Abstract
The cytochromes P450 are an important class of mono-oxygenases involved in xenobiotic metabolism and steroid biosynthesis in a diverse set of life forms. Discovery of CYP-119, a P450 from the archea Sulfolobus solfataricus has provided a means for understanding nature's method of stabilizing this important protein superfamily. To identify classes of stabilizing interactions used by CYP-119, we have generated a randomized library of point mutants and screened for mutants that are less thermostable than the wild type by monitoring the characteristic Soret band in the visible region of the cell lysis. The selected mutants were characterized by differential scanning calorimetry to compare the temperatures of the melting transitions of the various mutants. The identified mutations suggested that electrostatic interactions involving salt links and charge–charge interactions, as well as contributions from other interactions such as aromatic stacking, and side chain volume of hydrophobic residues contribute to enhanced thermostability in this cytochrome P450.
Keywords: Cytochrome P450, thermostability, random mutagenesis, salt links, electrostatic interactions
As the number of proteins isolated from organisms that grow at extreme temperatures increases, more is being understood about the properties of these proteins, which account for their inherent stability. Although protein stability has been the subject of numerous studies for several decades (Privalov 1979; Pace et al. 1996; Robertson and Murphy 1997), detailed examination of these naturally thermostable proteins provides insight on the mechanisms that nature has used to stabilize proteins in such extreme environments (Cowan 1995; Jaenicke 1998). A better understanding of these mechanisms could lead to important applications in biotechnology, where stability at high temperatures is advantageous (Zeikus et al. 1998).
For many years, investigators have been searching for a universal mechanism of protein stability that can be applied to any protein. Instead, emerging is the idea that there is not one general mechanism that can be applied to every protein, but rather a set of molecular mechanisms that play a role in conveying stability and are used in various combinations to increase stability in a particular protein (Vieille and Zeikus 1996; Lee and Vasmatzis 1997; Jaenicke and Bohm 1998). This set of mechanisms include increased hydrogen bonding, ionic stabilization of α-helixes, tighter packing of the protein core, increased number of salt links and salt-link networks, increased number of disulfide bonds, decreased conformational entropy of the unfolded state, and increased aromatic stacking interactions.
Several approaches have been taken to uncover these interactions responsible for stabilizing proteins at high temperatures. One approach, which has provided a substantial amount of information, is structural comparison of a protein family where a solved structure (X-ray or NMR) of both a mesophile and a thermophile exists (Tanner et al. 1996; Auerbach et al. 1998; Maes et al. 1999). Especially in cases with several protein structures of varying thermostability, structural trends are recognized corresponding to increasing stability of the proteins. However, this technique has some limitations to its general application. The first is the requirement of a solved structure of at least one mesophile and one thermophile in the protein class of interest. This greatly limits the current number of proteins for which structural analysis can be used. Even more significant is the degree of homology the members of the protein class have for each other. If the members of a protein family are not highly homologous, it is difficult to discern, even with structures, the specific differences attributed to the stability of the protein. The enzyme superfamily cytochrome P450 falls into this category.
Cytochrome P450 is a ubiquitous enzyme superfamily found throughout nature with >800 known members (Nelson 1999). These enzymes contain a heme prosthetic group that binds molecular oxygen and catalyzes a wide variety of regio and stereo-specific oxidations of organic substrates. Many of these reactions are difficult to achieve synthetically and produce products with great medical and industrial importance. For example, there are several P450s involved in the synthesis of the antibiotic erythromycin, including CYP-107A1 and CYP-107B1 (Shafiee and Hutchinson 1987; Andersen and Hutchinson 1992).
Erythromycin is commonly produced by fermentation of the bacteria Saccharopolyspora erythraea, the source of CYP-107A1 and CYP-107B1. However, if the enzymes responsible for the biosynthesis of erythromycin were modified to have greater stability and enhanced turnover rates, the same level of production could be obtained at reduced costs and time. This is just one of several applications that would greatly benefit the development of enhanced P450s. Thus, it would be useful to develop a modified cytochrome P450 system with improved physical parameters, such as stability and turnover rate.
Recently, the first known thermostable P450 (CYP-119) from the archea, Sulfolobus solfataricus was isolated and characterized (McLean et al. 1998). Initial characterization revealed that this P450 had a melting temperature ∼40°C higher than the mesophilic P450s (Uvarov et al. 1980; Anzenbacher et al. 1982; Pfeil et al. 1993). However, there was little information pertaining to the interactions responsible for this extreme stability.
Initial attempts to understand the molecular interactions giving rise to the thermostability were through the construction of homology models of CYP-119 (McLean et al. 1998; Chang and Loew 2000). The models indicated that increased aromatic stacking interactions and salt links might be important for stabilizing the protein. However, prediction of detailed interactions based on these models is difficult because of the low homology between P450s, often resulting in inaccuracies in the structure.
The crystal structure of CYP-119 was solved independently in two different laboratories (Yano et al. 2000; S.-Y. Park, K. Yamane, S.-i. Adachi, Y. Shiro, S.A. Maves, K.E. Weiss, and S.G. Sligar, in prep.). Structural comparison of CYP-119 with the five available mesophilic P450 structures indicated increased aromatic stacking interactions and increased salt-link networks as important contributors to the enhanced stability, similar to the predictions made from the homology models. However, the low homology between P450s and the existence of only one thermophilic structure makes these comparisons difficult. Because identity is low between the P450s, significant differences that contribute to thermostability as opposed to substrate specificity or redox partner recognition are difficult to recognize. Therefore, experimental evidence of interactions contributing to thermostability would help to confirm the interactions identified by structural analysis as well as identify additional interactions.
To identify an unbiased representation of stabilizing interactions, we chose an approach that does not require prior knowledge of the residues responsible for conveying stability, but rather allows the discovery of these residues based only on the selected phenotype. By screening a randomized library of CYP-119, we were able to extract those mutants that are less thermostable than wild type, thus revealing the type of interactions contributing to stability in CYP-119. Although this approach has typically been used to direct proteins toward a new desired function (Shao and Arnold 1996; Harris and Craik 1998; Arnold and Volkov 1999; Steipe 1999), it can also be used to look for mutations that disrupt the desired phenotype engineered by nature as a means for understanding these interactions. In this sense, we are able to use the information from the naturally thermostable P450 as a basis for engineering thermostability into other P450s instead of individually screening each P450 of interest for increased thermostability.
Results
The random library of CYP-119 was screened for mutants that were less thermostable than the wild type, thus isolating residues important for conveying the stability of the protein. This was accomplished in a 96-well format assay in which clones were grown and lysed in the plates and then optically screened for the presence of the low spin ferric P450 Soret at both low and high temperatures.
Those mutants that displayed the Soret band at the low temperature, but not at the high temperature, were selected as positive mutants and isolated for further characterization. From the first 3000 colonies screened in this manner, 13 were identified as less stable by this optical screen.
The sequences of the 13 identified mutants revealed 5 single mutants, 1 double mutant, 2 triple mutants, 3 quadruple mutants, and 2 insertion/deletion mutants.
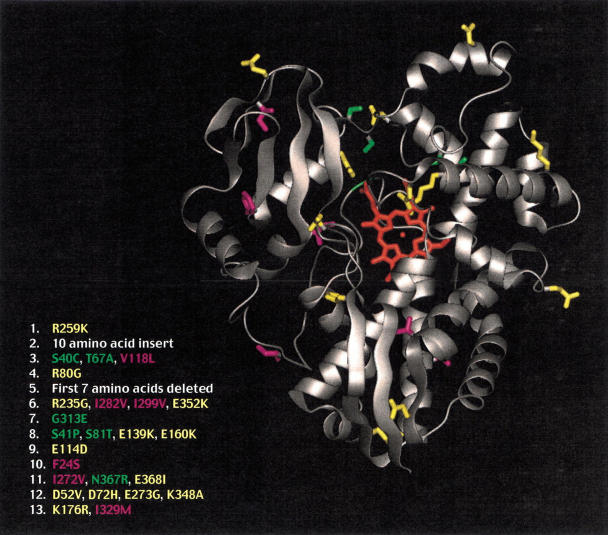
From the 11 noninsertion/deletion mutations, there were 13 charged residues, 6 polar, and 6 hydrophobic residues mutated. The mutants were mapped onto the structure of CYP-119 (S.-Y. Park, K. Yamane, S.-i. Adachi, Y. Shiro, S.A. Maves, K.E. Weiss, and S.G. Sligar, in prep.) as shown in Figure 1 ▶. From the structure, it is clear that the mutants are dispersed throughout the structure and not localized to one particular region. It is interesting to note that the majority of the mutants were found in solvent-accessible regions of the protein as opposed to buried residues in the core of the protein. Each of the selected mutants were expressed in Escherichia coli and purified to homogeneity for further characterization.
Fig. 1.
Homology model of CYP-119 (McLean et al. 1998) with residues that were identified from the screened library displayed. Charged residues are colored yellow, polar residues, green, and hydrophobic residues are purple. Listed are the 13 clones isolated from the screen with the mutations found in each clone.
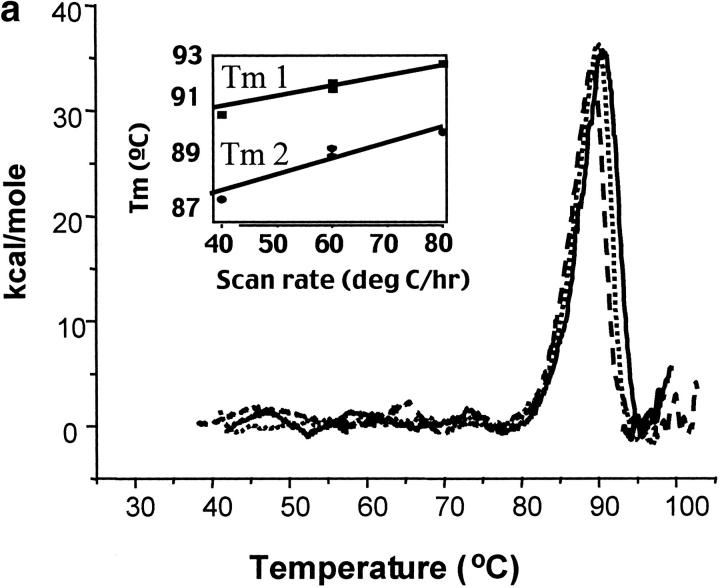
Initial characterization of the stability of these mutants was accomplished using differential scanning calorimetry to determine the thermodynamic stability of each of the mutants. However, to compare accurately mutant proteins, it was necessary to develop conditions for which the protein would be in equilibrium during the calorimetric scan. The thermal denaturation of wild-type CYP-119 was determined to be irreversible and scan rate dependent, as has been previously observed in the cytochrome P450s (Pfeil et al. 1993). Figure 2a ▶ shows the dependence of the Tm on the scan rate for wild type CYP 119 at pH 6.2. This dependence was thought to be due to formation of an aggregation state during unfolding as a result of the hydrophobic core of the protein, as observed in other heme proteins. This aggregation state is not an equilibrium state, but rather a kinetically controlled state, therefore, that is dependent on the scanning rate (Relkin 1994; Shnyrov et al. 1997).
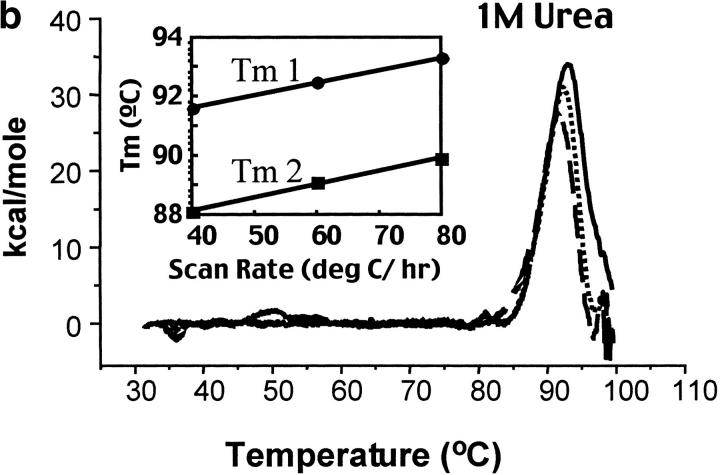
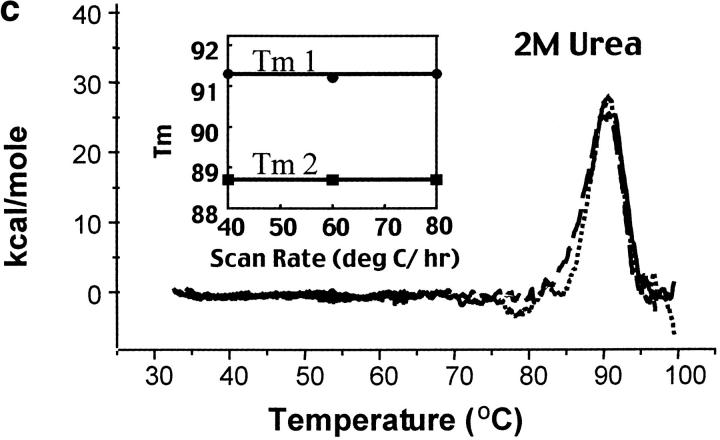
Fig. 2.
Calorimetric scan rate dependence of CYP-119. Differential scanning calorimetry thermograms (baseline subtracted) of CYP-119 in 100 mM potassium phosphate at pH 6.2 (a), 100 mM potassium phosphate at pH 6.2 and 1 M urea (b), and 100 mM potassium phosphate at pH 6.2 and 2 M urea (c). The solid line was taken with a scan rate of 80°C/h, the dotted line at 60°C/h, and the dashed line at 40°C/h. The inset shows the plot of the melting transition versus the scan rate for melting transitions one and two.
There has been some recent success fitting these nonequilibrium scans with a kinetic model (Sanchez-Ruiz et al. 1988; Sanchez-Ruiz 1992; Milardi et al. 1996, 1998). However, another method that has been used to analyze these systems is determining conditions in which the system is no longer scan rate dependent such as altering the pH, the ionic strength, or adding small amounts of chemical denaturant (Relkin 1994; Davoodi et al. 1998). By adding chemical denaturant to the buffer conditions, the production of the proposed aggregate intermediate can be inhibited, therefore, bringing the system into equilibrium. Under these conditions, standard thermodynamic equations can be used to fit the differential scanning calorimetry (DSC) traces to determine melting temperatures and related thermodynamic parameters.
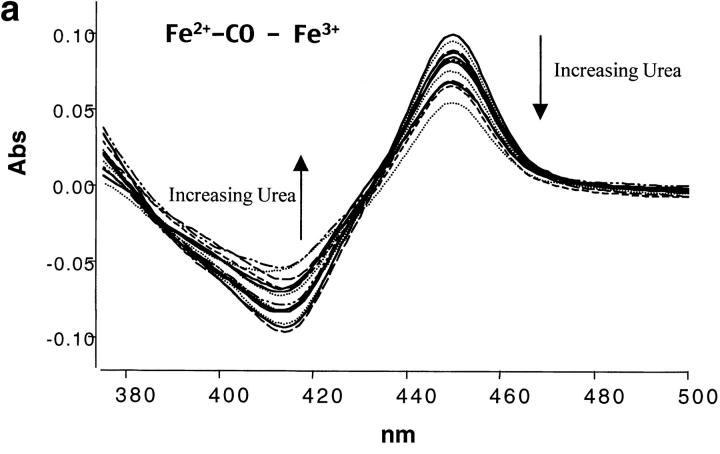
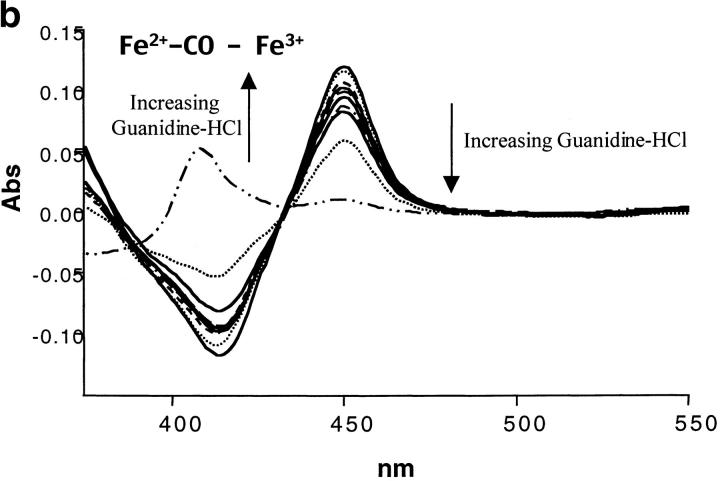
The scan rate dependence of CYP-119 was investigated using both guanidine hydrochloride and urea as cosolutes. The effect of these cosolutes on the protein was first determined optically by monitoring the spectra of reduced CO bound minus oxidized CYP-119 which gives the identifying Soret at 450 nm for cytochrome P450s. Shown in Figure 3 ▶ are the titrations of guanidine and urea on these spectra. From these titrations it was determined that CYP-119 is more stable to urea than guanidine. This was further validated by the 3-degree decrease in the peak maxima of a DSC scan in the presence of 1 M guanidine. This was not the case when urea was added to the solution. As shown in Figure 2b,c ▶, the Tm was dependent on scan rate in the presence of 1 M urea, but became independent in 2 M urea. The perturbation of the native structure of CYP-119 by urea was checked by circular dichroism in the near UV in the presence and absence of 2 M urea. The percent α-helix for CYP-119 with and without 2 M urea present was 44% calculated from the molar ellipticity at 222 nm.
Fig. 3.
The spectral titrations of urea from 0 to 8 M (a) and guanidine-HCl from 0 to 6 M (b) on reduced CO-bound CYP-119 in 50 mM potassium phosphate at pH 6.2. The spectra are difference spectra of ferrous carbon monoxide-bound CYP-119 minus ferric CYP-119.
The DSC thermograms all fit best to a nontwo-state model with two peaks using standard equilibrium equations (Freire 1994). The presence of two melting transitions was apparent from the asymmetric shape of the curve both with and without urea present. The curves are skewed toward the lower temperature and fit to two peaks with Tm for the wild type of 91.9° and 89.3°C in the presence of urea. The two peaks are about 60% and 40% of the total, respectively. The presence of multiple melting transitions has been previously noted for other P450s and attributed to multiple domains (Anzenbacher et al. 1982; Pfeil et al. 1993). However, definable domains are not obvious in the known structures of cytochrome P450s or in the model of CYP-119, therefore, the physical assignment of the two melting transitions is not obvious.
Using the equilibrium conditions determined for wild-type CYP-119, the isolated mutants from the screen were characterized by DSC to determine changes in the thermal stability of the protein. Table 1 lists the midpoint temperature of the first and second transitions (Tm1 and Tm2) for each mutant. The mutants decreased the calorimetric melting transition by as much as 10°C compared to wild type.
Table 1.
DSC melting transition temperatures of wild-type CYP 119 and selected mutants
| Mutation | Tm1 (°C) | Tm2 (°C) |
| Wt | 91.9 | 89.3 |
| K176R, I329M | 91.2 | 87.3 |
| D52V, D72H | ||
| E273G, K348R | 90.4 | 88.1 |
| I272V, N367R, E368I | 89.3 | 86.1 |
| E114D | 88.1 | 84.2 |
| R80G | 87.3 | 84.7 |
| R259K | 86.0 | 83.5 |
| S40C, T67A, V118L | 86.6 | 83.9 |
| R235G, I282V | ||
| I299V, E352K | 84.5 | 80.8 |
| G313E | 83.4 | 79.8 |
| F24S | 81.7 | 75.3 |
Discussion
Recent literature reveals that a large number of mechanisms have been used throughout nature to increase stability of proteins rather than one universal mechanism to stabilize all proteins (Vieille and Zeikus 1996; Jaenicke and Bohm 1998). Nevertheless, within a protein family trends have emerged where one or a couple of specific types of interactions have been used to optimize the stability of that protein family. This is best exemplified in cases where there are multiple structures in a highly homologous protein family, each with increasing thermal stabilities that show the progression of the optimized interactions, such as in the d-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) family (Tanner et al. 1996). In this family, comparison of five structures, with optimal temperatures ranging from 20° to 80°C, showed a correlation between increasing number of hydrogen bonds and salt links and the optimal temperatures, whereas other properties were not correlated. Such examples suggest that particular protein classes may favor certain types of interactions to increase stability. Using this rationale, it should be possible to engineer stability into any member of that protein family, once the fundamental mechanisms for increasing stability in that family are well understood.
The identification of a thermostable P450 provides the opportunity for determining classes of interactions that contribute to stabilizing cytochromes P450. However, with only one thermostable P450 known and very low sequence homology between the members of this superfamily, it remains difficult to discern the contributions to thermostability based on structural comparison alone. Thus, we used a randomized CYP-119 library to identify residues that may contribute to thermal stabilization. Clearly, this was not comprehensive in identifying every residue in CYP-119 contributing to thermostability, as a much larger number of mutant colonies would need to be screened to cover the sequence space. However, this initial set provides a starting point for understanding the type of interactions that may be important in cytochrome P450 stabilization.
The 13 clones with decreased stability had one to four point mutations per clone, with two clones resulting from an insertion or deletion. Both the insertion and deletion mutations are at the amino terminus of the protein. The insertion mutation resulted from the addition of 10 residues (IRIPFQENNK) between the first and second amino acid, which probably occurred from the DNA shuffling.
The deletion mutant arose because of a mutation to the start codon that caused an alternate start codon to be used, at what would normally be M8 in CYP-119. The addition or deletion of residues at the amino terminus most likely destabilize the protein by adding residues that may not fold into a well-defined structure or deleting the majority of the first helix. Therefore, these residues will not be discussed further.
The point mutations were found dispersed throughout the structure, suggesting that there is not a localized region conferring the stability. However, a majority of the mutations were found near the surface of the protein as opposed to the buried core. This seemed somewhat surprising as a large number of mutations that disrupt the packing of the hydrophobic core were expected. However, previous studies, which enhanced the thermostability of proteins through directed evolution, have also shown that most of the stabilizing interactions were found on the surface of the protein (Hoseki et al. 1999; Zhao and Arnold 1999).
Further analysis of the type of mutations identified reveals trends that may indicate the types of interactions used by cytochrome P450 for thermal stability.
Of the 25 point mutations, 13 of the identified mutations were changes of charged residues. Most of the charged residues indicated from our screen appear to either participate in a salt link or are located in a region with several charged residues. This suggests that individual salt links as well as optimization of overall charge–charge interactions, as seen in other proteins (Ibarra-Molero et al. 1999; Loladze et al. 1999; Rhode and Martin 1999; Spector et al. 2000; Strop and Mayo 2000), may play a role in stabilization of cytochrome P450.
Two single point mutants of charged residues retained the positive (R259K) or negative (E114D) charge, but the midpoint of the thermal transition decreased by 5.9° and 3.8°C, respectively. Both residues participate in salt links; therefore, the decrease in stability may be attributable to the weakening or breaking of this ionic interaction by the change in length of the side chain. R259 forms a salt link with the propionate group of the heme and thus, may play a role in both stability of the protein as well as stabilizing the heme group. Residue E114 participates in a salt-link network consisting of three residues, E114, R363, and E342. Interestingly, this salt-link network connects three different β-sheets—β3–1, β3–2, and β3–3—thus, stabilizing the structure by holding secondary structural elements together, which are not close in primary sequence. Although individual analogous salt links are present in some cases in the other P450s with known structure, the entire network is unique to CYP-119. This type of salt-link network appearing only in the thermophile, but not the mesophiles, has also been indicated to contribute to thermostability in the GAPDH family (Tanner et al. 1996).
From all of the mutants isolated, there was only one mutation to an aromatic residue. Clusters of aromatic residues participating in stacking interactions along one side of the molecule were proposed based on structural analysis to contribute to the stabilization of the protein (Yano et al. 2000; S.-Y. Park, K. Yamane, S.-i. Adachi, Y. Shiro, S.A. Maves, K.E. Weiss, and S.G. Sligar, in prep.). The one mutation of an aromatic residue was F24S, which is located in the β1–2 sheet and is part of the large cluster of aromatic residues observed in the structure. The melting transition of this mutant was decreased by 10.2°C from the wild type; however, this large decrease in stability is probably attributable to a combination of the disruption of the aromatic cluster as well as the replacement of a bulky hydrophobic group with a small polar group.
Other than the one aromatic residue, the rest of the hydrophobic residues isolated from the screen were all mutated to other hydrophobic groups and accompanied by one or more additional mutations in the clone in which they were found. Although the small changes in side chain volume (Ile to Val, Val to Leu, and Ile to Met) may have some affect on the stability of the protein, we do not see a strong indication of hydrophobic packing contributing to the increased stability of CYP-119.
Understanding the types of interactions contributing to the enhanced stability of CYP-119 provides a starting point for engineering other P450s for biotechnology application. The mutations isolated from this screen support suggestions made based on structural analysis for molecular interactions contributing to stability. These include charged residues participating in both salt links as well as optimization of charge–charge interactions and aromatic clusters. In addition, cumulative effects of small changes in the side chain volume of hydrophobic residues may contribute to the stability of the protein. Clearly, a larger library would need to be screened to isolate enough mutants to be able to determine statistically all of the interactions contributing to stability. However, the mutants isolated in this work provide experimental evidence that corresponds with the some of the interactions suggested from the structure, providing reasonable targets for engineering stability into other cytochrome P450s.
Materials and methods
Library formation
The plasmid pKS119, encoding the gene for CYP-119 (McLean et al. 1998), was transformed through the random mutator strain XL1-Red (Stratagene) five times to generate the initial library containing random point mutants. Further randomization of the gene was carried out using DNA shuffling techniques (Stemmer 1994a,b) and the random primer recombination method (Shao et al. 1998). Briefly, the initial randomized CYP-119 genes were excised using the PvuII restriction sites, which are found on each side of the gene outside the multiple cloning region of the plasmid. This was then used for the template of a polymerase reaction using a 100-fold excess of random 6-mer primers (Sigma) and Klenow large fragment polymerase. The fragments of DNA between 50 and 200 bp were then isolated using Microcon spin columns with molecular cutoff masses of 10,000 and 100,000 (Amicon) to remove the large template and small primers. These small fragments were then used as the templates of standard DNA shuffling reactions. After the shuffling reaction the resulting 1200-bp product, produced by amplification with the universal forward and reverse primers, which anneal to the plasmid just outside the multiple cloning region, was digested with EcoRI and BamHI, cloned into puc19, and transformed into DH5α E. coli strain to be used for screening.
Screening the library
Individual colonies of the randomized CYP-119 library were inoculated into 96-well microplates (2-mL volume) containing 1.2 mL of 2× YT media per well and grown with shaking at 800 rpm at 30°C for 22 h. Because of the variation in oxygen concentration, the rate of shaking was found to be very important for the level of protein expression. Replica plates were made, and then the microplates were centrifuged and the bacteria lysed using 200 μL of B-Per reagent (Pierce). The lysate was heated at 55°C for 20 min and then pelleted for 30 min at 4000 rpm to remove the precipitated protein. Absorbance spectra of the lysate were taken with the SpectraMax Plus Microplate reader (Molecular Devices) scanning from 350–500 nm. Two wells in every plate were inoculated with cells containing only vector, which were used as a blank to subtract off the high background of the lysate. The background appeared very consistent from well to well and plate to plate. The samples were then heated at 75°C for 20 min, pelleted, and the absorbance spectra read again. Those wells that contained spectra displaying a peak around 416 nm after heating to 55°C, but not after 75°C were selected as potential mutants with decreased thermostability.
Clones of the identified mutants were sequenced by the automated fluorescence-labeled dideoxy chain termination method at the University of Illinois Biotechnology Center.
Protein purification
Selected mutants and wild-type CYP-119 were expressed and purified as previously described (McLean et al. 1998) with the following changes for the mutants. The heat step previously executed at 75°C was reduced to 55°C for 20 min because of the decreased stability of the mutants. The lower temperature did not precipitate near the amount of unwanted proteins from solution as the wild-type protocol; therefore, an additional column was necessary for purification of the mutants. After the ammonium sulfate cut was carried out as previously described, the protein sample was loaded onto a Whatman DE-52 column equilibrated in 30 mM KPi at pH 7.2 and eluted with a gradient of 30–100 mM KPi at pH 7.2. The fractions containing the greatest A415–A280 ratio were pooled and the pooled fractions were further purified with a Bio-Rad P-100 column and a Waters DEAE HPLC column as previously described.
The protein concentrations were determined from the spectra of reduced CO bound minus oxidized protein using the previously determined extinction coefficient of ɛ450–490 = 91 mM−1 (McLean et al. 1998). The protein was reduced with an excess of dithionite in a CO-saturated 50 mM KPi buffer at pH 6.2. All spectra were taken on a Hitachi U3300 spectrophotometer with a spectral bandpass of 1 nm.
Differential scanning calorimetry
The scanning calorimetry was performed on the MCS DSC unit from Microcal (Northhampton, MA). Initial scans of the wild-type CYP-119 were performed in 100 mM KPi at pH 6.2 with protein concentrations between 30 and 100 μM and scan rates between 20° and 80°C /h. To determine the conditions at which the system was independent of scan rate, scans were repeated with the addition of 1 M guanidine, 1 M urea, or 2 M urea. All of the mutants were prepared in buffers containing 100 mM KPi at pH 6.2, 2 M urea, and 30 μM protein and scanned with a rate of 60°C/h.
The dependence of molar heat capacity on temperature was analyzed using the ORIGIN software (Microcal Ltd.). First, the data were fit to a chemical baseline that was then subtracted from the experimental data to remove any effects from changes in the molar heat capacity before and after the transition. The data were fit into several models including a two-state transition with one or two peaks and a non-two-state transition with one or two peaks. All of the experimental data including all of the mutants fit best to a non-two-state model with two transitions.
The difference between the two-state model and the non-two-state model is the addition of the van't Hoff heat parameter in the second model, because in the simplified two-state model the calorimetric heat and the van't Hoff heat are equal.
The midpoint of each transition (Tm) was determined from the fit for each of the mutants.
Circular dichroism
The circular dichroism spectra were taken on a Jasco J-720 Spectropolarimeter with a 0.1-cm pathlength in 10 mM Kpi buffer at pH 6.2, 4 μM protein, and either 1 M guanidine hydrochloride or 2 M urea.
Acknowledgments
This work is supported by National Institutes of Health (NIH) grants R01-GM 33775 and R01-GM 31756 as well as the support of a NIH Molecular Biophysics Training Grant. We thank Aretta Weber for excellent help in manuscript preparation.
The publication costs of this article were defrayed in part by payment of page charges.This article must therefore be hereby marked ``advertisement'' in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.17601.
References
- Andersen, J.F. and Hutchinson, C.R. 1992. Characterization of Saccharopolyspora erythraea cytochrome P-450 genes and enzymes, including 6-deoxyerythronolide B hydroxylase. J. Bacteriol. 174 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzenbacher, P., Hudecek, J., and Struzinsky, R. 1982. Study of thermal stability of cytochrome P450 by differential scanning calorimetry. FEBS Lett. 149 208–210. [Google Scholar]
- Arnold, F.H. and Volkov, A.A. 1999. Directed evolution of biocatalysts. Curr. Opin. Chem. Biol. 3 54–59. [DOI] [PubMed] [Google Scholar]
- Auerbach, G., Ostendorp, R., Prade, L., Korndorfer, I., Dams, T., Huber, R., and Jaenicke, R. 1998. Lactate dehydrogenase from the hyperthermophilic bacterium Thermotoga maritima—The crystal structure at 2.1 angstrom resolution reveals strategies for intrinsic protein stabilization. Structure 6 769–781. [DOI] [PubMed] [Google Scholar]
- Chang, Y.-T. and Loew, G. 2000. Homology modeling, molecular dynamics simulations, and analysis of CYP119, a P450 enzyme from extreme acidothermophilic archaeon Sulfolobus solfataricus. Biochemistry 39 2484–2498. [DOI] [PubMed] [Google Scholar]
- Cowan, D.A. 1995. Protein stability at high temperatures. Essays Biochem. 29 193–207. [PubMed] [Google Scholar]
- Davoodi, J., Wakarchuk, W.W., Surewicz, W.K., and Carey, P.R. 1998. Scan-Rate Dependence in protein calorimetry—The reversible transitions of Bacillus circulans xylanase and a disulfide-bridge mutant. Protein Sci. 7 1538–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire, E. 1994. Statistical thermodynamic analysis of differential scanning calorimetry data: Structural deconvolution of heat capacity function of proteins. Methods Enzymol. 240 502–530. [DOI] [PubMed] [Google Scholar]
- Harris, J.L. and Craik, C.S. 1998. Engineering enzyme specificity. Curr. Opin. Chem. Biol. 2 127–132. [DOI] [PubMed] [Google Scholar]
- Hoseki, J., Yano, T., Koyama, Y., Kuramitsu, S., and Kagamiyama, H. 1999. Directed evolution of thermostable kanamycin-resistance gene: A convenient selection marker for Thermus thermophilus. J. Biochem. 126 951–956. [DOI] [PubMed] [Google Scholar]
- Ibarra-Molero, B., Loladze, V.V., Makhatadze, G.I., and Sanchez-Ruiz, J.M. 1999. Thermal versus guanidine-induced unfolding of ubiquitin. An analysis in terms of the contributions from charge–charge interactions to protein stability. Biochemistry 38 8138–8149. [DOI] [PubMed] [Google Scholar]
- Jaenicke, R. 1998. What ultrastable globular proteins teach us about protein stabilization. Biochemistry-Moscow 63 312–321. [PubMed] [Google Scholar]
- Jaenicke, R. and Bohm, G. 1998. The stability of proteins in extreme environments. Curr. Opin. Structural Biol. 8 738–748. [DOI] [PubMed] [Google Scholar]
- Lee, B. and Vasmatzis, G. 1997. Stabilization of protein structures. Curr. Opin. Biotechnol. 8 423–428. [DOI] [PubMed] [Google Scholar]
- Loladze, V.V., Ibarra-Molero, B., Sanchez-Ruiz, J.M., and Makhatadze, G.I. 1999. Engineering a thermostable protein via optimization of charge–charge interactions on the protein surface. Biochemistry 38 16419–16423. [DOI] [PubMed] [Google Scholar]
- Maes, D., Zeelen, J.P., Thanki, N., Beaucamp, N., Alvarez, M., Thi, M.H.D., Backmann, J., Martial, J.A., Wyns, L., Jaenicke, R., and Wierenga, R.K. 1999. The crystal structure of triosephosphate isomerase (TIM) from Thermtotoga maritima: A comparative thermostability structural analysis of ten different TIM structures. Proteins 37 441–453. [PubMed] [Google Scholar]
- McLean, M.A., Maves, S.A., Weiss, K.E., Krepich, S., and Sligar, S.G. 1998. Characterization of a cytochrome P450 from the acidothermophilic archaea Sulfolobus solfataricus. Biochem. Biophys. Res. Comm. 252 166–172. [DOI] [PubMed] [Google Scholar]
- Milardi, D., Larosa, C., and Grasso, D. 1996. Theoretical basis for differential scanning calorimetric analysis of multimeric proteins. Biophys. Chem. 62 95–108. [DOI] [PubMed] [Google Scholar]
- Milardi, D., Larosa, C., Grasso, D., Guzzi, R., Sportelli, L., and Fini, C. 1998. Thermodynamics and kinetics of the thermal unfolding of plastocyanin. Eur. Biophys. J. 27 273–282. [Google Scholar]
- Nelson, D.R. 1999. Cytochrome P450 and the individuality of species. Arch. Biochem. Biophys. 369 1–10. [DOI] [PubMed] [Google Scholar]
- Pace, C.N., Shirley, B.A., McNutt, M., and Gajiwala, K. 1996. Forces contributing to the conformational stability of proteins. FASEB J. 10 75–83. [DOI] [PubMed] [Google Scholar]
- Pfeil, W., Nolting, B.O., and Jung, C. 1993. Apocytochrome P450cam is a native protein with some intermediate-like properties. Biochemistry 32 8856–8862. [DOI] [PubMed] [Google Scholar]
- Privalov, P.L. 1979. Stability of proteins: Small globular proteins. Adv. Prot. Chem. 33 167–241. [DOI] [PubMed] [Google Scholar]
- Relkin, P. 1994. Differential scanning calorimetry—A useful tool for studying protein denaturation. Thermochimica Acta 246 371–386. [Google Scholar]
- Rhode, D.J. and Martin, B.L. 1999. Localized structural effects of electrostatic interactions in a thermostable enzyme. Biochem. Biophys. Res. Comm. 258 179–183. [DOI] [PubMed] [Google Scholar]
- Robertson, A.D. and Murphy, K.P. 1997. Protein structure and the energetics of protein stability. Chem. Rev. 97 1251–1267. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ruiz, J.M. 1992. Theoretical analysis of Lumry-Eyring models in differential scanning calorimetry. Biophys. J. 61 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Ruiz, J.M., Lopez-Lacomba, J., Cortijo, M., and Mateo, P.L. 1988. Differential scanning calorimetry of the irreversible thermal denaturation of thermolysin. Biochemistry 27 1648–1652. [DOI] [PubMed] [Google Scholar]
- Shafiee, A. and Hutchinson, C.R. 1987. Macrolide antibiotic biosynthesis: Isolation and properties of two forms of 6-Deoxyerythronolide B hydroxylase from Saccharopolyspora erythraea (Streptomyces erythreus). Biochemistry 26 6204–6210. [DOI] [PubMed] [Google Scholar]
- Shao, Z.X. and Arnold, F.H. 1996. Engineering new functions and altering existing functions. Curr. Opin. Structural Biol. 6 513–518. [DOI] [PubMed] [Google Scholar]
- Shao, Z., Zhao, H., Giver, L., and Arnold, F.H. 1998. Random-priming in vitro recombination: An effective tool for directed evolution. Nucleic Acids Res. 26 681–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnyrov, V.L., Sanchezruiz, J.M., Boiko, B.N., Zhadan, G.G., and Permyakov, E.A. 1997. Applications of scanning microcalorimetry in biophysics and biochemistry. Thermochimica Acta 302 165–180. [Google Scholar]
- Spector, S., Wang, M., Carp, S.A., Robblee, J., Hendsh, Z.S., Fairman, R., Tidor, B., and Raleigh, D.P. 2000. Rational modification of protein stability by the mutation of charged surface residues. Biochemistry 39 872–879. [DOI] [PubMed] [Google Scholar]
- Steipe, B. 1999. Evolutionary approaches to protein engineering. Curr. Top. Microbiol. Immunol. 243 55–86. [DOI] [PubMed] [Google Scholar]
- Stemmer, W.P.C. 1994a. DNA shuffling by random fragmentation and reassembly: In vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. 91 10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1994b. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370 389–391. [DOI] [PubMed] [Google Scholar]
- Strop, P. and Mayo, S.L. 2000. Contribution of surface salt bridges to protein stability. Biochemistry 39 1251–1255. [DOI] [PubMed] [Google Scholar]
- Tanner, J.J., Hecht, R.M., and Krause, K.L. 1996. Determinants of enzyme thermostability observed in the molecular structure of Thermus aquaticus D-Glyceraldehyde-3-phosphate dehydrogenase at 2.5 A resolution. Biochemistry 35 2597–2609. [DOI] [PubMed] [Google Scholar]
- Uvarov, V.Y., Bachmanova, G.I., Archakov, A.I., Sukhomudrenko, A.G., and Myasoedova, K.N. 1980. Conformation and thermostability of soluble cytochrome P-450 and cytochrome P-450 incorporated into liposomal membrane. Biochemistry-USSR 45 1108–1113. [DOI] [PubMed] [Google Scholar]
- Vieille, C. and Zeikus, J.G. 1996. Thermozymes—Identifying molecular determinants of protein structural and functional stability. Trends Biotechnol. 14 183–190. [Google Scholar]
- Yano, J.K., Koo, L.S., Schuller, J.S., LI, H., Montellano, P.R., and Poulos, T.L. 2000. Crystal structure of a thermophilic cytochrome P450 from the archaeon Sulfolobus solfataricus. J. Biol. Chem. 275 31086–31092. [DOI] [PubMed] [Google Scholar]
- Zeikus, J.G., Vieille, C., and Savchenko, A. 1998. Thermozymes: Biotechnology and structure–function relationships. Extremophiles 2 179–183. [DOI] [PubMed] [Google Scholar]
- Zhao, H.M. and Arnold, F.H. 1999. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Engineer. 12 47–53. [DOI] [PubMed] [Google Scholar]