Abstract
Fragments of 419 bp of the UL16 open reading frame from 73 psittacid herpesviruses (PsHVs) from the United States and Europe were sequenced. All viruses caused Pacheco's disease, and serotypes of the European isolates were known. A phylogenetic tree derived from these sequences demonstrated that the PsHVs that cause Pacheco's disease comprised four major genotypes, with each genotype including between two and four variants. With the exception of two viruses, the serotypes of the virus isolates could be predicted by the genotypes. Genotypes 1 and 4 corresponded to serotype 1 isolates, genotype 2 corresponded to serotype 2 isolates, and genotype 3 corresponded to serotype 3 isolates. The single serotype 4 virus mapped to genotype 4. DNA from a virus with a unique serotype could not be amplified with primers that amplified DNA from all other PsHVs, and its classification remains unknown. Viruses representing all four genotypes were found in both the United States and Europe, and it was therefore predicted that serotypes 1, 2, and 3 were present in the United States. Serotype 4 was represented by a single European isolate that could not be genetically distinguished from serotype 1 viruses; therefore, the presence of serotype 4 in the United States could not be predicted. Viruses of genotype 4 were found to be the most commonly associated with Pacheco's disease in macaws and conures and were least likely to be isolated in chicken embryo fibroblasts in the United States. All four genotypes caused deaths in Amazon parrots, but genotype 4 was associated with Pacheco's disease only in Amazons in Europe. Genotypes 2, 3, and 4, but not 1, were found in African grey parrots. Although parrots from the Pacific distribution represent a relatively small percentage of the total number of birds with Pacheco's disease, all four genotypes were found to cause disease in these species.
The psittacid herpesvirus (PsHV), an alphaherpesvirus, causes Pacheco's disease (20). Pacheco's disease is an acute, generally fatal disease of psittacine birds (parrots) (3, 9, 12, 14, 16, 17). Many parrot species originating from multiple geographic regions are susceptible to Pacheco's disease. However, Amazon parrots (Amazona spp.) account for the majority of cases, followed by African grey parrots (Psittacus erithacus), macaws (Ara spp.), cockatoos (Cacatua spp.), and conures (Aratinga spp. and Pyrrhura spp.) (20). Pacheco's disease was first recognized in the late 1920s in Brazil (11) but was not seen again until the 1970s, when outbreaks of disease occurred in quarantined and recently-released-from-quarantine wild-caught parrots intended for the pet trade (14, 17). Although the trade in wild-caught parrots has diminished, outbreaks of Pacheco's disease continue to occur in quarantine stations and facilities where parrots are housed in close proximity (10, 13).
Increasing evidence suggests that one or more PsHV genotypes may also cause internal papillomatosis of parrots (8, 18, 19). This debilitating disease is common in imported and captive-raised Amazon parrots and macaws, in which it causes wartlike lesions in the oral cavity, upper digestive system, and cloaca.
At least three and possibly as many as five serotypes of PsHV are recognized. A study in Europe suggests that serotype 1 is the most common cause of Pacheco's disease, followed by serotype 2 and, less frequently, serotype 3. Serotypes 4 and 5 are represented only by single isolates (6). The serotypes causing Pacheco's disease in the United States and in other countries outside Europe are not known and would be difficult to determine as exchange of virus isolates and sera between countries is strictly regulated.
Within the PsHVs there appears to be a significant amount of genetic heterogeneity (1, 7, 15, 20). Restriction fragment length polymorphism (RFLP) analysis of the entire genome of 31 PsHVs of European origin produced eight major migration patterns (A through H). RFLP analysis with two additional restriction enzymes results in group A's being further divided into groups AA1A1, AA2A2, and AA3A3. Patterns B and C subdivide into BB1B1 and BB2B2 and CC1C and CC2C, respectively. There was a close, but incomplete, correlation between RFLP group A and serotype 1, with eight of nine serotype 1 viruses having the group A migration profile. However, one serotype 1 virus had a unique migration profile and a serotype 3 virus had a group A profile, as did the only serotype 4 virus. Other serotypes were not consistently defined by one migration profile. Four serotype 2 viruses were examined and had three migration profiles. A subset of viruses (PsHV 2/3) that are neutralized by antibodies raised to both serotypes 2 and 3 also had a unique migration profile. Three serotype 3 viruses were examined; two had unique migration profiles, and the third had an AA1A1 profile. Serotype 5 was the most difficult to grow and had the most divergent migration pattern (15).
A similar degree of genetic heterogenicity was found previously when 59 PsHVs from parrots of U.S. origin with Pacheco's disease were examined by PCR. In this study (20), five PCR primer sets produced 10 different patterns of amplification. DNA from most viruses (61.8%) could be amplified with all five primer sets, and the authors speculated that these viruses may be serotype 1 viruses (20).
The current methods of classifying the PsHVs have some significant limitations. Both serologic and RFLP analysis require that the viruses be isolated in cell culture, and this has not proved to be possible in a significant number of cases. Additionally, RFLP analysis requires that a significant amount of virus be grown and purified. PCR analysis is much more rapid and can be done with DNA extracted from infected tissue, bypassing the virus isolation step. However, unless the results of PCR are correlated with the specific serotypes or known genotypes, they have limited meaning.
A basic understanding of the molecular phylogeny of the PsHVs that includes multiple sequences from the known serotypes would be of great benefit. This would permit the correlation between genotype and serotype, allowing investigators from around the world to determine with which PsHVs they are working. It would also permit a more detailed understanding of the relationships between the genotypes and allow investigations into the possible relationship between genotype and pathogenicity. Finally, additional sequence data would facilitate the development of PCR primers that could detect all PsHVs in necropsy specimens and apparently uninfected birds and differentiate the viruses to genotype level.
In this investigation, we describe the molecular phylogeny of PsHVs from the United States and Europe that cause Pacheco's disease. We show that there is a distinct correlation between genotype and serotype and that the serotypes present in Europe are also present in the United States. We also show that within serotypes there is additional genetic diversity that may correlate with various levels of pathogenicity for different species of parrots and the viruses' ability to be isolated in chicken embryo fibroblasts (CEFs).
MATERIALS AND METHODS
Samples.
Two sets of samples were analyzed for this study. The first set consisted of PsHVs of known serotypes isolated from dead parrots in Europe with lesions consistent with those of Pacheco's disease (Table 1). These viruses were isolated in CEFs and shipped to the United States as lyophilized cell culture media (U.S. Department of Agriculture permit number 1999-3566). Viruses were classified serologically with cross-neutralization testing by using serum produced in rabbits against each virus (6). The second set of samples comprised tissues collected from whole, dead birds submitted for diagnostic necropsy to the Schubot Exotic Bird Center (Texas A&M University, College Station) and the Texas Veterinary Diagnostic Laboratory (College Station, Tex.) with a resulting diagnosis of Pacheco's disease (Table 2). The diagnosis of Pacheco's disease was based on the finding of characteristic histologic lesions and in many cases on the detection of herpesvirus virions by electron microscopy or the detection of herpesvirus-infected cells in impression smears of spleen or liver by use of a fluorescent antibody conjugate prepared against a PsHV isolated from a bird with Pacheco's disease. U.S. samples were included in this study based on previous PCR patterns of amplification and the species of bird from which they originated.
TABLE 1.
Isolate numbers, species of origin, serotypes, genotypes, and accession numbers of European PsHVs sequenced in this study
| Isolate no. | Species | Common name | Serotype | Genotype | Accession no. |
|---|---|---|---|---|---|
| KS144/79 | Amazona aestiva | Blue-fronted Amazon | 1a | 1 | AY282614 |
| 421/80 | Psittacus erithacus | African grey parrot | 1a | 4 | AY282660 |
| 80540/82 | Unknown | Parrot | 1a | 1 | AY282615 |
| 1688/87 | Pionus menstruus | Pionus parrot | 1b | 4 | AY282661 |
| 6840/87 | Amazona aestiva | Blue-fronted Amazon | 2a | 2 | AY282624 |
| 6851/87 | Amazona sp. | Amazon | 2a | 2 | AY282625 |
| 6899/87 | Psittacus erithacus | African grey parrot | 2a | 2 | AY282626 |
| 6919/87 | Psittacus erithacus | African grey parrot | 2a | 2 | AY282623 |
| 8326/87 | Pyrrhura rhodogaster | Crimson-bellied conure | 5a | ||
| 1622/88 | Amazona versicolor | St. Lucia Amazon | 3a | 3 | AY282635 |
| 3115/88 | Amazona ochrocephala | Yellow-crowned Amazon | 3a | 3 | AY282636 |
| 5906/89 | Amazona versicolor | St. Lucia Amazon | 3a | 3 | AY282637 |
| 1978/90 | Amazona aestiva | Blue-fronted Amazon | 1a | 4 | AY282662 |
| 5180/90 | Amazona aestiva | Blue-fronted Amazon | 1a | 4 | AY282663 |
| 132/91 | Amazona ochrocephala | Yellow-crowned Amazon | 3a | 4 | AY282664 |
| 5045/91 | Psittaculirostris sp. | Large fig parrot | 1a | 1 | AY282616 |
| 730/II/92 | Ara ararauna | Blue and gold macaw | 1a | 4 | AY282666 |
| 451 I/92 | Amazona aestiva | Blue-fronted Amazon | 1a | 4 | AY282665 |
| 1043/92 | Amazona farinosa | Mealy Amazon | 1a | 4 | AT282667 |
| 1483/92 | Amazona leucocephala | Cuba Amazon | 4a | 4 | AY282668 |
| 1585/92 | Amazona sp. | Amazon | 1a | 4 | AY282669 |
| 1803/92 | Amazona farinosa, Amazon ochrocephala, Pionus menstruus, Aratinga pertinax | Pooled sample (Amazons, pionus, and conure) | 2c | 2 | AY282627 |
| 2371/92 | Amazona ochrocephala | Yellow-crowned Amazon | 3a | 3 | AY282638 |
| 1070/93 | Amazona amazonica | Orange-winged Amazon | 1a | 2 | AY282628 |
| 4739/93 | Amazona ochrocephala | Yellow-crowned Amazon | 3a | 3 | AY282639 |
| 2864/94 | Amazona versicolor | St. Lucia Amazon | 2/3c | 3 | AY282640 |
| 3787/94 | Cacatua alba | White cockatoo | 2c | 2 | AY282629 |
| 4294/94 | Amazona versicolor | St. Lucia Amazon | 2/3c | 3 | AY282641 |
| 1255/95 | Ara chloroptera | Green-winged macaw | 1c | 4 | AY282670 |
| 632/96 | Amazona viridigenalis | Green-cheeked Amazon | 2c | 2 | AY282630 |
TABLE 2.
Virus designations, species of origin, PCR groups, genotypes, abilities to be isolated in CEFs, and accession numbers of U.S. PsHVs sequenced in this study
| Virus | Species | Common name | PCR groupa | Replication in CEFsb | Genotype | Accession no. |
|---|---|---|---|---|---|---|
| 87-0282 | Amazona autopalliata | Yellow-naped Amazon | 2 | Positive | 3 | AY282642 |
| 88-0064 | Cacatua moluccensis | Moluccan cockatoo | 1 | Positive | 1 | AY282617 |
| 88-0097 | Ara macao | Scarlet macaw | 1 | Positive | 4 | AY282671 |
| 88-0476 | Amazona aestiva | Blue-fronted Amazon | 6 | Negative | 3 | AY282643 |
| 88-0637 | Amazona autopalliata | Yellow-naped Amazon | 6 | 3 | AY282644 | |
| 88-0723 | Amazona amazonica | Orange-winged Amazon | 6 | Positive | 3 | AY282645 |
| 88-1069 | Eclectus roratus | Eclectus | Negative | 4 | AY282672 | |
| 89-0326 | Amazona autopalliata | Yellow-naped Amazon | 6 | Positive | 3 | AY282646 |
| 89-1117 | Cacatua moluccensis | Moluccan cockatoo | Negative | 4 | AY282673 | |
| 89-1314 | Amazona oratrix | Yellow-headed Amazon | 1 | 1 | AY282618 | |
| 90-0032 | Eolophus roseicapillus | Galah | 1 | Positive | 3 | AY282647 |
| 90-0753 | Ara chloroptera | Green-winged macaw | 1 | Negative | 4 | AY282674 |
| 91-0451 | Ara macao | Scarlet macaw | 1 | Negative | 4 | AY282675 |
| 91-1171 | Psittacus erithacus | African grey parrot | 1 | Positive | 4 | AY282676 |
| 91-1185 | Amazona sp. | Yellow-necked Amazon | 7 | Positive | 3 | AY282648 |
| 92-0115 | Amazona farinosa | Mealy Amazon | 2 | Negative | 3 | AY282649 |
| 92-0365 | Amazona autumnalis | Red-lored Amazon | 1 | Negative | 2 | AY282631 |
| 92-0581 | Amazona viridigenalis | Green-cheeked Amazon | 1 | Negative | 3 | AY282650 |
| 92-0980 | Psittacus erithacus | African grey parrot | 10 | Negative | 4 | AY282677 |
| 92-1289 | Psittacus erithacus | African grey parrot | 9 | Negative | 4 | AY282678 |
| 93-0039 | Nymphicus hollandicus | Cockatiel | 1 | Positive | 4 | AY282679 |
| 93-0338 | Psittacus erithacus | African grey parrot | 2 | Negative | 3 | AY282651 |
| 93-1190 | Amazona oratrix | Yellow-headed Amazon | 1 | Positive | 1 | AY282619 |
| 93-1304 | Psittacus erithacus | African grey parrot | 1 | Negative | 4 | AY282680 |
| 96-0036 | Aratinga canicularis | Orange-fronted conure | 1 | 4 | AY282681 | |
| 97-0050 | Psittacara erythrogenys | Cherry-headed conure | 7 | 4 | AY282682 | |
| 97-0001 | Amazona sp. | Amazon | 1 | Positive | 1 | AF261756 |
| 98-0114 | Platycerus sp. | Rosella | 1 | 1 | AY282620 | |
| 98-0020 | Ara sp. | Macaw | 1 | 4 | AY282683 | |
| 98-0025 | Aratinga sp. | Conure | 1 | 4 | AY282684 | |
| 98-0026 | Psittacus erithacus | African grey parrot | 2 | 2 | AY282632 | |
| 98-0027 | Amazona sp. | Amazon | 8 | 2 | AY282633 | |
| 98-0032 | Unknown | Parrot | 4 | 2 | AY282634 | |
| 99-0009 | Psittacus erithacus | African grey parrot | 4 | 3 | AY282652 | |
| 99-0019 | Aratinga sp. | Conure | 3 | AY282652 | ||
| 99-0020 | Nymphicus hollandicus | Cockatiel | 1 | 1 | AY282621 | |
| 99-0021 | Ara sp. | Macaw | 6 | 3 | AY282654 | |
| 99-0025 | Ara macao | Scarlet macaw | 1 | 4 | AY282685 | |
| 99-0027 | Amazona finschi | Lilac-crowned Amazon | 1 | AY282622 | ||
| 99-0028 | Callocephalon fimbriatum | Gang-gang cockatoo | 3 | AY282655 | ||
| 99-0042 | Amazona aestiva | Blue-fronted Amazon | 2 | 3 | AY282656 | |
| 01-0033 | Amazona oratrix | Yellow-headed Amazon | 3 | AY282657 | ||
| 02-0040 | Amazona sp. | Amazon | 3 | AY282658 | ||
| 02-0056 | Tanygnathus megalorhynchos | Great-billed parrot | 3 | AY282659 |
PCR group as presented in Tomaszewski et al. (20).
Indicates positive or negative replication in cell culture (unpublished data).
Isolation of herpesviruses from birds from the United States.
Virus isolation attempts were made with tissues from 22 birds with Pacheco's disease (Table 1) (12). CEFs derived from 11-day-old specific-pathogen-free embryos (SPAFAS; Charles River, Wilmington, Mass.) were grown until 75% confluent. The fibroblast monolayer was incubated with a 10% (wt/vol) homogenate of liver or combined liver and spleen in cell culture medium for 1 h (12). The homogenate was aspirated from the cells, the cells were washed, and the monolayers were observed daily for 7 days. The presence of a herpesvirus in the CEF monolayers was confirmed by characteristic cytopathic effects and the presence of eosinophilic intranuclear inclusion bodies within CEFs. Up to four blind passages were undertaken before it was concluded that the virus would not grow in this culture system.
PCR procedures.
Lyophilized cell culture medium was rehydrated overnight in 500 μl of sterile H2O. A 100-μl aliquot was boiled (5 min), and denatured proteins were pelleted by centrifugation. The supernatant was added directly to the PCR mix. Additionally, 100 μl of rehydrated isolate 8326/87 (serotype 5) was subjected to DNA purification by using the Puregene DNA isolation kit (Gentra Systems), with a final elution volume of 10 μl of H2O. DNA from tissues was purified using the Puregene DNA isolation kit.
The PsHV UL17/16 open reading frame (AF261756) was aligned with the UL17/16 open reading frames of other alphaherpesviruses from the GenBank database to identify conserved regions that could be used to develop a PCR primer set that could amplify DNA from all PsHVs. Forward primer 5′-TGCGTGGGGTTAAACTCGGAAC-3′ and reverse primer 5′-CGACTACACGAGCCTAACATC-3′ were selected. PCR was performed as described by Tomaszewski et al. (20). Briefly, a 25-μl reaction mix containing 100 ng of genomic DNA (or 1 μl of cell culture supernatant), 25 pmol of each primer, four 0.1 mM (each) deoxynucleotide triphosphates, 2.5 mM magnesium chloride, 0.75 U of Taq, and 1× buffer A (Promega) was used with the following protocol: an initial denaturation step of 94°C for 5 min, followed by 40 cycles of 60°C for 45 s, 72°C for 90 s, and 94°C for 30 s, followed by a final extension cycle of 72°C for 5 min.
Amplification products were separated by electrophoresis on a 1% agarose gel. Amplicons were purified with QIAquick gel extraction (Qiagen) prior to sequencing. An amplicon was produced from each sample except isolate 8326/87 (serotype 5). PsHV primer sets 9F, 9R, 11F, 11R, and 23F (20) and DNA polymerase consensus degenerate primers DFA and KGI followed by nested primers ILK and IYG were subsequently used in an effort to amplify DNA from three different samples of this isolate (21).
DNA sequencing and sequence analysis.
PCR amplicons were sequenced directly with Big Dye Terminator DNA sequencing kit and ABI 377 DNA sequencer (Applied Biosystems) by using the amplification primers and forward primers 5′-CGATCCTCTATTGACCATCCTTAC-3′ and 5′-TACCTAACCAAGCCCAAACGTAG-3′ as needed. Sequences were submitted to GenBank (Tables 1 and 2). UL16 gene sequences (nucleotides 1 to 419) were aligned by Sequencher 3.1.2 (Gene Codes Corporation). Sequences were imported to Paup* 4.0 for analysis. Phylogenetic trees were constructed using bootstrap resampling (1,000 replicates) with neighbor joining. Parsimony with heuristic search was used to construct trees determining phenotypic relationships.
Statistical analysis.
The chi-square test of independence was used to test the hypotheses that the percentage of macaws affected by genotype 4 was significantly higher than that of macaws affected by other genotypes, that genotype 4 viruses from the United States were less likely to be isolated in CEFs than other genotypes, that genotype 4 was less likely than all other genotypes combined to be found in Amazon parrots in the United States, and that genotype 1 was less likely than all other genotypes combined to be found in African grey parrots (2, 13). Two samples for which the species of parrot was not identified and one sample in which the virus was isolated from a pool of three species were not included in these calculations. Results were considered significant at P values of ≤0.05.
Nucleotide sequence accession numbers.
Sequences were deposited in GenBank under the accession numbers listed in Tables 1 and 2.
RESULTS
PCR amplification of PsHV DNA.
Viral DNA was amplified from all of the PsHVs except isolate 8326/87 (serotype 5) by using the consensus primers. Primer sets 9F, 9R, 11F, 11R, and 23F derived from the sequence of a known PsHV and the degenerate DNA polymerase primers DFA and KGI followed by nested primers ILK and IYG were used with PCR in an attempt to amplify DNA from the 8326/87 isolate. None of these primer sets resulted in DNA amplification.
Phylogenetic analysis.
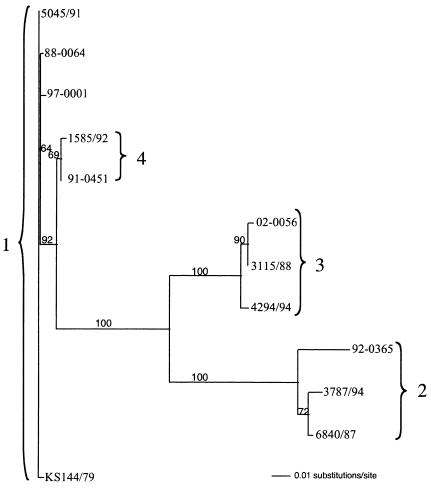
Sequences of 419 bp comprising the end of UL16 corresponding to the amino terminus from 73 PsHVs were aligned. A total of 91 point mutations occurred at 88 loci. There was also a 6-bp deletion (bases 173 to 178). Sequence analysis showed 12 genetic variants from the 73 isolates, with each sequence occurring 1 to 24 times. With the use of bootstrap resampling with neighbor joining, a phylogenetic tree was determined (Fig. 1). Ten viruses mapped to genotype 1, and within genotype 1, three point mutations defined four variants. Twelve viruses mapped to genotype 2, and within genotype 2, 17 point mutations defined three variants. Twenty-five viruses mapped to genotype 3, and within genotype 3, five point mutations defined four variants. Twenty-seven viruses mapped to genotype 4, and one point mutation defined the genotype's two variants. Genotype 4 was distinguished from all other genotypes by the 6-bp deletion.
FIG. 1.
Phylogenetic tree of representatives of the 12 unique PsHV UL16 partial sequences created by distance (Kimura two-parameter) analysis with 103 neighbor-joining bootstrap replicates by using PAUP* 4.0. Bootstrap values above 50 are displayed.
Correlation of molecular phylogeny with RFLP patterns.
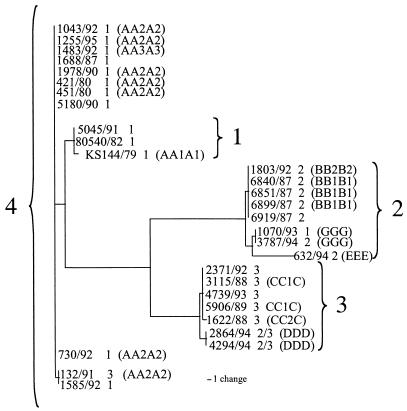
With the use of RFLP analysis of the entire virus genome, it was predicted that there would be eight major PsHV genotypes (A through H) with additional subgroups AA1A1, AA2A2, AA3A3, BB1B1, BB2B2, CC1C, and CC2C. The single AA1A1 isolate sequence mapped to genotype 1. All seven AA2A2 isolates mapped to genotype 4 and could be separated into two variants. The AA3A3 isolate also mapped to genotype 4. This isolate could not be distinguished with our data from the most common AA2A2 variant. The BB1B1 and BB2B2 isolates had identical sequences and mapped to genotype 2. CC1C and CC2C differed by a single point mutation and mapped to genotype 3. The DDD viruses, both of which were serotype 2/3, mapped to a unique branch of genotype 3. The EEE, FFF, and GGG viruses all mapped to a distinct branch of genotype 2. FFF and GGG, however, could not be distinguished from each other (Fig. 2).
FIG. 2.
Phylogenetic tree of sequenced European isolates created using the maximum-parsimony method of PAUP* 4.0 with serotypes and RFLP patterns added (15).
Correlation of molecular phylogeny with patterns of PCR amplification.
Of the 73 viruses examined in this study, 34 PsHVs were previously characterized by PCR. In the previous study, five sets of PCR primers were used to screen each herpesvirus and 10 amplification patterns were detected (20). Viruses representing eight of these patterns (1,2, 4, and 6 to 10) were examined in our present study. There were 18 PCR group 1 viruses, and 15 (83%) were either genotype 1 or 4, the two most closely related genotypes. Genotype 1 was not found among other PCR groups. The single viruses in PCR groups 9 and 10 and one of two representatives in PCR group 7 also mapped to genotype 4. Genotype 2 was found once among four PCR groups (1, 2, 4, and 8), and genotype 3 was found among five PCR groups (1, 2, 4, 6, and 7). All five PCR group 6 viruses were genotype 3.
Correlation of molecular phylogeny with serotypes.
The serotype 1 viruses mapped to genotypes 1 and 4, with the exception of isolate 1070/93 (FFF), which mapped to genotype 2. The six serotype 2 viruses mapped to the three branches of genotype 2. The three serotype 3 viruses mapped to one branch of genotype 3. The two serotype 2/3 viruses mapped to a second unique branch of genotype 3. A single serotype 3 virus mapped to genotype 4. It had the same RFLP pattern as several other viruses in group 4 and could not be distinguished with our data from other viruses in this group. The only serotype 4 virus had an RFLP pattern of AA2A2, and it mapped to a branch of genotype 4 with another AA2A2 virus (Fig. 2). Representatives of all genotypes were found among U.S. viruses, and these viruses are predicted to have the same serotypes as the European isolates with the same sequences.
Abilities of U.S. PsHVs of different genotypes to grow in CEFs.
Attempts at isolation in CEFs were made with 23 PsHVs from the United States. Three of three genotype 1 viruses, five of nine genotype 3 viruses, and two of 10 genotype 4 viruses were successfully isolated. The one genotype 2 virus was not isolated. The percentage of genotype 4 viruses from the United States that could be isolated in CEFs was statistically significantly smaller than that of U.S. viruses of the three other genotypes combined (P ≤ 0.05).
Geographic distribution of genotypes.
All four genotypes were found in both the United States and Europe. However, three viruses from the United States formed a unique branch in genotype 1, one branch of genotype 2 that had an internal node corresponded only to European isolates, and the two European serotype 2/3 viruses mapped to a unique branch in genotype 3.
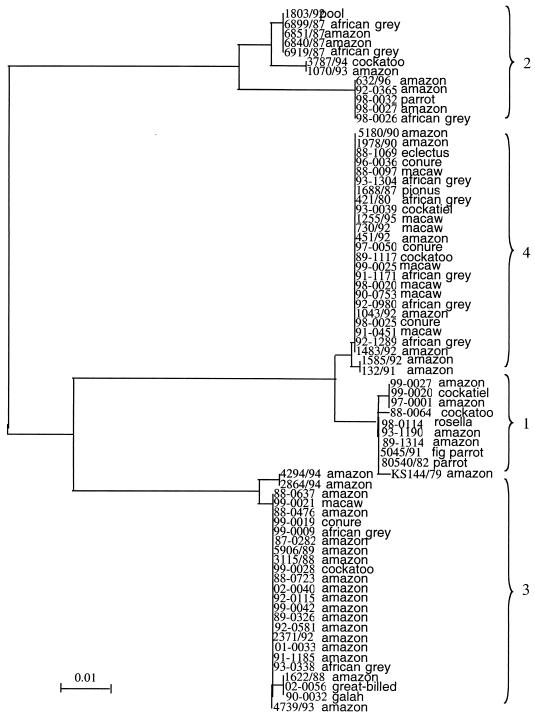
Species distribution among genotypes.
Amazon parrots are the parrots most commonly diagnosed with Pacheco's disease. They were also the most common parrot in this study, representing 48.8% of the parrots from which viruses were analyzed (Table 1 and 2). All four genotypes were identified among Amazon parrots. However, no genotype 4 viruses (0 of 17) recovered from birds from the United States were from Amazons, while 15 of 29 viruses of the other three genotypes recovered from birds from the United States were from Amazons (P ≤ 0.05). Seven of 19 of the European genotype 4 samples were derived from Amazon parrots. Viruses from eight macaws were analyzed. Six were from the United States and two were from Europe. Both the viruses from Europe and five of the viruses from the United States had identical sequences and mapped to genotype 4. The other virus from a macaw mapped to genotype 3. The probability that a virus from a macaw would be of genotype 4 was significantly higher than the probability that it would be of the other three genotypes (P ≤ 0.5). Three viruses from conures mapped to genotype 4, and one mapped to genotype 3. PsHVs from African grey parrots mapped to genotypes 2, 3, and 4, but not to 1 (Fig. 3). It could not be ruled out statistically that the absence of genotype 1 among African grey parrots was the result of chance. PsHVs of all four genotypes were found among birds from the Pacific distribution.
FIG. 3.
Phylogenetic tree of all isolates identified by parrot type by using 103 neighbor-joining bootstrap replicates with Clustal X (similar results were obtained by using the maximum-parsimony method of PAUP* 4.0).
DISCUSSION
In this study, we document and compare the genotypes and phenotypes of 73 PsHVs originating from Europe and the United States. Comparing the sequences of these viruses, we identified four major genotypes comprising 12 variants. Naming the specific genotypes is problematic as previous PsHV designations were based on serotype and the two most closely related genotypes share the same serotype (serotype 1). To keep the names of the genotypes and serotypes as consistent as possible, we propose that the genotype of the original PsHV1 reference strain KS144/79 be called genotype 1 and that genotype 2 and genotype 3 be those of the reference strains for serotype 2 (6840/87) and serotype 3 (3115/88), respectively. Genotype 4 corresponds to viruses that are most closely related to those of genotype 1 but can be distinguished from genotype 1 by both point mutations and a 6-bp deletion. All the viruses of genotype 4 are serotype 1, with the exception of the unique serotype 4 virus (1483/92).
Subdivisions of the four PsHV genotypes allowed resolution of all of the previously reported RFLP patterns with the exception of those of three viruses (15). A virus classified as AA3A3 cannot be differentiated from an AA2A2 viruses, the BB2B2 virus cannot be differentiated from a BB1B1 virus, and the FFF virus cannot be differentiated from the GGG virus. Additional sequence data are needed to further differentiate these viruses and will undoubtedly define additional branches within each genotype.
The correlation between genotype and serotype is excellent. All but one serotype 1 virus map to either genotype 1 or 4, all but one serotype 3 virus map to genotype 3, and all serotype 2 viruses map to genotype 2. The serotype 4 virus maps to genotype 4 and cannot be differentiated from several serotype 1 viruses with our data. However, it has a unique RFLP pattern, indicating that it represents a subpopulation of the genotype 4 viruses and contains mutations that resulted in its becoming a unique serotype. The two viruses that were neutralized by antiserum to both serotype 2 and 3 viruses should be considered a genetically distinct variant of genotype 3 as they are identical to each other but represent unique variants of this genotype. Isolate 8326/87 (serotype 5) was identified as a herpesvirus based on its cytopathic effects and sensitivity to chloroform, inhibition of replication by 5-iodo-2′-deoxyuridine, and electron microscopic morphology. However, it was difficult to grow in CEFs (E. F. Kaleta, unpublished data), had a unique RFLP pattern, and was serologically distinct from all of the other PsHVs (6, 15). We were unable to amplify its DNA from three separate samples by using six different PsHV primer sets and additional degenerate primer sets that have successfully amplified DNA from highly divergent herpesviruses (21). We must therefore conclude that the so-called PsHV serotype 5 is either a highly divergent virus and should not be considered a PsHV or was not present in the samples we analyzed.
Two European viruses, 1070/93 and 132/91, are problematic. Virus 1070/93 mapped to genotype 2 but was reported to be serotype 1 and virus 132/91 mapped to genotype 4 but was reported to be serotype 3 (5). Both viruses also had sequences and RFLP patterns identical to those of another virus within each genotype, viruses that have the predicted serotypes. It is possible that these two viruses have mutations that have resulted in a change of serotype. It is also possible that these viruses represent a recombination event that occurred in a bird infected simultaneously with two serotypes of the PsHV. We cannot rule out this possibility; however, coinfection of parrots with multiple serotypes has yet to be demonstrated and the RFLP patterns of these viruses do not support this possibility. Finally, we cannot completely rule out the possibility of laboratory error during the isolation or serologic typing process.
Previously reported PCR amplification patterns of the PsHVs are only moderately predictive of genotypes. All viruses with PCR amplification pattern 6 are genotype 3. However, not all genotype 3 viruses have the same amplification pattern. The amplification pattern 1 viruses are predominately those of genotypes 1 and 4, but three (13.6%) are of other genotypes.
A major objective of this study was to determine whether the viral serotypes and genotypes found in Europe are representative of those found in the United States. Largely they are. Representatives of all genotypes are present in both the United States and Europe. Variants of some of the genotypes, however, are not always present in both Europe and the United States. Whether these finding are the result of a small sample size or an actual difference in prevalences of these viruses will require additional investigation. Some differences in the prevalences of the genotypes observed among the sequenced U.S. and European viruses are noted. In particular, genotype 2 viruses made up 27% of the European viruses sequenced and only 9% of the U.S. viruses sequenced. However, conclusions cannot be drawn as to the actual prevalences of these viruses in birds dying from Pacheco's disease as all European viruses are of cell culture origin and the ability to isolate each genotype in CEFs may vary. Additionally, neither the European isolates nor U.S. viruses examined in this study are selected randomly. The likelihood of the presence of all three PsHV serotypes in the United States is significant, as it is thought that a polyvalent vaccine may be necessary to protect against these three serotypes (10, 15) and the only vaccine currently available is monovalent.
Our data suggest that different genotypes are more pathogenic to some species of parrots than others. Specifically, genotype 4 appears to be pathogenic to both macaws and conures, genotype 3 rarely is, and genotypes 1 and 2 may not cause Pacheco's disease in these species at all. Similarly, genotype 4 is not found to cause Pacheco's disease in Amazon parrots in the United States, and no African grey deaths are attributed to infections with genotype 1. The Pacific species appear to be equally susceptible to all genotypes. Our data also suggest that genotype 4 viruses have a reduced ability to grow in CEFs derived from SPAFAS eggs. Alternately, this virus may grow to lower titers in the infected host. Given the close correlation between genotype and serotype and the different pathogenicities of these genotypes for different species and CEFs, we suggest that each genotype can also be considered a specific pathotype.
Herpesviruses are highly host adapted, causing little or no disease but lifelong infections in the species that they typically infect (4). Disease results when infection occurs in nonadapted species. It is thought that Pacheco's disease is the manifestation of persistently infected, virus-adapted parrots' infecting naïve species (5). Our data do not define which species may ultimately be carriers of the observed genotypes, but our data do have the potential to be used to address this question. Previously, it has been shown that PsHVs can be detected in swabs of mucous membranes from many persistently infected parrots by PCR (13). However, this assay did not detect PsHVs in every bird in this study that was thought to be infected. Retrospectively, this may have been the result of the choice of primers and the inability of one or both primers to detect all genotypes (E. K. Tomaszewski, unpublished observations). In the present study, the primer set used in the initial amplification step appears to amplify DNA from all known PsHV genotypes. This primer set therefore has the potential to be used with PCR to screen swabs of mucosal surfaces from a range of parrot species to identify which ones are persistently infected with PsHVs. Point mutations and deletions within the four genotypes potentially allow for the design of PCR primer sets that can differentiate between the four genotypes, permitting the identification of the specific genotype infecting a bird. Alternately, genotypes can be defined by sequencing of amplicons. These tools could also be used to search for and identify genotypes of PsHVs associated with mucosal papillomas.
Acknowledgments
We acknowledge the following for their financial support of this research: the Department of Large Animal Medicine and Surgery and the Schubot Exotic Bird Health Center, Texas A&M University, the Association of Avian Veterinarians, the Midwestern Avian Research Exhibition, the Geraldine R. Dodge Foundation, Central Indiana Cage-Bird Club, the Alaska Bird Club, North County Aviculturalists, Central Jersey Bird Club, Long Island Bird Club, Semiconductor Equipment and Materials International, Miami Valley Bird Club, Kathryne and Richard Thorpe, Barbara A. Brinker, Charles and Margaret Bloodworth, Paul T. and Jacqueline L. Frederickson, Nancy Miller, Martha Gravlee, Mary Lee Leinneweber, Mary Yerardi, Gail Padgett, Michael Ambrose, Rodica Stoicoiu, Joanie Doss, Elizabeth Wilson, Sally Spencer, Lyne Dicker, and Ronald and Linda Wilson.
REFERENCES
- 1.Aini, L., L. M. Shih, A. E. Castro, and Y. X. Zee. 1993. Comparison of herpesvirus isolates from falcons, pigeons, and psittacines by restriction endonuclease analysis. J. Wildl. Dis. 29:196-202. [DOI] [PubMed] [Google Scholar]
- 2.Brown, B. W., and M. Hollander. 1977. Statistics. A biomedical introduction, p. 185-187. John Wiley & Sons, New York, N.Y.
- 3.Chartwright, M., T. R. Spraker, and D. McCluggage. 1985. Psittacine inclusion body hepatitis in an aviary. J. Am. Vet. Med. Assoc. 187:1045-1046. [PubMed] [Google Scholar]
- 4.Davison, A. J. 2002. Evolution of herpesviruses. Vet. Microbiol. 86:69-88. [DOI] [PubMed] [Google Scholar]
- 5.Gaskin, J., B. Raphael, A. Major, and G. Hall. 1981. Pacheco's disease: the search for the elusive carrier bird, p. 24-28. In Proceedings of the American Association of Zoo Veterinarians. AAZV, Seattle, Wash.
- 6.Gravendyck, M., S. Tritt, H. Spenkoch-Piper, and E. F. Kaleta. 1996. Antigenic diversity of psittacine herpesviruses: cluster analysis of antigenic differences obtained from cross-neutralization tests. Avian Pathol. 25:345-357. [DOI] [PubMed] [Google Scholar]
- 7.Günther, B. M. F., B. G. Klupp, M. Gravendyck, J. E. Lohr, T. C. Mettenleiter, and E. F. Kaleta. 1997. Comparison of the genomes of 15 avian herpesvirus isolates by restriction endonuclease analysis. Avian Pathol. 26:305-316. [DOI] [PubMed] [Google Scholar]
- 8.Johne, R., A. Knorath, M.-E. Krautwald-Junghanns, E. F. Kaleta, H. Gerlach, and H. Müller. 2002. Herpesviral, but no papovaviral sequences, are detected in cloacal papillomas of parrots. Arch. Virol. 147:1869-1880. [DOI] [PubMed] [Google Scholar]
- 9.Kaleta, E. F., U. Heffels, U. Neumann, and T. Mikami. 1980. Nachweis eines Herpesvirus bie Amazonen (Amazona aestiva und Amazona ochrocephala). Zentbl. Vetmed. 27:405-411. [PubMed] [Google Scholar]
- 10.Magnino, S., G. Conzo, A. Fiorietti, L. F. Menna, T. Rampin, G. Sironi, M. Fabbi, and E. F. Kaleta. 1996. An outbreak of Pacheco's parrot disease in psittacine birds recently imported to Campania, Italy: isolation of psittacid herpesvirus 2. Zentbl. Vetmed. Reihe B 43:631-637. [DOI] [PubMed] [Google Scholar]
- 11.Pacheco, G., and O. Bier. 1930. Epizootie chex les perroquets du Brésil. Relations avec le psittacose. C. R. Soc. Biol. (Paris) 105:109-111. [Google Scholar]
- 12.Panigrahy, B., and L. C. Grumbles. 1984. Pacheco's disease in psittacine birds. Avian Dis. 28:808-812. [PubMed] [Google Scholar]
- 13.Phalen, D. N., E. L. Tomaszewski, C. S. Radabaugh, and B. Dahlhausen. 2000. Psittacid herpesviruses and herpesvirus disease in psittacine birds, p. 259-262. In Proceedings of the Association of Avian Veterinarians. AAV, Portland, Oreg.
- 14.Randall, D. J., M. D. Dagless, H. G. R. Jones, and J. W. MacDonald. 1979. Herpesvirus infection resembling Pacheco's disease in Amazon parrots. Avian Pathol. 8:229-238. [DOI] [PubMed] [Google Scholar]
- 15.Schröder-Gravendyck, A. S., E. F. Kaleta, R. E. Marschang, and M. Gravendyck. 2001. Differentiation of psittacine herpesvirus field isolates by restriction endonuclease analysis. Avian Pathol. 30:551-558. [DOI] [PubMed] [Google Scholar]
- 16.Senne, D. A., J. E. Pearson, L. D. Miller, and G. A. Gustaeson. 1983. Virus isolation from pet birds submitted for importation into the United States. Avian Dis. 27:731-744. [PubMed] [Google Scholar]
- 17.Simpson, D. F., J. E. Hanley, and J. M. Gaskin. 1975. Psittacine herpesvirus infection resembling Pacheco's parrot disease. J. Infect. Dis. 131:390-396. [DOI] [PubMed] [Google Scholar]
- 18.Styles, D. K., D. N. Phalen, and E. K. Tomaszewski. 2002. Elucidating the etiology of avian mucosal papillomatosis in psittacine birds, p. 175-178. In Proceedings of the Association of Avian Veterinarians. AAV, Monterey, Calif.
- 19.Tomaszewski, E., D. N. Phalen, and V. G. Wilson. 1999. Synchronicity, papillomas, and herpes disease, p. 219-221. In Proceedings of the Association of Avian Veterinarians. AAV, New Orleans, La.
- 20.Tomaszewski, E., V. G. Wilson, W. L. Wigle, and D. N. Phalen. 2001. Detection and heterogeneity of herpesviruses causing Pacheco's disease in parrots. J. Clin. Microbiol. 39:533-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.VanDevanter, D. R., P. Warrener, L. Bennett, E. R. Schultz, L. Coulter, R. L. Garber, and T. M. Rose. 1996. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 34:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]