPC-FA structure. A and B were prepared with MOLSCRIPT (Kraulis 1991) and Raster3D (Merritt and Murphy 1994). C and D were prepared with the program DINO (Philippsen 1999).
Comparing the relative electrostatic properties of the five peroxidases does not reveal any larger differences in their substrate access channel properties; however, the side chain properties seem to give rise to topological differences that might influence peroxidase specificity.
In all structures, the F′/F″ segment is forming a ledge below the substrate access channel (Fig. 1C ▶). In HRPC, SBP, and PNP, a depression is found in the ledge just below the active site entrance, while the A. thaliana peroxidases (ATP A2 and ATP N) have much less pronounced curvature in this area. The residues responsible for this phenomenon are residues 142/143 (HRPC numbers). The hydrophobicity of the residue at position 143 seems to determine the orientation of the hydrophobic residue in position 142. When 143 is hydrophobic (e.g., HRPC, SBP, and PNP) the hydrophobic 142 will pack towards it and open a depression in the F′/F″ ledge. In the A. thaliana peroxidases a hydrophilic residue at position 143 results in closing of the depression.
Another notable topological feature is the equatorial cleft found in SBP, PNP, and ATP A2. In ATP N and HRPC, the cleft is not running all the way across the substrate access channel front of the molecule, but is blocked by residues from the loop region following the F″ helix. These differences in topology are likely to be of relevance in determining the branching direction in a polymerization reaction and/or the orientation of a polymeric substrate. If the oxidized monolignol reacts with its target molecule while still bound in the substrate access channel of the peroxidase (Fig. 1D ▶), then a depression in the ledge will allow the reaction to take place between the target and the terminal carbon of the propenoic acid substituent. A limitation in the equatorial cleft will, on the other hand, prevent a reaction from taking place between the target and the third carbon of the propenoic acid substituent.
Haem propionyl-protein interactions
The possible role of the hydrogen bonding interactions between the haem propionyls and the protein matrix for the susceptibility of the protein to haem loss has been discussed recently (Mirza et al. 2000). SBP have been reported to be less susceptible to haem loss than HRPC (McEldoon and Dordick 1996). The structurally characterized plant peroxidases only differ in one haem propionyl-apoprotein interaction. Both SBP and HRPA2 have one of their haem propionates hydrogen bonded to the side chain of an arginine, while the corresponding residue in HRPC is a glutamine (Q176, HRPC numbers). PNP also have a glutamine in this position, while ATP N, a plant peroxidase that readily looses haem at room temperature and pH 9.0 (K.G. Welinder, unpubl.) have a lysine residue. In all the structurally characterized plant peroxidases, the same propionyl also is hydrogen bonded to the side chain of a serine (S73, HRPC numbers). The other haem propionyl is hydrogen bonded to an arginine side chain (R31, HRPC numbers), and to a serine side chain (S35, HRPC numbers). In LIP, the haem propionates do not have hydrogen bonds to a residue corresponding to Q176 (HRPC numbers), but have hydrogen bonds to serine, aspartic acid, and to the active site arginine. No data on haem binding have been published for PNP and ATP A2. It is likely that propionyl-arginine and propionyl-serine hydrogen bonds are of importance for haem binding in the active site of plant peroxidases, but from the present data we cannot draw any definitive conclusions.
The active site
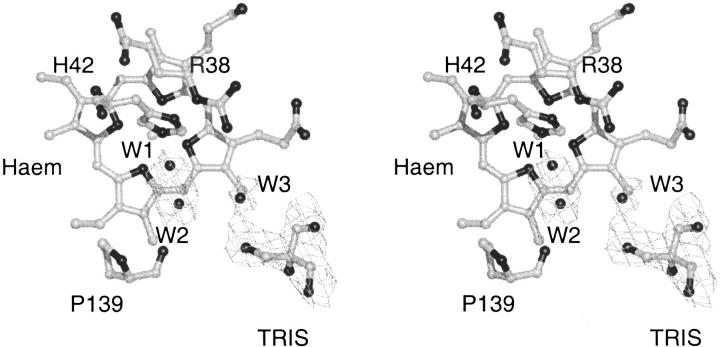
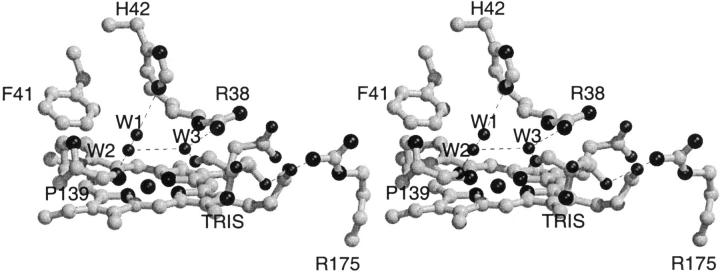
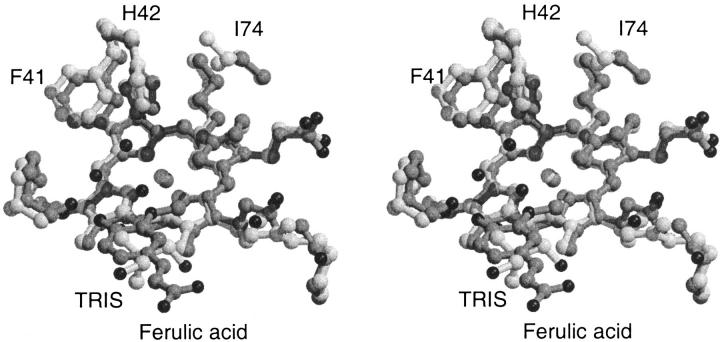
Because of the relative poor quality (2.8 Å resolution, 88.2 % overall completeness) of the data, only the four water molecules located in the active site, at positions corresponding to water molecules found in the active site of other plant peroxidase structures, have been included in the SBP structure. The 2Fo-Fc electron density for the two water molecules closest to the haem iron (W1 and W2) is not properly resolved and the water density is most likely corresponding to a single disordered water molecule (Fig. 2 ▶). This interpretation also is consistent with the relative short 2.3 Å distance between W1 and W2. W1 has a hydrogen bond to the catalytic histidine, H42 (3.0 Å). W2 has a hydrogen bond (2.7 Å) to the backbone oxygen of P139 and to the third active site water molecule, W3 (3.4 Å) that has a long-range interaction to the catalytic arginine, R38 (3.9 Å) and a hydrogen bond to a hydroxy group of the TRIS molecule (2.8 Å). The TRIS molecule has an additional long-range interaction to R175NH2 (3.8 Å), an arginine within hydrogen bonding distance of a haem propionate (R175NE 3.0 Å, R175NH2 3.0 Å(Fig. 3 ▶). W3 is found at a position corresponding to the position of the phenol oxygen in the ternary HRPC-CN-FA complex and/or in the binary HRPC-FA1 complex (Henriksen et al. 1999) and the TRIS molecule is located in the same position as the aromatic portion of FA in the HRPC-FA2 complex (FA has two binding modes in the HRPC-FA complex, FA1 and FA2) (Henriksen et al. 1999) (Fig. 4 ▶). The observation of the TRIS molecule in this position confirms the relevance of the HRPC-FA2 binding mode as a more exterior plant peroxidase substrate binding site with a longer hydrogen and electron transfer pathway relative to the HRPC-FA1 binding mode. Alternatively, the site could be a shuttling terminal for reducing substrates allowing water molecules and hydrogen peroxide to diffuse to the haem center. The crystallographic HRPC-FA complex study and the present study also suggest that a large variety of substances and buffer molecules are potential inhibitors of the peroxidase reaction.
Fig. 2.
The 1σ 2Fo-Fc electron density in the active site cavity of SBP. The two water molecules closest to the haem iron are not resolved properly and probably are corresponding to a single disordered water molecule.
Fig. 3.
Stereo drawing of the hydrogen bonding pattern in the active site of SBP.
Fig. 4.
Stereo drawing of the superposition of the active sites of SBP (light grey) and HRPC:FA2 complex (dark grey). The TRIS molecule that is found in the SBP structure is hydrogen bonded to the active site arginine through a water molecule. H42 is rotated 45° about χ2 in SBP relative to HRPC and as result, F41 is displaced ∼0.5 Å. The substitution of A74 in HRPC with an isoleucine in SBP is likely to give rise to the H42 rotation. The overall result is a more solvent, exposed δ-meso haem edge in SBP.
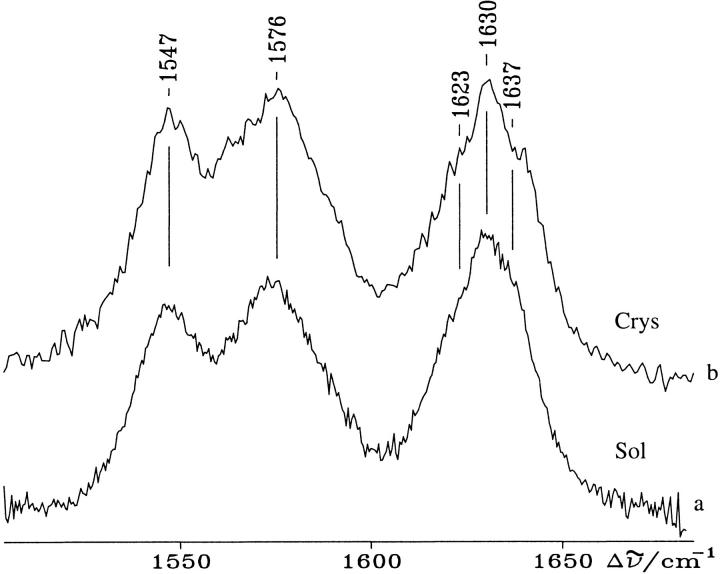
The distance between the haem iron and the closest water molecule is 4.0 Å. This is considerably longer than the distances found in the mainly 5-c QS HRPC (3.2 Å) and in PNP (3.1 Å) and could be influenced by the presence of the positively charged TRIS molecule at the exterior substrate binding site. Figure 5 ▶ compares the resonance Raman spectra (RR) of solution (without TRIS) and singe crystal samples of SBP (with TRIS) both obtained at room temperature. The RR spectra of SBP in solution previously have been interpreted as mainly being a result of a quantum mechanically mixed-spin state (QS) resulting from the admixture of intermediate spin (S = 3/2) and high spin (S = 5/2) configurations, coexisting with five and six coordinate high spin haems (5-c and 6-c HS) (Nissum et al. 1998). The present RR spectra obtained on single crystal and solution samples with visible excitation does not differ markedly from the previously recorded spectra of SBP in solution (Nissum et al. 1998). They are characterized mainly by the 5-c QS haem core size marker bands at 1547 cm-1 (|v11), 1576 cm-1 (|v19) and 1637 cm-1((|v10). Moreover, two bands at 1623 and 1630 cm-1 are observed: the former resulting from the |v(C=C) stretching mode of one vinyl substituent and the latter from the overlap of the second vinyl mode at 1632 cm-1 and the |v10 of the 5-c HS species at 1629 cm-1 (Nissum et al. 1998). The spectra from single crystal and solution are identical and in accordance with the active site geometry as determined in the crystal structure. However, previous spectroscopic studies (RR, UV-vis and EPR) of SBP in solution at 5–12 K indicates that the haem crevice undergo conformational changes induced by lowering the temperature (Indiani et al. 2000). At low temperature, three haem species have been identified corresponding to a 5-c QS form and two new 6-c low-spin haems. These two latter forms are observed already at 180 K. As the crystal structure was determined at 100 K, it would be expected that this conformational change could be observed in the crystal structure. But because the crystal was soaked in a solution containing 35% glycerol prior to freezing and data collection, it is possible that the presence of glycerol is preventing the conformational change as previously observed for CCP in solution at 10 K (Smulevich et al. 1989). However, it also is possible that TRIS itself, present in the secondary substrate binding site, prevents the formation of the low-spin species.
Fig. 5.
Resonance Raman spectra at room temperature of SBP in 0.1 M bicine buffer pH 7.0, 1 mM CaCl2 (a), and of a single crystal (b). Experimental conditions: 5 cm-1 spectral resolution (a) 514.5 nm excitation: 70 mW laser power at the sample, 45 s/0.5 cm-1 accumulation time; (b) 50 μW laser power on the crystal surface, 1560 s accumulation time.
The distal cavity of SBP contains the active site residues F41 and H42 in slightly but significantly different orientations than those found in HRPC, ATP A2 and ATP N, but similar to those found in PNP. H42 in SBP and PNP are rotated by 45° and 30°, respectively, about χ2 relative to H42 in HRPC. The corresponding movement (∼0.5 Å) of F41 in both PNP and SBP probably is a result of this rotation. The substitution of A74 of HRPC with a bulky isoleucine in SBP and PNP probably is causing the rotation of H42 (Fig. 4 ▶). This substitution is observed commonly in plant peroxidases and hence the F41/H42 orientation observed in SBP and PNP is likely to be found in many other plant peroxidases.
As the second order rate constant for the reaction of SBP and HRPC with H2O2 has been reported to be very similar at neutral pH (Nissum et al. 2000) the slight rotation of H42 does not seem to be of major importance for the first step in the peroxidase reaction.
Haem accessibility
A likely consequence of the rotation of H42 and the movement of F41 in SBP and PNP is a more solvent accessible δ-meso haem edge in these peroxidases. The solvent accessible surface of haem C20 in SBP, PNP, HRPC, ATP A2 and ATP N is 10.4 Å2, 11.1 Å2, 5.6 Å2, 5.3 Å2, and 6.2 Å2, respectively (calculated with the program NACCESS (Hubbard and Thornton, 1993), with a 1.4 Å probe radius). As the δ-meso edge is the part of the haem group where the electron transfer has been proposed to take place (Ator and Ortiz de Montellano 1987), a more solvent accessible δ-meso edge must be expected to have importance for the reactivity of the enzyme. However, except for the SBP-veratryl alcohol studies (McEldoon et al. 1995; Nissum et al. 2000), no detailed kinetic data for the reaction of SBP or PNP with reducing substrate are published.
The large δ-meso haem edge accessibility might be an important factor for the decreased compound I stability observed for SBP relative to HRPC (Nissum et al. 2000). The haem group could be expected to be more prone to exogenous reduction (e.g., by inorganic or organic buffer molecules or impurities) if the surface area of the δ-meso edge is enlarged.
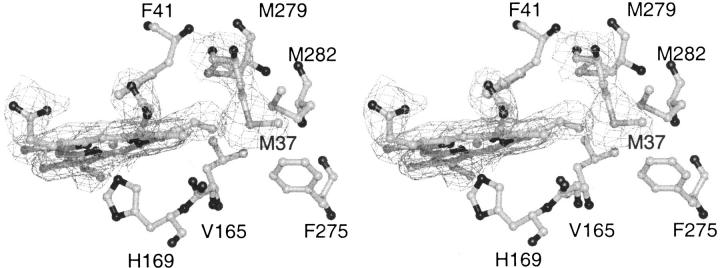
An inspection of the SBP haem electron density shows a coherent 2Fo-Fc electron density between the haem group and M37 (HRPC numbers) at the 1σ level (Fig. 6 ▶). Valine, leucine, and isoleucine are the most common residues at this position in other plant peroxidases. The M37SD atom is 3.9 Å from the haem C8 vinyl substituent, and this is close enough for van der Waals interactions between the two atoms. M279, M282, and M37 are forming a methionine cluster around the C8 vinyl of SBP. The p orbitals of the sulphur atoms could allow SBP to transfer electrons from M37 to the π cation haem radical of compound I, and hereby mimicking the radical delocalization observed in LIP, where the radical is transferred to a tryptophan residue in the periphery of haem (Doyle et al. 1998; Choinowski et al. 1999). These interactions will explain why SBP is less susceptible to haem loss and has a higher thermal stability than HRPC. They also could increase the probability of endogenous compound I reduction. Directed evolution of HRPC (Morawski et al. 2000) has resulted in a L37I mutation that influenced both the specific activity and the thermal stability of the enzyme, indicating that position 37 is a key residue in these respects.
Fig. 6.
Stereo picture of the 1σ 2Fo-Fc electron density surrounding the haem group and M37 in SBP. M37 has a closest distance of 3.9 Å to the C8 haem vinyl substituent.
Conclusions
We have reported the x-ray crystal structure of soybean peroxidase with the buffer molecule TRIS bound in the active site. The x-ray structure together with the single crystal RR spectra show that the iron is mainly in the 5-c state. The haem spin states, including the quantum-mechanically mixed-spin (QS) state of the crystal are identical to those observed in solution. Compared to the structure of HRPC in its binary complex with ferulic acid, the SBP-TRIS complex confirms the position of a secondary exterior substrate binding site in the plant peroxidases, a site that could be a shuttle station for reducing substrates. The structure has shown that Ile74 is a residue likely to influence δ-meso haem edge solvent accessibility. Haem-apoprotein interaction between M37 and the C8 haem vinyl substituent has not been observed previously in plant peroxidases and it could have an effect on the thermal stability of the enzyme and its susceptibility to haem loss.
Materials and methods
Recombinant SBP production
A fragment encoding mature SBP (determined by protein sequencing, K.G. Welinder, unpubl.) was produced by PCR on a full-length cDNA clone encoding SBP (Gijzen 1997). The resulting fragment was cloned into the NdeI and BamH I restriction sites of the pET-3a vector. E. coli BL21(DE3) was transformed with the resulting plasmid, pETSBP. Cells were grown in TB at 37°C and the expression induced by 0.4 μM IPTG at OD600 = 0.8. Cells were harvested 4 h after induction. Inclusion bodies were isolated according to Teilum et al. (1999), and SBP activity was gained by folding in 20 mM Tris-HCl, pH 8.7, 2.25 M urea, 0.4 mM GSH, 0.6 mM GSSG, 1.5 μM haem, 5 mM CaCl at a protein concentration of 0.4 mg/mL overnight at 4°C.
Purification of folded and active SBP
The folding mixture was concentrated using a Minitan II concentrator (Millipore) and dialyzed thoroughly against 10 mM BisTris-HCl, pH 6.5. After dialysis the solution was centrifuged and the supernatant applied to a Q-Sepharose HP 26/10 anion-exchange column (Pharmacia Biotech) and eluted with a gradient of 0–0.1 M NaCl in 10 mM BisTris-HCl, pH 6.5 over six column volumes. The peak fractions were pooled and concentrated using an Ultrafree NMWL 10,000 centrifugal ultrafiltration device (Millipore).
Crystallization and data collection
Ammonium sulphate precipitated recombinant SBP was brought in solution by dialysis against 10 mM Tris-HCl buffer, pH 7.5 and concentrated by ultrafiltration to a concentration of 1.9 mg/mL. The crystals of SBP were prepared by hanging drop vapor diffusion at 4°C by mixing 2 μL of protein solution with 2 μL of precipitant. NH4H2PO4 in a concentration between 0.2 and 0.6 M was used as precipitant and the solution was buffered with 0.1 M Tris-HCl buffer pH 7.5. Crystals appeared within 2 weeks. The SBP crystal quality was relatively poor (Table 1) and optimization of crystallization conditions was attempted including buffer, precipitant, and additive screening. Some carbohydrate additives produced crystals but the quality of the crystals did not improve. TRIS and NH4H2PO4 were present in all successful crystallization experiments. SBP crystals are rodlike and suffer from a pronounced tendency to form clusters. The clusters are formed at the tip of even the nicest single crystal rods and have to be removed during crystal mounting. The crystals diffracted to 2.8 Å at 100 K, at the MAXLAB II synchrotron facility, beam line 711, University of Lund. Only using the synchrotron source, it was possible to obtain data of a quality sufficient for structure determination. Prior to data collection, the crystals were transferred to a cryo-protectant consisting of 0.26 M NH4H2PO4, 35% glycerol for less than 1 minute and flash cooled in liquid nitrogen. The space group was determined to be P31 or the enantiomorphic P32, with cell dimensions a = b = 106.4 Å, c = 105.0 Å with at least three molecules in the asymmetric unit related by noncrystallographic symmetry as judged from the self-rotation functions and Patterson maps. Seventy successive frames of 1.0° oscillation were recorded with a MAR345 image plate in the 150 μm/180 mm mode. Data integration and data processing were done with HKL (Otwinowski and Minor 1997). The data collection statistics are listed in Table 1.
Table 1.
Statistics for crystallographic structure determination of SBP
| Data collection | |
| Wavelength (Å) | 0.996 |
| Resolution range (Å) | 11–2.80 |
| Observations | 175095 |
| Unique reflections | 26929 |
| Completeness (%)a | 88.2 (69.3) |
| Rsym (%)a,b | 9.6 (28.7) |
| Refinement | |
| Resolution range (Å) | 11–2.80 |
| Unique reflections (I > 0) | 25330 |
| Rcryst (%)c | 26.7 |
| Rfree (%)d | 29.5 |
| Number of nonhydrogen atoms | |
| Protein | 4658 |
| Solvent | 12 |
| Heterogen atoms | 45 |
| R.m.s. deviations from ideal values | |
| Bond length (Å) | 0.006 |
| Bond angle (°) | 1.1 |
| Estimated coordinate error (Å) | |
| Luzzati plote | 0.45 |
| Ramachandran plotf | |
| Residues in most favoured (disallowed: Asp256) regions (%) | 82.3 (0.8) |
| mean B-factor (Å2) | |
| Protein | 18.3 |
| Haem | 11.1 |
| Tris | 41.1 |
| Solvent | 11.18 |
a Numbers in parenthesis are for the highest resolution shell, 2.85–2.80 Å.
b Rsym = Σhkl(Σi(|Ihkl,i − <Ihkl>|)) / Σhkl,i<Ihkl>, where Ihkl,i is the intensity of an individual measurement of the reflection with Miller indices h, k, and l, and <Ihkl> is the mean intensity of that reflection.
c Rcryst = Σhkl(∥Fo,hkl| − |Fc,hkl∥) / Σhkl|Fo,hkl|, where |Fo,hkl| and |Fc,hkl| are the observed and calculated structure factor amplitudes.
dRfree is equivalent to the R-factor, but calculated with reflections omitted from the refinement process (5% of reflections omitted).
e Calculated with the program CNS (Brünger et al. 1998).
f Calculated with the program PROCHECK (Laskowski et al. 1993).
Phasing was done by molecular replacement using AMoRe (Navaza 1994) and the structure of HRPC as search model (pdb entry 1atj). The space group was determined by comparison of molecular replacement solutions in the space groups P3, P31 and P32. Only in the space group P31 the translation function solutions gave significant better correlation coefficient and R-factor. Three molecules were found in the asymmetric unit of which two had the same rotational solution. Rigid body refinement was done with X-PLOR (Brünger 1992) while the subsequent refinement was done in CNS (Brünger et al. 1998) with model rebuilding done in o (Jones et al. 1991). Refinement was carried out applying strict NCS to the molecules and refining initially against the mli target function (Brünger et al. 1998). The strict NCS constraints were used throughout the refinement to avoid overfitting the data. No initial B-factor correction was used, as the resolution of the data was too poor to justify this. The relative low fraction of residues in the most favored region of the Ramachandran plot is a consequence of the weight applied to the X-ray term. A lower weight for the X-ray term resulted in a better geometry of the structure but also in an increase of Rfree. The chosen weight for the X-ray term was half the weight suggested by CNS during the initial annealing, which resulted in a compromise between low Rfree and good geometry. Four water molecules were in the active site cavity where the sigma level and position of 2Fo-Fc density justified it. Individual but restrained B-factors were applied during refinement, because it gave a reduction of the Rfree. The refinement and stereochemistry statistics are listed in Table 1.
The SBP crystals were mounted in a small amount of mother liquor in 0.6 mm capillaries that were sealed for the resonance Raman experiment. Ammonium sulphate precipitated recombinant SBP was dialyzed extensively against 2 mM CaCl2 and diluted 1:1 in bicine or phosphate buffer to a final concentration of 0.1 M and 1 mM CaCl2, pH7.0 for the RR spectra of SBP in solution. The RR spectra in solution were obtained using a system previously described (Nissum et al. 1998). For the experiments on single crystals, a Raman microprobe apparatus previously reported (Smulevich et al. 1999) was modified further. It consists of an Olympus BHSM2 microscope equipped with an extra-long working distance Nikon M Plan achromatic objective capable of 60× magnification with a 0.70 numerical aperture. The laser beam waist in the focus of the objective is estimated to be about 2 μm. The spectrometer was a HR460 Jobin-Yvon monochromator with a grating of 1800 grooves/mm and an ultimate resolving power exceeding 104 at 550 nm. The detector was a liquid-nitrogen–cooled Spectraview 2D CCD head of 578×375 pixels, which enabled the simultaneous recording of a spectral region of ∼450 cm-1 in our spectrometer. To obtain a reasonable signal to noise ratio, a 200-μm slit was used, giving a 5-cm-1 spectral resolution. The RR spectra were obtained with the 514.5 nm line of an Ar+ laser (Coherent Innova/90) at room temperature. The spectra were calibrated with indene as standard. The frequencies were accurate to ± 1 cm-1 for the intense isolated bands and about ± 2 cm-1 for the overlapped bands and shoulders.
Acknowledgments
This work was supported by the Danish Natural Research Council through the DANSYNC grant. MAXLABII, beam line 711, University of Lund supplied the radiation used for structure determination. EU contract BIO4–97–2031, ``Towards designer peroxidases'' (to GS, KGW, and MG) supported stipends to AH and KT. We thank the European Laboratory for nonlinear Spectroscopy, LENS (Firenze, Italy) for provision of the microRaman facilities for the experiment on the single crystal of SBP and Dr. M. Becucci (LENS) for expert assistance. M. Gijzen, Agriculture and Agri-Food Canada, London, Ontario, kindly provided the SBP cDNA clone.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ``advertisement'' in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
The structure of SBP has been deposited in the RCSB Protein Data Bank with accession number 1FHF.
Abbreviations
ATP A2, Arabidopsis thaliana peroxidase A2
ATP N, Arabidopsis thaliana peroxidase N
CCP, cytochrome c peroxidase
CIP, Coprinus cinereus peroxidase
FA, ferulic acid
HRPC, horseradish peroxidases C
HS, high spin
LIP, Lignin peroxidase
LS, low spin
PNP, peanut peroxidase
QS, quantum mechanically mixed-spin state
RR, resonance Raman
SBP, soybean peroxidase
TRIS, tris(hydroxymethyl)aminomethane.
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.37301.
References
- Ator, M.A. and Ortiz de Montellano, P.R. 1987. Protein control of prosthetic heme reactivity. Reaction of substrates with the heme edge of horseradish peroxidase. J. Biol. Chem. 262 1542–1551. [PubMed] [Google Scholar]
- Barton, G.J. 1993. ALSCRIPT. A tool to format multiple sequence alignments. Protein Engineering 6 37–40. [DOI] [PubMed] [Google Scholar]
- Bernards, M.A., Fleming, W.D., Llewellyn, D.B., Priefer, R., Yang, X., Sabatino, A., and Plourde, G.L. 1999. Biochemical characterization of the suberization-associated anionic peroxidase of potato. Plant Physiol. 121 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger, A.T. 1992. X-PLOR Version 3.1. A system for x-ray crystallography and NMR, Yale University Press, New Haven, CT.
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., Read, R.J., Rice, L.M., Simonson, T., and Warren, G.L. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54 905–921. [DOI] [PubMed] [Google Scholar]
- Choinowski, T., Blodig, W., Winterhalter, K.H., and Piontek, K. 1999. The crystal structure of lignin peroxidase at 1.70 Å resolution reveals a hydroxy group on the Cβ of tryptophan 171: A novel radical site formed during the redox cycle. J. Mol. Biol. 286 809–827. [DOI] [PubMed] [Google Scholar]
- Doyle, W.A., Blodig, W., Veitch, N.C., Piontek, K., and Smith, A.T. 1998. Two substrate interaction sites in lignin peroxidase revealed by site-directed mutagenesis. Biochemistry 37 15097–15105. [DOI] [PubMed] [Google Scholar]
- Dunford, H.B. 1999. Heme peroxidases, John Wiley & Sons, New York.
- Gajhede, M., Schuller, D.J., Henriksen, A., Smith, A.T., and Poulos, T.L. 1997. Crystal structure of horseradish peroxidase C at 2.15 Å resolution. Nat. Struct. Biol. 4 1032–1038. [DOI] [PubMed] [Google Scholar]
- Gijzen, M. 1997. A deletion mutation at the ep locus causes low seed coat peroxidase activity in soybean. Plant J. 12 991–998. [DOI] [PubMed] [Google Scholar]
- Gillikin, J.W. and Graham, J.S. 1991. Purification and developmental analysis of the major anionic peroxidase from the seed coat of Glycine max. Plant Physiol. 96 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen, A., Smith, A.T., and Gajhede, M. 1999. The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates. J. Biol. Chem. 274 35005–35011. [DOI] [PubMed] [Google Scholar]
- Henriksen, A., Welinder, K.G., and Gajhede, M. 1998. Structure of barley grain peroxidase refined at 1.9-Å resolution. A plant peroxidase reversibly inactivated at neutral pH. J. Biol. Chem. 273 2241–2248. [DOI] [PubMed] [Google Scholar]
- Hubbard, S.J., Thornton, J.M. 1993. NACCESS v. 2.1.1. Department of Biochemistry and Molecular Biology, University College London.
- Indiani, C., Feis, A., Howes, B.D., Marzocchi, M.P., and Smulevich, G. 2000. Effect of low temperature on soybean peroxidase: spectroscopic characterization of the quantum-mechanically admixed spin state. J. Inorg. Biochem. 79 269–274. [DOI] [PubMed] [Google Scholar]
- Jones, A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these maps. Acta Crystallogr. A47 110–119. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- McEldoon, J.P., and Dordick, J.S. 1996. Unusual thermal stability of soybean peroxidase. Biotechnol. Prog. 12 555–558. [Google Scholar]
- McEldoon, J.P., Pokora, A.R., and Dordick, J.S. 1995. Lignin peroxidase-type activity of soybean peroxidase. Enzyme Microb. Technol. 17 359–365. [Google Scholar]
- Merritt, E.A. and Murphy, M.E.P. 1994. Raster3D version 2.0: A program for photorealistic molecular graphics. Acta Crystallogr. D50 869–873. [DOI] [PubMed] [Google Scholar]
- Mirza, O., Henriksen, A., Østergaard, L., Welinder, K.G., and Gajhede, M. 2000. Arabidopsis thaliana peroxidase N. Structure of a novel neutral peroxidase. Acta Crystallogr. D56 372–375. [DOI] [PubMed] [Google Scholar]
- Morawski, B., Lin, Z., Cirino, P., Joo, H., Bandara, G., and Arnold, F.H. 2000. Functional expression of horseradish peroxidase in Saharomyces cervisiae and Pichia pastoris. Protein Eng. 13 377–384. [DOI] [PubMed] [Google Scholar]
- Munir, I.Z. and Dordrick, J.S. 2000. Soybean peroxidase as an effective bromination catalyst. Enzyme Microb. Technol. 26 337–341. [DOI] [PubMed] [Google Scholar]
- Nissum, M., Schiødt, C.B., and Welinder, K.G. 2000. Reactions of soybean peroxidase and hydrogen peroxide pH 2.4–12.0, and veratryl alcohol at pH 2.4. Submitted. [DOI] [PubMed]
- Nissum, M., Feis, A., and Smulevich, G. 1998. Characterization of soybean seed coat peroxidase: resonance Raman evidence for a structure-based classification of plant peroxidases. Biospectroscopy 4 355–364. [DOI] [PubMed] [Google Scholar]
- Østergaard, L., Pedersen, A.G., Jespersen, H.M., Brunak, S., and Welinder, K.G. 1998. Computational analyses and annotations of the Arabidopsis peroxidase gene family. FEBS Lett. 433 98–102. [DOI] [PubMed] [Google Scholar]
- Østergaard, L., Teilum, K., Mirza, O., Mattsson, O., Petersen, M., Welinder, K.G., Mundy, J., Gajhede, M., and Henriksen, A. 2000. Arabidopsis ATP A2 peroxidase. Expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Mol. Biol. 44 231–243. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of x-ray diffraction data collected in oscillation mode. Meth. Enz. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Philippsen, A. 1999. DINO: Visualizing structural biology. http://www.bioz.unibas.ch/∼xray/dino.
- Poulos, T.L. and Kraut, J. 1980. The stereochemistry of peroxidase catalysis. J. Biol. Chem. 255 8199–8205. [PubMed] [Google Scholar]
- Quiroga, M., Guerrero, C., Botella, M.A., Barceló, A., Amaya, I., Medina, M.I., Alonso, F.J., de Forchetti, S.M., Tigier, H., and Valpuesta, V. 2000. A tomato peroxidase involved in the synthesis of lignin and suberin. Plant Physiol. 122 1119–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, E. and Kolattukudy, P.E. 1989. Molecular cloning, nucleotide sequence, and abscisic acid induction of a suberization-associated highly anionic peroxidase. Mol. Gen. Genet. 217 223–232. [DOI] [PubMed] [Google Scholar]
- Russell, R.B. and Barton, G.J. 1992. Multiple protein sequence alignment from tertiary structure comparison. Proteins: Struct. Funct. Genet. 14 309–323. [DOI] [PubMed] [Google Scholar]
- Schuller, D.J., Ban, N., Huystee, R.B., McPherson, A., and Poulos, T.L. 1996. The crystal structure of peanut peroxidase. Structure 4 311–321. [DOI] [PubMed] [Google Scholar]
- Smith, C.G., Rodgers, M.W., Zimmerlin, A., Ferdinando, D., and Bolwell, G.P. 1994. Tissue and subcellular immunolocalisation of enzymes of lignin synthesis in differentiating and wounded hypocotyl tissue of French bean (Phaseolus vulgaris L.). Planta 192 155–164. [Google Scholar]
- Smulevich, G., Feis, A., Indiani, C., Becucci, M., and Marzocchi, M.P. 1999. Peroxidase-benzhydroxamic acid complexes: Spetroscopic evidence that a Fe-H2O distance of 2.6 A can correspond to hexa-coordinate high-spin heme. J. Biol. Inorg. Chem. 4 39–47. [DOI] [PubMed] [Google Scholar]
- Smulevich, G., Mantini, A.R., English, A.M., and Mauro, J.M. 1989. Effects of temperature and glycerol on the resonance Raman spectra of cytochrome c peroxidase and selected mutants. Biochemistry 28 5058–5064. [DOI] [PubMed] [Google Scholar]
- Teilum, K., Østergaard, L., and Welinder, K.G. 1999. Disulfide bond formation and folding of plant peroxidases expressed as inclusion body protein in Escherichia coli thioredoxin reductase negative strains. Protein Expr. Purif. 15 77–82. [DOI] [PubMed] [Google Scholar]
- Welinder, K.G., Mauro, J.M., and Nørskov Lauritsen, L. 1992. Structure of plant and fungal peroxidases. Biochem. Soc. Trans. 20 337–340. [DOI] [PubMed] [Google Scholar]