Abstract
The RNase H activity of retroviral reverse transcriptases (RTs) degrades viral genomic RNA after it has been copied into DNA, removes the tRNA used to initiate negative-strand DNA synthesis, and generates and removes the polypurine tract (PPT) primer used to initiate positive-strand DNA synthesis. The cleavages that remove the tRNA and that generate and remove the PPT primer must be specific to generate linear viral DNAs with ends that are appropriate for integration into the host cell genome. The crystal structure of human immunodeficiency virus type 1 (HIV-1) RT in a complex with an RNA/DNA duplex derived from the PPT revealed that the 5′ end of the PPT deviates from traditional Watson-Crick base pairing. This unusual structure may play a role in the proper recognition of the PPT by HIV-1 RT. We made substitution mutations in the 5′ end of the PPT and determined their effects on virus titer. The results indicated that single and double mutations in the 5′ end of the PPT had modest effects on virus replication in a single-cycle assay. More complex mutations had stronger effects on virus titer. Analysis of the two-long-terminal-repeat circle junctions derived from infecting cells with the mutant viruses indicated that the mutations affected RNase H activity, resulting in the retention of PPT sequences on viral DNA. The mutants tested preferentially retained specific segments of the PPT, suggesting an effect on cleavage specificity. These results suggest that structural features of the PPT are important for its recognition and cleavage in vivo.
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) is the virally encoded enzyme that converts the viral genome, which is single-stranded RNA, into double-stranded DNA (3, 8, 33). This conversion involves a collaboration between the two enzymatic activities of RT: a DNA polymerase that can use either RNA or DNA as a template and an RNase H that cleaves RNA if (and only if) it is present in an RNA/DNA duplex. Both the polymerase and the RNase H are required for the conversion of the viral RNA genome into DNA; mutations that inactivate either the polymerase or RNase H activities block viral replication (24, 27, 29, 30).
HIV-1 RT is a heterodimeric protein consisting of p66 and p51 subunits (4). Both subunits are derived from the Gag-Pol polyprotein by cleavage with viral protease. The p66 subunit is 560 amino acids long; the p51 subunit contains the first 440 amino acids of the p66 subunit. The p66 subunit has two domains, polymerase and RNase H; the C-terminal portion of p66 forms the RNase H domain. Based on the crystal structure, the polymerase domain of HIV-1 RT has been likened to a right hand, composed of fingers, palm, thumb, and connection subdomains (14). The p51 subunit corresponds closely, but not exactly, to the polymerase domain of p66. The folding within each of the subdomains is similar for both subunits, but the overall spatial arrangement of the subdomains differs between p66 and p51 (11, 14). The p66 subunit contains both the polymerase and RNase H active sites. The triad of aspartic acid residues at positions 110, 185, and 186 that form the polymerase active site are present in p51; however, the smaller subunit does not contribute directly to the polymerase activity of RT (17). Based on crystallographic analysis, the RNase H active site is located ca. 17 to 18 nucleotides away from the polymerase active site; this distance is supported by biochemical experiments (7, 9, 11, 18, 34).
HIV-1 reverse transcription is initiated from a tRNALys3 base paired near the 5′ end of the viral genome at the primer-binding site (pbs) (reviewed in references 3 and 33). The RNA genome is positive strand. Viral DNA synthesis is initiated from a host tRNA whose 3′ end is base paired to the viral RNA genome. DNA synthesis creates an RNA/DNA duplex that is a substrate for RNase H. The viral genome is terminally redundant, which permits the transfer of negative-strand DNA to the 3′ end of viral RNA, allowing negative-strand synthesis to continue. This creates an RNA/DNA duplex that is cleaved by RNase H. In general, degradation of the viral RNA is not sequence specific, and a range of different-sized products is generated. However, the RNase H of HIV-1 RT does make some specific cleavages. The removal of the tRNALys3 primer by HIV-1 RT RNase H occurs one nucleotide from the RNA-DNA junction (6, 21, 28, 32). This cleavage defines the right end of the unintegrated viral DNA. The cleavages that generate and remove the polypurine tract (PPT) primer used to initiate positive-strand DNA synthesis are also specific. The PPT primer is completely removed by RNase H, which defines the left end of the unintegrated viral DNA. The specificity of the RNase H cleavages that define the ends of the linear viral DNA are important for viral replication; these ends are the substrates for the integration of the DNA into the host genome, a reaction that is carried out by the viral enzyme integrase.
The crystal structure of HIV-1 RT in a complex with an RNA/DNA primer template whose sequence was derived from the PPT has provided information on the interactions of RT and its template primer (26). The complex of HIV-1 RT and an RNA/DNA duplex is similar to the complex of HIV-1 RT and a DNA/DNA duplex; however, there are differences (5, 26). In a complex with RT, both DNA/DNA and RNA/DNA duplexes have a bend of ca. 40° 5 to 9 bp from the polymerase active site. In the DNA/DNA-containing complex, the bend in the nucleic acid is associated with a transition from A-form to B-form geometry; a similar transition has been observed in DNA/DNA substrates bound to other DNA polymerases. The majority of the contacts between the enzyme and the nucleic acid are near the polymerase active site, where the nucleic acid geometry is closer to A-form. Farther from the polymerase active site the geometry of the DNA/DNA duplex is closer to B-form. The RNA/DNA duplex adopts a geometry between A form and B form (H form). In the RT/RNA/DNA complex there are additional contacts between the nucleic acid and the protein relative to the RT/DNA/DNA complex. HIV-1 RT interacts with several of the 2′ hydroxyl groups of the RNA template. Near the RNase H active site a network of amino acids interacts with the DNA primer strand. These amino acids, designated the RNase H primer grip, play a role in positioning the primer strand relative to the RNase H active site, which helps determine the specificity of RNase H cleavage. There are mutations in the RNase H primer grip that affect the specificity of RNase H cleavage in vitro and in vivo (13, 23). Altering the specificity of RNase H cleavage affects the generation and removal of the PPT primer used to initiate positive-strand DNA synthesis. Consequently, some of the linear viral DNAs generated by these mutant RTs are not appropriate substrates for integration into the host cell genome. A portion of the linear viral DNAs that do not integrate are ligated by host enzymes to form two-long-terminal-repeat (2-LTR) circles (16). The circle junction contains the ends of the linear viral DNAs that were ligated to form the 2-LTR circles. We analyzed the 2-LTR circle junction as a surrogate for the ends of the linear DNA and showed that mutations in the RNase H primer grip affect the specificity of the generation (and removal) of the PPT primer used for positive-strand DNA synthesis.
During reverse transcription the PPT must remain resistant to RNase H degradation long enough to serve as the primer for positive-strand DNA synthesis. The PPT primer is generated by two specific cleavages: one at the U-tract-PPT junction and the other at the PPT-U3 junction (3). A specific cleavage reaction removes the PPT primer from the viral DNA; this cleavage occurs at the PPT-U3 junction (RNA-DNA junction). At least in the complex with HIV-1 RT, the crystal structure of the PPT portion of the RNA/DNA duplex is unusual (26). The 5′ end of the HIV-1 PPT consists of a run of four A residues and then a G residue; the last three nucleotides (two A's and one G) in this region do not exhibit traditional (Watson-Crick) base pairing in the RNA/DNA duplex bound to HIV-1 RT. The 4 bp adjacent to the RNase H active site are properly base paired; however, there are two unpaired bases in the RNA/DNA duplex: one in the primer strand and one in the template strand. These two unpaired bases shift the base pairing out of and then back into the normal register. This unusual structure may be recognized by RT and facilitate the specific cleavages that generate or remove the PPT primer.
The RNase H cleavage reactions that generate and remove the PPT have been well characterized in vitro (15, 19, 20, 22, 23). Mutations in the PPT can affect the specificity of RNase H cleavage in vitro; however, the data are complex (15, 19, 20, 22). In some of the in vitro experiments, the specificity of PPT-U3 cleavage by HIV-1 RT was affected by mutations in the 3′ end of the HIV-1 PPT (a run of six G's) (15). However, another laboratory reported that mutations in the 5′ end of the PPT did not affect the specificity of RNase H cleavage at the PPT-U3 junction (19). Because the mutations were different in the two studies, the data are difficult to compare directly. We wanted to determine whether mutations in the 5′ end of the PPT would affect the generation and/or removal of the PPT primer in vivo. We introduced single transversion mutations (A to C) or double transversion mutations into the 5′ end of the PPT of an HIV-1-based retroviral vector. We also constructed a vector that has multiple mutations throughout the 5′ end of the PPT (extending through the G in the fifth position of the PPT which is in the middle of the eight A's that form the 5′ end of the HIV-1 PPT; see Fig. 1). Single and double mutations in the 5′ end of the PPT modestly affected virus titer in a single cycle of retroviral replication. The virus containing multiple mutations in the first half of the PPT had a much lower titer (5% of the wild-type level) than any of the viruses with single or double mutations in the four distal bases of the PPT. Analysis of 2-LTR circle junctions revealed that all of the mutations in the PPT caused a dramatic increase in the proportion of 2-LTR circle junctions that contain PPT insertions compared to the wild type. In mutants whose 2-LTR circle junctions were analyzed, a particular segment of the PPT was retained; the specific segment was different in the three cases. These results demonstrate that the sequences in the 5′ end of the PPT are important determinants for the proper generation and removal of the PPT primer during viral replication.
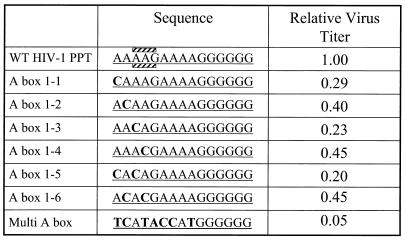
FIG. 1.
Mutations in the PPT affect viral titer. The left column shows the name of the different PPT mutants; the middle column shows the sequences. The top row shows the sequence of the wild-type PPT. The boxes containing stripes designate the nucleotides that deviate from traditional Watson-Crick base pairing in the crystal structure of HIV-1 RT in a complex with an RNA/DNA duplex derived from the PPT. Transversion mutations (A to C) were introduced into the 5′ end of the HIV-1 PPT. The mutated bases are shown in boldface. Virions containing these mutant PPTs were generated as described in Materials and Methods. The effects of the mutations on the relative virus titer (normalized to p24) are shown in the rightmost column. The relative titer was determined by averaging at least two independent experiments for each mutant.
MATERIALS AND METHODS
Construction of the BspMI cassettes.
The previously described HIV-1-based plasmid pNLNgoMIVR-E-.HSA was used as a template for PCR amplification (12). The amplified segment went from the XhoI site located ca. 200 bp upstream of the PPT (forward primer) to just upstream of the PPT (reverse primer). The reverse primer introduces an EcoRI site at the 5′ end of the primer and a BspMI site adjacent to the viral sequences. The sequence of the forward primer is 5′-GACTTGCTCGAGACCTAGAAAAACATGGAG-3′, and the sequence of the reverse primer is 5′-CTGATGAATTCGCGACCTGCCTAAGATCTACAGCTGCCTTG-3′ (containing the EcoRI and BspMI sites). A second PCR was performed from just downstream of the PPT (forward primer) to a site just beyond the PmlI site in the LTR (reverse primer). The forward primer included an EcoRI site at the 5′ end and a BspMI site near the viral sequences. The sequence of the forward primer is 5′-CTAAGGAATTCGCGACCTGCGGGCTAATTCACTCCCAAAG-3′ (containing the EcoRI and BspMI sites). The reverse primer spans the PmlI site present in the LTR; the sequence of the reverse primer is 5′-GATGGCCACGTGATGAAATGCTAGGCGG-3′. The PCR products were gel purified, digested with the appropriate set of restriction endonucleases (XhoI-EcoRI or EcoRI-PmlI), and used in a three-piece ligation reaction with a cloning vector containing XhoI and PmlI sites. Sequence analysis of the plasmid containing the BspMI cassette was performed to ensure that no unwanted mutations were introduced during PCR amplification. This plasmid is called pSKPPT.
Construction of the HIV-1 PPT mutants.
Complementary oligonucleotides containing the desired PPT mutations were synthesized (Biosource International). The oligonucleotides were treated with T4 polynucleotide kinase at 37°C for 1 h, and then NaCl was added to a final concentration of 50 mM. The mixture was placed in a boiling water bath and allowed to cool to room temperature. After an annealing step, the oligonucleotides were ligated into the pSKPPT cassette vector that had been digested with BspMI and gel purified. Ligations were performed overnight at room temperature; the ligation mixture was digested with EcoRI to linearize any residual pSKPPT circular DNA and used to transform DH5α competent cells. The resulting plasmids contained the desired mutant PPTs in a segment of the HIV-1 genome that includes the XhoI site upstream of the PPT and the PmlI site in the LTR. The mutant PPTs were introduced into the HIV-1-based vector pNLNgoMIVR-E-.HSA by using a three-piece ligation strategy. The three pieces are from the XhoI site to the PmlI site, the PmlI site to an AatII site in the vector, and from the AatII site to the XhoI site. The resulting plasmids were analyzed by restriction endonuclease digestion and DNA sequencing.
Cells.
The human embryonal kidney cell line 293 was obtained from the American Type Culture Collection. The human osteosarcoma cell line HOS was obtained from Richard Schwartz (Michigan State University, Lansing). 293 and HOS cells were maintained in Dulbecco modified Eagle medium (Invitrogen) supplemented with 5% fetal bovine serum, 5% newborn calf serum, and penicillin (50 U/ml) plus streptomycin (50 μg/ml) (Quality Biologicals).
Transfection, infection, and phenotyping protocol.
293 cells were transfected with 3 μg of pNLNgoMIVR-E-.HSA and 1.5 μg of pHCMV-g (obtained from Jane Burns, University of California at San Diego) by the calcium phosphate method. 293 cells were plated in 100-mm-diameter dishes at a density of 1.0 × 106 cells per plate on the day prior to transfection. At this plating density, the cells were ca. 25% confluent on the day of transfection. The precipitate was added to the 293 cells dropwise. Fresh medium was added 16 h after transfection. The 48-h supernatants were harvested and clarified by low-speed centrifugation, and an aliquot was used to infect HOS cells. The amount of p24 in the supernatant was determined by using the HIV-1 p24 antigen capture assay kit (AIDS Vaccine Program, SAIC, Frederick, Md.); the p24 concentration was used to control for the amount of virus in the samples. HOS cells were plated in 60-mm-diameter dishes at a density of 1.5 × 105 cells per plate on the day prior to infection. The virus was allowed to absorb to the cells for 4 h, and then fresh medium was added. At 48 h after infection, the cells were harvested from the plate by treatment with 1.0 ml of EDTA (Invitrogen), an additional 3 ml of phosphate-buffered saline (PBS) was added, and then the cells were collected by centrifugation, washed, and resuspended in 200 μl of PBS. The cells were labeled with phycoerythrin-conjugated rat anti-mouse CD24 monoclonal antibody (Pharmingen) by standard procedures, fixed with paraformaldehyde, and subjected to fluorescence-activated cell sorting (FACS) to determine the viral titer.
Transfections, infections, and nucleic acid extraction for 2-LTR circle junction analysis.
293 cells were transfected with 5 μg of pNLNgoMIVR-E-.HSA vector and 3 μg of pHCMV-g by the calcium phosphate method. The medium on the cells was changed 16 h after infection. The 48-h supernatants were harvested and clarified by centrifugation, and 4 ml of virus-containing supernatant was used to infect HOS cells. The supernatants were left on the cells for 4 h, and then fresh medium was added. The total DNA was isolated from the HOS cells ca. 36 h after infection by using a viral blood DNA kit (Qiagen).
PCR amplification, cloning, and sequencing of 2-LTR circle junctions.
The 2-LTR circle junctions were amplified in 100-μl reactions by using an upstream PCR primer that anneals near the RU5 junctions and a downstream primer that anneals in the U3 region of the LTR. The sequence of the upstream primer was 5′-CGATGAATTCGCTAACTAGGGAACCCACTGCT-3′; the sequence of the downstream primer was 5′-GCCATTCTAGAGTTCTCTCCTTTATTGGCCTC-3′. We used 10 μl of DNA and 0.25 μl each of the forward and reverse primers (100 nM final concentration) with 90 μl of Platinum PCR Supermix (Invitrogen) in each reaction. The expected product is ca. 350 bp long and has EcoRI and XbaI cleavage sites introduced by the primers. The PCR products were digested with EcoRI and XbaI and cloned into SK (Stratagene). The 2-LTR circle junction clones were analyzed by using restriction enzyme digestion and DNA sequence analysis.
RESULTS
Mutant PPTs and the effects of mutations on virus titer.
BspMI cassette mutagenesis was used to generate mutations in the PPT of the HIV-1 vector pNLNgoMIVR-E-.HSA (12). This vector expresses the retroviral gag-pol gene; it also expresses the cell surface marker CD24 (HSA) from the nef reading frame (31). The sequences of the mutant PPTs are compared to the wild-type PPT sequence in Fig. 1. Three bases (two A's and a G) are mispaired in the crystal structure of HIV-1 RT in a complex with a RNA/DNA template primer derived from the PPT. The mispaired bases are indicated in the figure. Transversion mutations (A to C) were introduced into each of the four A's at the 5′ end of the PPT (A box 1-1 to A box 1-4). This type of mutation should provide a strong base pair and disrupt the mispairing in the PPT that was seen in the HIV-1 RT complex. Two mutants were generated that had two A-to-C transversions (A box 1-5 and 1-6); one mutant was generated that contained multiple mutations throughout the nine 5′-most bases of the PPT (multi-A box). Viruses with genomes containing the mutant PPTs were generated by cotransfecting plasmids that contain the HIV-1 vector and a plasmid (pHMCV-g) that expresses the VSV-g envelope glycoproteins (2, 35). The virions were used to infect HOS cells and the cells were stained with anti-CD24 to determine virus titer by using FACS; the titers were normalized relative to the amount of p24. All of the mutations in the PPT decreased virus titer in a one-round replication assay. The single and double mutations in the first A box decreased the virus titer from 20 to 45% of the wild-type level (see Fig. 1). A mutant containing multiple mutations throughout the 5′ end of the PPT decreased virus titer to about 5% of the wild-type level.
Analysis of 2-LTR circle junctions obtained from infections with wild-type virus and PPT mutants.
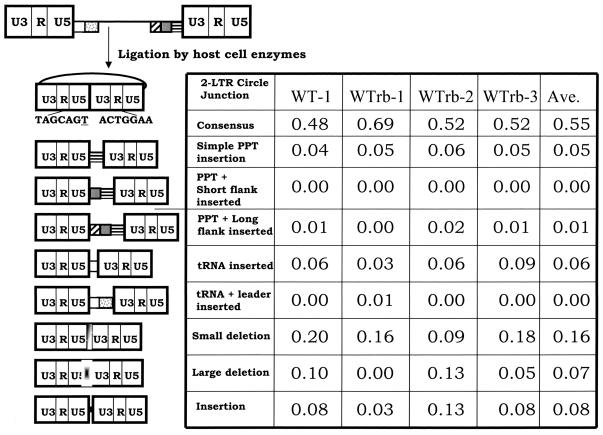
Previously, we used the sequence of the 2-LTR circle junction as a surrogate for the ends of the linear viral DNA (13). To demonstrate that the percentage of aberrant 2-LTR circle junctions is reproducible, we compared the populations obtained from independent experiments with vectors containing the wild-type PPT. The analysis of the 2-LTR circle junctions is shown in Fig. 2. A Pearson's chi-square test showed that these populations of circle junctions are homogeneous. The averages for each category of the 2-LTR circle junction populations are shown in the rightmost column of the figure. When the 2-LTR circle junction contains the entire sequence of both ends of the linear viral DNA, this is the consensus 2-LTR circle junction. The 2-LTR circle junctions displayed a consensus sequence at a frequency of 0.55. Simple PPT insertions were present at a frequency of 0.05. tRNA insertions were present at a frequency of 0.06. This is slightly higher than the results we obtained in our initial experiments; however, the difference is not statistically significant (13).
FIG. 2.
2-LTR circle junctions obtained after infecting cells with wild-type virus. The top of the figure shows the linear viral DNA that is the product of reverse transcription. The first column shows the results obtained with the original vector pNLNgoMIVR-E-.HSA. The wild-type PPT sequence was reconstructed by using the same BspMI mutagenesis protocol used to construct the PPT mutants, and the reconstructed vector was sequenced to show, it was identical to the original vector. Three independent experiments were performed that using a rebuilt vector containing the wild-type PPT; these are designated WTrb in the top of the columns. The pbs is indicated by a white box, the leader sequence downstream of the pbs is shown with gray dots, the PPT is a box with black horizontal bars, the U-tract is a gray box, and the sequences immediately upstream of the U-tract are indicated by the box with diagonal bars. The linear viral DNA can be ligated by cellular enzymes in the nucleus of an infected cell to form a 2-LTR circle. A consensus circle junction is shown at the top. Different categories of aberrant 2-LTR circle junctions are shown. The underlined “T” is derived from the riboA located at the end of the negative strand; it is derived from the tRNALys3 primer used by HIV-1 RT to initiate negative-strand DNA synthesis. The numbers of samples analyzed were 80 for the parental vector and 64, 64, and 77, respectively, for the three experiments with the rebuilt vector.
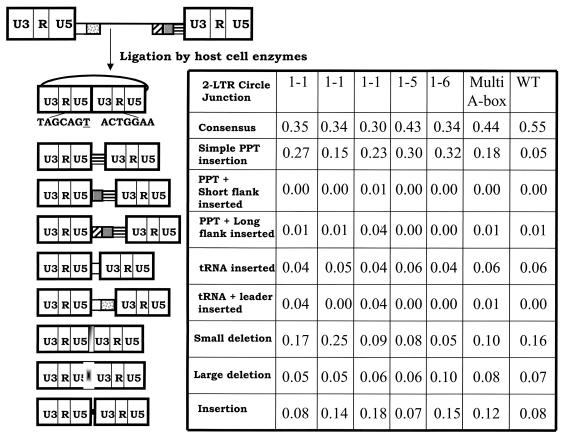
Analysis of the 2-LTR circle junctions from the mutants is shown (Fig. 3). Three independent experiments for the mutant A box 1-1 are shown to demonstrate the reproducibility of the assay. 2-LTR circle junctions analysis of A box 1-5, A box 1-6, and multi-A box are shown. In general, all of the mutants decreased the proportion of 2-LTR circle junctions containing a consensus sequence compared to the wild type. The increase is specifically in the insertion of PPT sequences. A global chi-square analysis of the mutant populations demonstrated that they are similar to each other; however, the mutant populations are different from the wild-type population. The proportion of junctions containing a consensus sequence is reduced in all of the mutants, and the retention of simple PPTs is significantly increased in all mutant populations with respect to the wild type (P < 0.01). The populations of tRNA sequences at the 2-LTR circle junctions are not statistically different between the mutants and the wild type.
FIG. 3.
2-LTR circle junctions derived from infecting cells with PPT mutants. The pbs is indicated by a white box, the leader sequence downstream of the pbs is shown with gray dots, the PPT is a box with black horizontal bars, the U-tract is a gray box, and the sequences immediately upstream of the U-tract are indicated by the box with diagonal bars. This linear viral DNA can be ligated by cellular enzymes in the nucleus of an infected cell to form a 2-LTR circle. A consensus circle junction is shown at the top. Different categories of aberrant 2-LTR circle junctions are shown. Three independent experiments were performed with PPT mutant A box 1-1 to demonstrate the reproducibility of the assay. The results obtained from one experiment are shown for mutants A box 1-5, A box 1-6, and multi-A box.
Mutations in the PPT cause different preferred sites of RNase H cleavage.
The sequences of the mutant PPTs are shown (Fig. 4). The bases that have been changed in the PPT are indicated by a negative image of the altered base. The arrows represent the 5′ end of the retained PPT segment; and the height of the arrow is proportional to the number of observations of that PPT segment in the 2-LTR circle junctions. Mutant A box 1-6 and multi-A box retained a specific PPT segment that was present in nearly 50% of circles containing a PPT insertion. A common PPT segment was present 29% of the time and 37% of the time for mutant A box 1-1 and 1-5, respectively. Even the wild type had a preferred segment of the PPT that was retained in 25% of the 2-LTR circle junctions containing PPT insertion. The probability of retention of these specific PPT segments was analyzed by mathematical simulation where 20 items (the number of wild-type PPTs retained in our data set) were randomly assigned to 15 bins (the length of the PPT) to determine whether the retention of the preferred segment could be explained by chance. The analysis demonstrated that for the wild-type PPT, a specific site of retention would be retained by chance in 5 of 20 observations (for the wild-type PPT, the preferred segment was retained in 5 of the circle junctions that had a PPT segment), only 7.25 times in 1,000 experiments. The probability is much smaller for the mutants, in which a specific PPT segment was retained between 29 and 47%. This suggests that RT has a preferred site of cleavage within the wild-type PPT and that the mutations we introduced into the PPT affect the preferred site(s) of PPT cleavage.
FIG. 4.
Sequence of PPTs retained at the 2-LTR circle junction from vectors containing wild-type PPT or PPT mutants. The sequence of the PPT for each mutant is shown. The arrows represents the 5′ end of the retained PPT segment. The segment retained most frequently is indicated by an asterisk, and the frequency of that particular PPT segment is given in the column at the right of the figure. The height of the arrow is proportional to the frequency of observation of each given PPT segment. The sequences derived from wild-type PPTs also contain the results obtained in previous studies (which were included to increase the number of observations). Statistical analysis of the probability of retention of a PPT at each site marked as a preferred cleavage site indicates that these frequencies of retention at each preferred site did not occur by chance, the probability for the site of preferred retention for the wild type was 7.25 in 1,000; the probability for each of the mutants shown was less (see the text).
DISCUSSION
HIV-1 vectors containing mutant PPTs were used to determine the role of sequences in the 5′ end of the PPT in the generation and/or removal of the PPT primer used for positive-strand DNA synthesis by RT. We are particularly interested in this segment of the PPT because it is mispaired when an RNA/DNA duplex containing the PPT is in a complex with HIV-1 RT. We made transversion mutations (A to C) that should generate a strong base pair and affect the mispairing observed in the complex with RT. Single mutations and double mutations in the first four bases (A boxes 1-1 through 1-6 in Fig. 1) of the PPT had relatively modest effects on titer (<10-fold). However, a complex mutation (multi-A box) in the first nine bases of the PPT substantially impaired infection of HOS cells (about 20-fold). Analysis of the 2-LTR circle junctions derived from infecting cells with these A box mutants demonstrated that the mutant PPTs are retained at a much higher frequency than the wild-type PPT. However, all of the mutant PPTs do lead to the generation of 2-LTR circles with the consensus sequence, showing that even the multi-A box mutations do not prevent some correct cleavage of the PPT.
The PPT is a highly specialized nucleic acid element in the retroviral genomes. Most of the RNase H cleavages made by RT are thought to be nonspecific, generating a range of different-sized products. The cleavages that generate (and remove) the PPT primer are precise. Since this type of cleavage specificity is not observed in the general degradation of the RNA genome, some structural feature of the nucleic acid forming the PPT must play an important role determining cleavage specificity. The 5′ end of the PPT is mispaired in the crystal structure of RT in complex with nucleic acid (26), and mutations in the 5′ end of the PPT affect RNase H cleavage both in vivo (as shown in the present study) and in vitro (15). This suggests that RNase H recognizes some special features of this region of the PPT and that the mutations affect the ability of RT to appropriately recognize these features and allow the proper RNase H cleavages to occur. The PPT primer is (properly) generated by RNase H cleavage at the U-tract-PPT junction and at the PPT-U3 junction (see Fig. 5A). RT then extends the PPT primer, using the negative-strand DNA as the template for polymerization. Normally, the PPT primer is removed at the RNA-DNA junction; this cleavage must be specific to generate the correct end of the linear viral DNA that serves as the substrate for integration into the host cell genome.
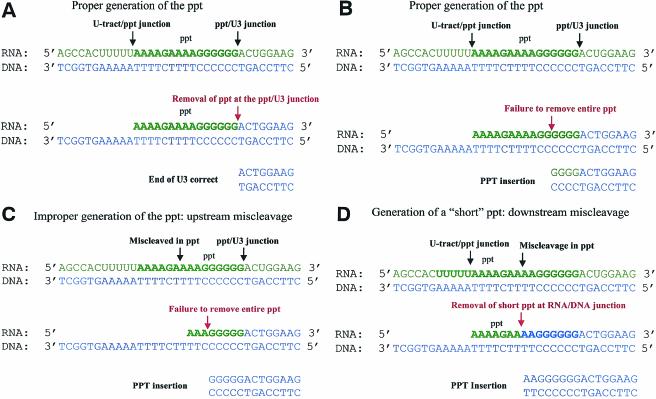
FIG. 5.
Mechanisms of retention of PPT sequences. RNAs are shown in green. DNA is shown in blue. Black arrows represent RNase H cleavages that generate the PPT; red arrows represent RNase H cleavages that remove the PPT. (A) The correct generation and removal of the PPT produce a DNA with the proper end. The RNase H cleavages that generate the PPT are made at the U-tract-PPT junction and the PPT-U3 junction (black arrows). The 3′ end of the PPT RNA segment is used to initiate positive-strand DNA synthesis. RNase H removes the PPT at the PPT-U3 junction (red arrow). (B) Failure to remove the entire PPT leads to a complete (or partial) PPT insertion at the 2-LTR circle junction. The RNase H cleavages that generate the PPT occur at the U-tract-PPT junction and the PPT-U3 junction (black arrows). RNase H fails to remove the PPT at the PPT-U3 junction but cleaves within the PPT itself (red arrow). (C) Improper generation of the 5′ end of the PPT can lead to a PPT insertion. The cleavage reactions that generate the PPT occur downstream of the U-tract-PPT junction (black arrow). As a consequence, RNase H fails to remove the entire PPT (red arrow). (D) Generation of a short PPT (3′ miscleavage) leads to PPT insertion at the 2-LTR circle junction. The 5′ end of the PPT is generated at the U-tract-PPT junction; however, the 3′ end is cleaved upstream of the PPT-U3 junction. The primer is shorter than the correct primer. The short primer is used to initiate positive-strand DNA synthesis and is then removed at the RNA-DNA junction (red arrow), leading to a PPT insertion.
There are three different ways that a PPT sequence can be retained in the 2-LTR circle junction. RT can generate the proper PPT (see Fig. 5B) and use it to initiate positive-strand DNA synthesis. If RNase H fails to remove part or all of the PPT, then the PPT sequences remain attached to the U3 DNA, leading to the retention of PPT sequences at the 2-LTR circle junction. In this mechanism, a partial PPT represents a failure to make a cleavage at the PPT-DNA junction.
Alterations in the cleavage specificity of RNase H (miscleavages) can lead to the presence of a PPT sequences at the 2-LTR circle junction. If the upstream cleavage reaction occurred within the PPT, rather than at the U-tract-PPT junction, a short PPT would be generated (see Fig. 5C). This short PPT could have the correct 3′ end (from a correct cleavage at the PPT-U3 junction), but after the initiation of DNA synthesis the RNA-DNA junction adjacent to the shorter PPT might not be an efficient substrate for RNase H cleavage, leading to retention of part or all of this shorter PPT. RT could also generate a short PPT through miscleavage at the 3′ end of the PPT (see Fig. 5D). RT could use this primer to initiate positive-strand DNA synthesis. Even if RT removes this primer at the RNA-DNA junction, a PPT insertion would be generated because DNA synthesis begins in the PPT.
Model substrates have been used to study the specificity of RNase H cleavage in vitro (15, 19, 20, 22, 23). Many of these experiments have focused on the generation of the start site for DNA synthesis and consequently only monitor the PPT-U3 cleavage step. Although there are experiments that suggest that mutations in the A box do not alter the specificity of the PPT-U3 cleavage in vitro, one study shows that multiple mutations in the 5′ end of the PPT did affect cleavage at the PPT-U3 junction (15). It is difficult to compare the in vitro experiments with our data, both because the mutations are different and because we do not know whether an alteration in the PPT-U3 cleavage is responsible for the retention of the PPT segment in our experiments; however, the 5′ end of the PPT appears to be important for one (or more) of the specific cleavages that HIV-1 RNase H must make.
Not only do mutations in the 5′ end of the PPT cause preferential retention of PPT sequences, the segments that are retained are different for the various mutants. The minor sites (nonpreferred) of cleavage (Fig. 4) are dispersed for both the wild-type and mutant PPTs. However, there is a preferred PPT segment that is retained from the wild-type PPT. The mutations we studied also lead to the retention of a preferred segment; however, the various mutations lead to the retention of different PPT segments. This suggests that RT recognizes a feature (presumably the structure) of the nucleic acid. Mutations change the structure of the PPT-containing duplex, which alters the specificity (preferred sites) of RNase H cleavage. Although the PPT mutations do lead to the retention of specific PPT segments, the available data are not sufficient to allow us to understand the underlying rules. In two cases where there is retention of a specific segment (A box 1-5 and A box 1-6) the 5′ end of the piece that is preferentially retained is two nucleotides 3′ of one of the two C's introduced into the PPT (see Fig. 4). However, this is not sufficient to explain how mutations in the PPT affect PPT cleavage in vivo. The data do suggest that the mispaired segment seen in the PPT in an RNA/DNA duplex in a complex with HIV-1 RT is not essential for the proper generation and removal of the PPT primer. All of the mutants, including multi-A box, can generate the proper ends of the viral DNA, albeit at a lower frequency than does the wild type. It is possible that the mispaired segment contributes to the proper generation and removal of the PPT in vivo.
Techniques that probe or alter the structure of the nucleic acid have been used to examine the role of structural distortions in an RNA/DNA duplex containing the PPT in vitro. The structure of the PPT (in the absence of RT) was analyzed by potassium permanganate footprinting (15). These experiments revealed that structural distortions preexist in the nucleic acid and that distortions are present not only at the first base of U3 but also within the PPT itself. The importance of the unpaired bases for RNase H cleavage was determined by the incorporation of non-base-pairing pyrimidine isosteres, which destabilize specific positions in the RNA/DNA duplex (J. W. Rausch, J. Qu, H. Y. Yi-Brunozzi, Eric T. Kool, and S. F. Le Grice, unpublished data). The positions of RNase H cleavage were strongly affected by the substitution of nonpairing isosteres in the upstream A-tract in the 5′ end of the PPT. Disruption of other base-paired regions also impaired PPT processing. Taken together, these data suggest that sequences within (and adjacent to) the PPT contribute to the structure of the nucleic acid and to the specificity of RNase H cleavage by RT. This is consistent with the effects of mutations in the 5′ end of the PPT affect cleavage specificity in vivo: although the mutations caused an increase in the miscleavages, there were correct cleavages of the mutant PPTs, suggesting that multiple regions of the PPT contribute to the specificity of PPT cleavage.
The rules for PPT selection and/or cleavage may differ between different retroviruses, but common themes appear to exist. The PPTs of murine leukemia virus (MLV) and HIV both contain 6 G's at the 3′ terminus of the PPT. Mutations in this G box in the HIV-1 PPT strongly affect the specificity of RNase H cleavage (19). When mutations in the run of G's at the 3′ end of the PPT were present in a pool of MLVs, there was a strong selection for the maintenance of the run of G's (25). Other elements outside the PPT may also be important for optimal PPT utilization. Most, if not all, retroviruses have a U-tract upstream of the PPT. Mutations in this U-tract affect the replication kinetics of MLV, and deletions in this region upstream of the PPT lead to the generation of viruses with aberrant left ends, indicating an affect on RNase H cleavage specificity (1). Similarly, mutations in the U-tract of simian immunodeficiency virus impair virus replication in vivo (10). Since the sequences of the PPTs are similar, it would appear there are common structural features in these PPTs that retroviral RTs recognize. These various elements must allow RNase H to properly position itself on the RNA/DNA duplex and make the precise cleavages. Taken together, the in vivo and in vitro data suggest that structural features of the PPT play an important role in the specificity of RNase H cleavage and, by extension, in the replication of retroviruses.
Acknowledgments
We thank David Munroe and Claudia Stewart for DNA sequencing, Louise Finch for FACS analysis, and Hilda Marusiodis for help in preparing the manuscript.
Research in the laboratory of S.H.H. was supported by the National Cancer Institute and by the National Institute for General Medical Sciences. Research in the laboratory of E.A. was supported by grants AI 27690 and GM55609. S.G.S. was supported by an NIH-NIAID NRSA fellowship (AI 09578).
REFERENCES
- 1.Bacharach, E., J. Gonsky, D. Lim, and S. P. Goff. 2000. Deletion of a short, untranslated region adjacent to the polypurine tract in Moloney murine leukemia virus leads to formation of aberrant 5′ plus-strand ends in vivo. J. Virol. 74:4755-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337-342. [DOI] [PubMed] [Google Scholar]
- 3.Coffin, J. M., S. H. Hughes, and H. E. Varmus (ed.). 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 4.Di Marzo Veronese, F., T. D. Copeland, A. L. DeVico, R. Rahman, S. Oroszlan, R. C. Gallo, and M. G. Sarngadharan. 1986. Characterization of highly immunogenic p66/p55 as the reverse transcriptase of HTLVIII/LAV. Science 231:1289-1291. [DOI] [PubMed] [Google Scholar]
- 5.Ding J., S. H. Hughes, and E. Arnold. 1997. Protein-nucleic acid interactions and DNA conformation in a complex of human immunodeficiency virus type 1 reverse transcriptase with a double-stranded DNA template-primer. Biopolymers 44:125-138. [DOI] [PubMed] [Google Scholar]
- 6.Furfine, E. S., and J. E. Reardon. 1991. Human immunodeficiency virus reverse transcriptase ribonuclease H: specificity of tRNA-lys-primer excision. Biochemistry 30:7041-7046. [DOI] [PubMed] [Google Scholar]
- 7.Furfine, E. S., and J. E. Reardon. 1991. Reverse-transcriptase-RNase H from human immunodeficiency virus: relationship of the DNA polymerase and RNA hydrolysis activities. J. Biol. Chem. 266:406-412. [PubMed] [Google Scholar]
- 8.Gilboa, E., S. W. Mitra, S. Goff, and D. Baltimore. 1979. A detailed model of reverse transcription and tests of crucial aspects. Cell 18:93-100. [DOI] [PubMed] [Google Scholar]
- 9.Gopalakrishnan, V., J. A. Peliska, and S. J. Benkovic. 1992. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc. Natl. Acad. Sci. USA 89:10763-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ilyinskii, P. O., and R. C. Desrosiers. 1998. Identification of a sequence element immediately upstream of the polypurine tract that is essential for replication of simian immunodeficiency virus. EMBO J. 17:3766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, A. Hizi, S. H. Hughes, and E. Arnold. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Julias, J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julias, J. G., M. J. McWilliams, S. Sarafianos, E. Arnold, and S. H. Hughes. 2002. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc. Natl. Acad. Sci. USA 99:9515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 15.Kvaratskhelia, M., S. R. Budihas, and S. F. Le Grice. 2002. Pre-existing distortions in nucleic acid structure aid polypurine tract selection by HIV-1 reverse transcriptase. J. Biol. Chem. 277:16689-16696. [DOI] [PubMed] [Google Scholar]
- 16.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3273-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Grice, S. F. J., T. Naas, B. Wohlgensinger, and O. Schatz. 1991. Subunit elective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 10:3905-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Post, K., J. Guo, E. Kalman, T. Uchida, R. J. Crouch, and J. G. Levin. 1993. A large deletion in the connection subdomain of murine leukemia virus reverse transcriptase or replacement of the RNase H domain with Escherichia coli RNase H results in altered polymerase and RNase H activities. Biochemistry 32:5508-5517. [DOI] [PubMed] [Google Scholar]
- 19.Powell, M. D., and J. G. Levin. 1996. Sequence and structural determinants required for priming of plus-strand DNA synthesis by the human immunodeficiency virus type 1 polypurine tract. J. Virol. 70:5288-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pullen, K. A., and J. J. Champoux. 1990. Plus-strand origin for human immunodeficiency virus type 1: implications for integration. J. Virol. 64:6274-6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pullen, K. A., L. K. Ishimoto, and J. J. Champoux. 1992. Incomplete removal of the RNA primer for minus-strand DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 66:367-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rattray, A. J., and J. J. Champoux. 1989. Plus-strand priming by Moloney murine leukemia virus. The sequence features important for cleavage by RNase H. J. Mol. Biol. 208:445-456. [DOI] [PubMed] [Google Scholar]
- 23.Rausch, J. W., D. Lener, J. T. Miller, J. G. Julias, S. H. Hughes, and S. F. Le Grice. 2002. Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry 41:4856-4865. [DOI] [PubMed] [Google Scholar]
- 24.Repaske, R., J. W. Hartley, M. F. Kavlick, R. R. O'Neill, and J. B. Austin. 1989. Inhibition of RNase H activity and viral replication by single mutations in the 3′ region of Moloney murine leukemia virus reverse transcriptase. J. Virol. 63:1460-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robson, N. D., and A. Telesnitsky. 2000. Selection of optimal polypurine tract region sequences during Moloney murine leukemia virus replication. J. Virol. 74:10293-10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, Jr., J. Ding, J. Whitcomb, M. Gait, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20:1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schatz, O., F. V. Cromme, T. Naas, D. Lindemann, J. Mous, and S. F. J. Le Grice. 1990. Inactivation of the RNase H domain of HIV-1 reverse transcriptase blocks viral infectivity, p. 293-404. In T. Papas (ed.), Gene regulation and AIDS. Portfolio, Houston, Tex.
- 28.Smith, J. S., and M. Roth. 1992. Specificity of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H in removal of the minus-strand primer, tRNALys3. J. Biol. Chem. 267:15071-15079. [PubMed] [Google Scholar]
- 29.Telesnitsky, A., S. W. Blain, and S. P. Goff. 1992. Defects in Moloney murine leukemia virus replication caused by a reverse transcriptase mutation modeled on the structure of Escherichia coli ribonuclease H. J. Virol. 66:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tisdale, M., T. Schultze, B. A. Larder, and K. Moelling. 1991. Mutations within the RNase H domain of HIV-1 reverse transcriptase abolish viral infectivity. J. Gen. Virol. 72:59-66. [DOI] [PubMed] [Google Scholar]
- 31.Wegner, R. H., J. M. Rochelle, M. F. Seldin, G. Kohler, and P. J. Nielsen. 1993. The heat stable antigen (mouse CD24) gene is differentially regulated but has a housekeeping promoter. J. Biol. Chem. 268:23345-23352. [PubMed] [Google Scholar]
- 32.Whitcomb, J. M., R. Kumar, and S. H. Hughes. 1990. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J. Virol. 64:4903-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitcomb, J. M., and S. H. Hughes. 1992. Retroviral reverse transcription and integration: progress and problems. Annu. Rev. Cell Biol. 8:275-306. [DOI] [PubMed] [Google Scholar]
- 34.Wohrl, B. M., and K. Moelling. 1990. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry 29:10141-10147. [DOI] [PubMed] [Google Scholar]
- 35.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43:99-112. [DOI] [PubMed] [Google Scholar]