Abstract
The hantavirus G1 protein contains a long C-terminal cytoplasmic tail of 142 residues. Hantavirus pulmonary syndrome-associated hantaviruses contain conserved tyrosine residues near the C terminus of G1 which form an immunoreceptor tyrosine activation motif (ITAM) and interact with Src and Syk family kinases. During studies of the G1 ITAM we observed that fusion proteins containing the G1 cytoplasmic tail were poorly expressed. Expression of G1 cytoplasmic tail constructs were dramatically enhanced by treating cells with the proteasome inhibitor ALLN, suggesting that the protein is ubiquitinated and degraded via the 26S proteasome. By using a 6-His-tagged ubiquitin, we demonstrated that the G1 cytoplasmic tail is polyubiquitinated and degraded in the absence of proteasome inhibitors. Expression of only the ITAM-containing domain also directed protein ubiquitination and degradation in the absence of upstream residues. Deleting the C-terminal 51 residues of G1, including the ITAM, stabilized G1 and blocked polyubiquitination and degradation of the protein. Site-directed mutagenesis of both ITAM tyrosines (Y619 and Y632) to phenylalanine also blocked polyubiquitination of G1 proteins and dramatically enhanced G1 protein stability. In contrast, the presence of Y627, which is not part of the ITAM motif, had no effect on G1 stability. Mutagenesis of just Y619 enhanced G1 stability, inhibited G1 ubiquitination, and increased the half-life of G1 by threefold. Mutating only Y632 had less of an effect on G1 protein stability, although Y619 and Y632 synergistically contributed to G1 instability. These findings suggest that Y619, which is conserved in all hantaviruses, is the primary signal for directing G1 ubiquitination and degradation. Collectively these findings indicate that specific conserved tyrosines within the G1 cytoplasmic tail direct the polyubiquitination and degradation of expressed G1 proteins and provide a potential means for down-regulating hantavirus G1 surface glycoproteins and cellular proteins that interact with G1.
Hantaviruses infect human endothelial and immune cells, causing two human diseases, hemorrhagic fever with renal syndrome (HFRS) and hantavirus pulmonary syndrome (HPS) (7, 24, 33, 65). Hantaviruses cause severe morbidity with long-term recovery periods, and a 45% mortality rate is associated with HPS-causing hantaviruses. Hantaviruses do not lyse human endothelial cells, and there is little recruitment of immune cells to the endothelium of infected patients (24, 33, 54, 61, 65). In contrast to human infection, hantaviruses persistently infect their small mammal hosts with no apparent deleterious effects (50). There is little understanding of how hantaviruses establish persistence in their hosts, cause disease in humans, or regulate immune and endothelial cell functions that could contribute to viral pathogenesis or persistence.
Hantaviruses are enveloped negative-stranded RNA viruses with a tripartite genome (50). Hantaviruses encode four proteins: a polymerase, a nucleocapsid protein, and two virion surface glycoproteins, G1 and G2, that are recognized by neutralizing antibodies. G1 and G2 are synthesized as a polyprotein which is cotranslationally cleaved during translocation into the endoplasmic reticulum. G1 and G2 contain 56 cysteines that contribute to highly ordered structures of heterodimeric lumenal domains of G1 and G2. G1 and G2 are both C-terminally anchored during translocation with short G2 (8 residues) and long G1 (142 residues) cytoplasmic tails. G1-G2 heterodimers are trafficked to the cis-Golgi, and hantaviruses bud into the lumen of the Golgi instead of at the cell surface. Virion release is consistent with a vesicular secretory process (21, 39, 40). In contrast to the hantavirus nucleocapsid protein, G1 and G2 are expressed at low levels in infected cells and recombinant G1 proteins are also poorly expressed in a variety of expression systems (40, 49). However, it is unclear what regulates G1 protein expression levels and what role limiting glycoprotein expression may play in pathogenesis, immunologic surveillance, or clearance of hantaviruses.
The long G1 cytoplasmic tail of HPS-associated hantaviruses contains an immunoreceptor tyrosine activation motif (ITAM) which directs its association with Src and Syk family kinases (15). Cellular B- and T-cell receptors contain conserved ITAMs [two tandem Yxx(L/I) sequences] within their cytoplasmic tails which recruit Src and Syk family kinases and activate intracellular signaling pathways (45). Syk interactions have also been shown to play critical roles in integrin signaling and endothelial cell functions (20, 53, 60). Src and Syk family signaling was recently shown to be modulated by the cellular protein Cbl (19, 28, 31, 42-44, 63). Cbl is an adapter protein that binds Src and Syk family kinases and directs their ubiquitination and degradation and as a result down-regulates cell surface receptor activation signals (23, 28, 35, 42-44, 47).
Ubiquitination is a posttranslational protein modification that requires ATP and three different proteins, a ubiquitin activating enzyme (E1), a ubiquitin conjugating enzyme (E2), and a ubiquitin ligase (E3) (3, 17). Briefly, free ubiquitin is recruited to E1 and transferred to E2. E2 ubiquitin conjugases bind to RING finger domains on a multitude of E3 ligases. E3 ligases specify protein targets by recruiting cellular proteins, through specific adapter domains, and by recruiting E2 conjugases that direct their ubiquitination. The polyubiquitin tag is recognized by the 26S proteasome, which unfolds and degrades the targeted protein (3, 17). It is now clear that ubiquitination specifically regulates a variety of cellular processes, including cell cycle progression, cell signaling, and apoptosis (3). Cbl is also an E3 ligase whose role in down-regulating key cell signaling proteins has demonstrated that immune cell activation responses are also through the ubiquitin proteasome pathway (18, 42, 43, 55, 63, 66).
In this report we show that the cytoplasmic tail of the hantavirus G1 protein is a target for polyubiquitination and that conserved tyrosines within the G1 tail direct this interaction. Deletional analysis demonstrated that the ITAM tyrosines present in HPS-associated hantaviruses are required for G1 polyubiquitination, since mutagenesis or deletion of these residues blocked G1 cytoplasmic tail ubiquitination and degradation. Mutagenesis of Y619, which is conserved in all hantaviruses, had the greatest effect on the stability of expressed G1 protein. These findings demonstrate a functional role for tyrosines within the G1 protein cytoplasmic tail in directing protein ubiquitination. The ability of the ITAM motif in NY-1V to interact with Src and Syk family kinases and the ability of ITAM tyrosines to direct G1 polyubiquitination further suggests that the G1 cytoplasmic tail may play roles in regulating immunosurveillance or modulating immune and endothelial cell signaling responses during hantavirus infection.
MATERIALS AND METHODS
Cell lines and antibodies.
COS7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% calf serum, 0.2 mM glutamate, 50 U of penicillin per ml, and 50 μg of streptomycin per ml. L40 yeast cells were grown on SC media (0.67% yeast nitrogen base without amino acids, 2% glucose, and 2 μg of amino acid supplements/ml). Anti-Gal4 (SC-510) and anti-ubiquitin (SC-8017) antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.). Anti-LexA antibody (06-719) was from Upstate Biotechnology Inc.
Plasmids.
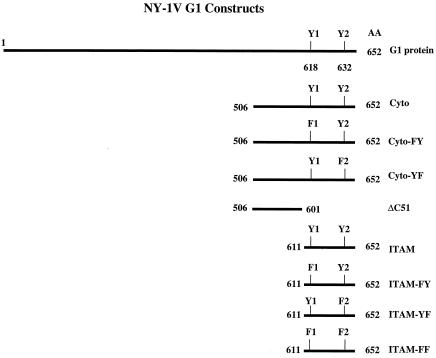
A series of constructs containing deletions or mutations of the strain NY-1V G1 cytoplasmic tail (Fig. 1) was C-terminally fused to Gal4 by insertion into the pBind plasmid (Promega) using G1 primers containing BamHI and XbaI restriction sites. Tyrosines 619 and 632 within ITAM and Cyto constructs were individually mutagenized to phenylalanine (FY, F619-Y632; YF, Y619-F632) by oligo-directed mutagenesis (QuickChange mutagenesis; Stratagene). Similarly, both ITAM tyrosines within the ITAM-Gal4 construct were mutated to phenylalanine (FF) by site-directed mutagenesis (Fig. 1). For yeast expression studies, ITAM and Cyto constructs described above (Fig. 1) were inserted downstream and in frame of the LexA protein in plasmid pBTM116. NY-1V G1 primers containing BamHI and PstI restriction sites were used to generate LexA-ITAM (amino acids 611 to 651) and LexA-Cyto (amino acids 506 to 652) fusion proteins.
FIG. 1.
Schematic representation of NY-1V G1 protein constructs.
Yeast transformation and protein expression.
Transformation of L40 yeast cells was performed by the lithium-acetate method (2, 48). Ten micrograms of transforming DNA and 50 μg of salmon sperm DNA (boiled prior to use) were used for transformation. Transformants were resuspended in the end in 300 μl of YPD (1% yeast extract, 2% peptone, 2% glucose) and were incubated 2 to 3 days at 30°C on trp− plates. Colonies were picked and grown in 5 ml of trp− SC media (0.67% yeast nitrogen base without amino acids, 2% glucose, and a 2-μg/ml concentration of amino acid supplements without tryptophan) overnight at 30°C with shaking to saturation. Approximately 108 cells were pelleted from cultures, supernatants were removed, and pellets were resuspended in 50 μl of sodium dodecyl sulfate (SDS) loading buffer with equal amounts of glass beads and vortexed for 5 min. Samples were boiled for 5 min, spun for 5 min, and resolved on SDS-polyacrylamide gel electrophoresis (PAGE). Western blots were probed with anti-LexA antibodies as described above.
Transfection.
COS7 cells were transfected by using an enhanced calcium phosphate method as previously described (34). COS7 cells (5 × 105) were plated at 40 to 50% confluency in 6-well plates and were transfected for 16 h with a constant amount of plasmid DNA (5 to 10 μg). Medium was replaced, and cell lysates were collected 36 to 48 h posttransfection. Where indicated, 24 h after transfection the proteasome inhibitor ALLN (Calbiotech) was applied to cells at a final concentration of 25 μM overnight.
Immunoprecipitation and Western blots.
Cells were lysed in 0.1% NP-40 lysis buffer for immunoprecipitation and were clarified by centrifugation (46). Lysates were precleared with protein A/G, incubated with indicated antibodies for 2 h or overnight at 4°C, and precipitated with protein A/G agarose. Immunoprecipitated proteins were separated by SDS-10% PAGE and were transferred to nitrocellulose for Western blotting. Western blot analysis was performed as previously described with indicated antibodies (10, 46) and was developed with species-specific horseradish peroxidase-conjugated antibodies (Amersham Pharmacia Biotech) (1:2,000 dilution in 2% bovine serum albumin and TBST [10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Tween 20]). Proteins were detected by chemiluminescence using the ECL reagent (Amersham Pharmacia Biotech).
Ubiquitin labeling.
To detect ubiquitinated proteins we used a 6-His-tagged ubiquitin method (5). COS7 cells were transfected as described above with 2 μg of G1 plasmid DNA in the presence or absence of 1.25 μg of the pMT107 plasmid, expressing ubiquitin with 6 histidines at its N terminus (gift of D. Bohmann, University of Rochester). Media were replaced, and 36 h posttransfection cells were harvested and 6-His-tagged proteins were precipitated by using nickel-nitrilotriacetic acid (NTA)-agarose (Qiagen) as previously described and were Western blotted as described above (5).
Pulse-chase analysis of G1 fusion proteins.
COS7 cells were transfected with ITAM or ΔC51 constructs (10 μg per 30-mm-diameter dish) as described above. Where indicated, 24 h posttransfection the proteasome inhibitor ALLN was applied to cells at a final concentration of 25 μM overnight. Cell monolayers were washed twice with phosphate-buffered saline and were starved of methionine and cysteine by incubation for 1 h at 37°C in methionine/cysteine-free Dulbecco's modified Eagle medium (Gibco/Invitrogen). Cellular proteins were pulse labeled with 400 μCi of [35S]Translabel (ICN) for 1 h at 37°C. Media were removed, and cells were washed twice with PBS and were chased with Dulbecco's modified Eagle medium (containing 2 mM methionine and cysteine) for indicated times. Cells were collected and lysed in a solution of 20 mM Tris-HCl (pH 7.6), 150 mM NaCl, 2 mM EDTA, 1% deoxycholate, 1% Triton X-100, 1× protein inhibitor cocktail (P8340; Sigma Chemical). Proteins were immunoprecipitated by using anti-Gal4 antibodies, and samples were separated by SDS-PAGE, visualized by autoradiography, and quantitated densitometrically by using Adobe Photoshop.
RESULTS
Expression of hantavirus NY-1V G1 protein cytoplasmic tail in yeast.
In the course of studying G1 interactions with cellular proteins we observed that fusion of the G1 cytoplasmic tail (residues 506 to 652) to expressed proteins dramatically decreased their recombinant protein expression levels. Yeast expressing LexA or LexA-G1 fusion proteins (Cyto or ITAM; Fig. 1) were selected and analyzed for fusion protein expression (15). An equal amount of total cellular protein was analyzed by SDS-PAGE and was Western blotted for LexA. Figure 2 demonstrates the low-level expression of LexA when fused to the G1 cytoplasmic tail or ITAM-containing G1 domains. These findings suggested that the G1 cytoplasmic tail might contribute to the instability and degradation of the G1 protein.
FIG. 2.
Expression of G1 fusion proteins in yeast. L40 yeast strain was transformed with LexA (lane 1), LexA-ITAM (lane 2), and LexA-Cyto (lane 3). Equal amounts of total protein were separated by SDS-PAGE, were Western blotted, and were probed with anti-LexA antibody. Arrows indicate the respective positions of indicated LexA or LexA fusion proteins.
The hantavirus G1 cytoplasmic tail directs protein ubiquitination.
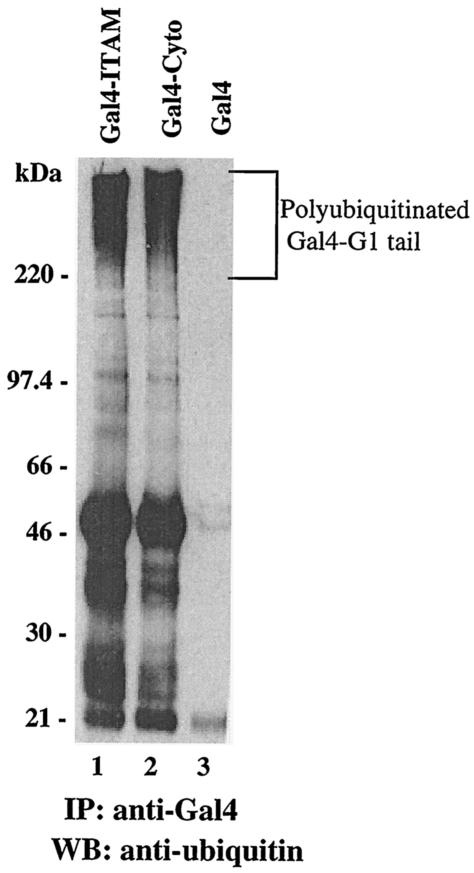
One means by which proteins are targeted for degradation within cells is through their ubiquitination and degradation in the 26S proteasome. In this pathway, proteins are conjugated to multiple copies of ubiquitin that serve as recognition sequences for 26S proteasomal degradation (3, 17, 18). In order to determine if the G1 cytoplasmic tail directs ubiquitination, we fused G1 cytoplasmic tails to the C terminus of Gal4 and assayed for protein ubiquitination within transfected cells. COS7 cells were transfected with Gal4-G1-ITAM and Gal4-G1-Cyto constructs, and Gal4 proteins were immunoprecipitated and Western blotted with an anti-ubiquitin antibody. Both full-length and ITAM-containing G1 cytoplasmic tail fusion proteins were polyubiquitinated, as revealed by the presence of a high-molecular-weight smear (Fig. 3, lanes 1 and 2). In contrast, Gal4 expression alone (lane 3) did not result in the polyubiquitination of the recombinant protein. These results indicate that both the full-length G1 cytoplasmic tail and the ITAM domain within G1 directed polyubiquitination of the expressed proteins.
FIG. 3.
Hantavirus G1 protein is polyubiquitinated. COS7 cells were transfected with Gal4-ITAM (lane 1), Gal4-Cyto (lane 2), and Gal4 (lane 3). Cells were lysed and proteins were immunoprecipitated with anti-Gal4 antibody. Immunoprecipitates (IP) were resolved by SDS-PAGE and were Western blotted (WB) with anti-ubiquitin antibody.
Proteasome inhibitor ALLN stabilizes G1 fusion protein expression.
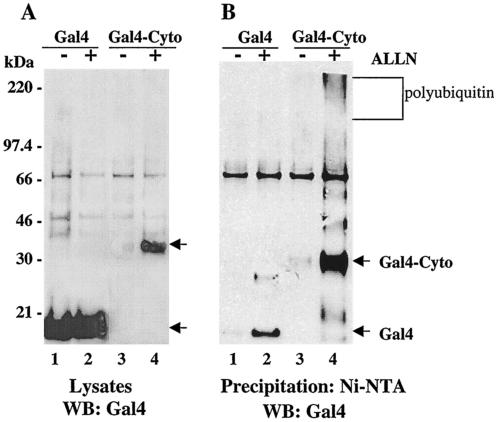
Polyubiquitinated proteins are recognized by the 26S proteasome and are degraded within cells. In order to determine if G1 fusion proteins are degraded by the proteasome, we analyzed G1 protein expression in the presence or absence of the proteasome inhibitor ALLN. COS7 cells were transfected with Gal4 or Gal4-G1-Cyto constructs along with a plasmid expressing a 6-His-tagged ubiquitin. Figure 4A (lanes 1 and 2) indicates that Gal4 is expressed in cell lysates at nearly the same level in the presence or absence of a proteasome inhibitor. In contrast, the Gal4-G1 cytoplasmic tail fusion protein was detected when cells were treated with the proteasome inhibitor but was nearly absent without ALLN (Fig. 4, lanes 3 and 4). In Fig. 4B, ubiquitinated proteins containing 6-His tags were precipitated from lysates by using Ni-NTA resin and were Western blotted for Gal4. Comparison of Fig. 4A and B demonstrates that the majority of Gal4 present in lysates is not ubiquitinated following expression and that Gal4 is not present in polyubiquitinated forms in the presence or absence of ALLN. In contrast, the Gal4-G1 cytoplasmic tail is barely detectable in the absence of the proteasome inhibitor, while in the presence of ALLN the Gal4-G1 fusion protein is highly expressed and is present in polyubiquitinated forms. Additional bands in the ALLN-treated lanes are consistent with the sequential addition of 8.5-kDa ubiquitin monomers. These findings demonstrate that the G1 cytoplasmic tail directs polyubiquitination and degradation of Gal4 fusion proteins via the proteasome.
FIG. 4.
Proteasome inhibitor stabilizes G1 protein expression. COS7 cells were transfected with Gal4 (lanes 1 and 2) or Gal4-Cyto (lanes 3 and 4) constructs simultaneously with His6-ubiquitin in the presence (+) or absence (−) of the proteasome inhibitor ALLN. Cells were treated for 16 h with ALLN or dimethyl sulfoxide (control) and were harvested. Equivalent amounts of total protein from straight lysates (A) or samples from Ni-NTA agarose pull-downs (B) were run on SDS-PAGE gels and were Western blotted (WB) with Gal4 antibody.
ITAM tyrosines direct G1 ubiquitination and degradation.
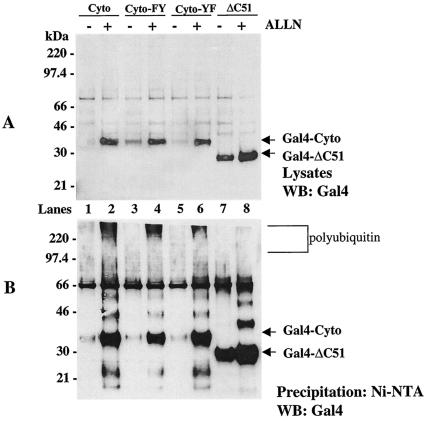
Our findings suggested that elements within the G1 cytoplasmic tail direct protein polyubiquitination and degradation. We previously reported that the NY-1V G1 cytoplasmic tail contains two tandem YxxL sequences which form an ITAM (15). Since the G1 ITAM directs interactions with cellular kinases that are themselves regulated by polyubiquitination and degradation, we hypothesized that ITAM tyrosines may be required for G1 ubiquitination and degradation. In order to study the role of the ITAM in polyubiquitination and degradation, we mutated ITAM tyrosines present in the G1 cytoplasmic tail (Cyto-FY, Cyto-YF) or deleted the ITAM domain entirely (ΔC51) (Fig. 1). Figure 5A shows that mutating the first ITAM tyrosine to phenylalanine partially enhanced the stability of the Cyto-FY protein in cell lysates in the absence of the proteasome inhibitor (Fig. 5A, lane 3). However, most of the Cyto-FY protein was still polyubiquitinated and degraded like the wild-type protein (Fig. 5B, lanes 1 to 4). Mutation of the second ITAM tyrosine (Cyto-YF) had less of an effect on the stability of the protein or on its ubiquitination and degradation (Fig. 5A and B, lanes 5 and 6). In contrast, deleting the C-terminal 51 amino acids of G1 (ΔC51), removing both ITAM tyrosines, dramatically enhanced the stability of the Gal4-ΔC51 fusion protein and blocked polyubiquitination of the protein (Fig. 5A and B, lane 8). These findings suggested that the G1 ITAM domain determines the stability of expressed G1 proteins.
FIG. 5.
The C-terminal domain of the G1 protein cytoplasmic tail directs polyubiquitination. COS7 cells were transfected with Cyto (lanes 1 and 2), Cyto-FY (lanes 3 and 4), Cyto-YF (lanes 5 and 6), and ΔC51 (lanes 7 and 8) constructs simultaneously with His6-ubiquitin in the presence (+) or absence (−) of the proteasome inhibitor ALLN. Cells were lysed, and equal amounts of total protein from straight lysates (A) or samples from Ni-NTA agarose precipitations (B) were run on SDS-PAGE and were Western blotted (WB) with Gal4 antibody.
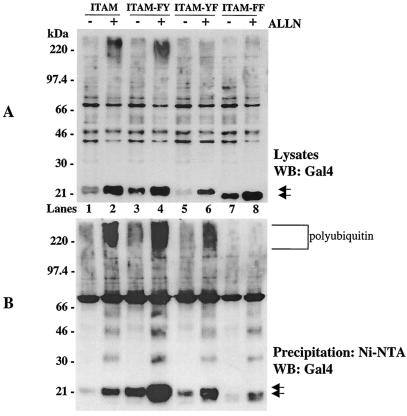
In order to define the role of ITAM elements on G1 stability, we analyzed the role of tyrosines within the C-terminal G1 ITAM domain on protein ubiquitination and degradation (Fig. 6). Mutation of the first ITAM tyrosine (ITAM-FY; Fig. 6A, lane 3) resulted in an even more dramatic enhancement of G1 protein stability in lysates than was apparent by expressing the comparable full-length cytoplasmic tail construct (Cyto-FY; Fig. 5A, lane 3). Similarly, ALLN enhanced the stability of the ITAM-FY protein and its polyubiquitination (Fig. 6B, lane 4). Although mutating the second ITAM tyrosine had little apparent effect on protein stability or polyubiquitination, mutation of both ITAM tyrosines (ITAM-FF) resulted in a dramatic effect on protein stability and polyubiquitination (Fig. 6B, lanes 3 to 6 versus lanes 7 and 8). The ITAM-FF protein was stable in cells in the absence of ALLN (Fig. 6A, lane 7), similar to deleting the ITAM (Fig. 5), and was not present in polyubiquitinated forms (Fig. 6B, lanes 7 and 8). These findings indicate that ITAM tyrosines within the NY-1V G1 cytoplasmic tail direct protein ubiquitination and degradation.
FIG. 6.
ITAM tyrosines direct G1 protein polyubiquitination and degradation. COS7 cells were transfected with ITAM (lanes 1 and 2), ITAM-FY (lanes 3 and 4), ITAM-YF (lanes 5 and 6), and ITAM-FF (lanes 7 and 8) constructs simultaneously with His6-ubiquitin in the presence (+) or absence (−) of the proteasome inhibitor ALLN. Cells were lysed, and equal amounts of total protein from straight lysates (A) or samples from Ni-NTA agarose pull-downs (B) were run on SDS-PAGE and were Western blotted (WB) with Gal4 antibody.
Pulse-chase analysis of G1 fusion protein stability.
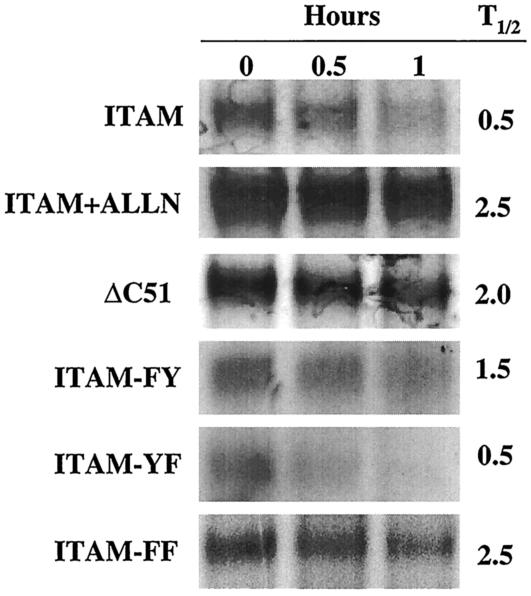
As another measure of G1 fusion protein stability we analyzed the half-life of expressed G1 fusion proteins by pulse-chase analysis. The stability of expressed proteins was analyzed 48 h posttransfection by pulsing transfected cells with [35S]methionine for 1 h and chasing with excess cold methionine. Gal4 was immunoprecipitated from samples and was analyzed by SDS-PAGE and autoradiography. Densitometric analysis of bands was used to determine the half-life of proteins (Fig. 7). ITAM fusion proteins were degraded, with a half-life of 0.5 h, while ALLN treatment extended the half-life of the protein by at least fivefold (≥2.5 h). Deleting the ITAM (ΔC51) resulted in a half-life of 2 h, nearly the same as that achieved by treating cells with the proteasome inhibitor. Mutations of the first ITAM tyrosine also increased the half-life of the protein by threefold, confirming above results that this tyrosine plays an important role in G1 stability (Fig. 7). Mutating the second ITAM tyrosine had no apparent effect on the half-life of the protein. However, mutation of both ITAM tyrosines resulted in extending the protein half-life to a level identical to that of ALLN treatment of cells. Collectively these findings suggest that the downstream ITAM tyrosine contributes to the instability of the G1 fusion protein by enhancing the destabilizing effect of tyrosine 619. As a result, tyrosines 619 and 632 have synergistic effects on the stability of expressed G1 protein and contribute to rapid G1 protein turnover.
FIG. 7.
Pulse-chase analysis of G1 fusion protein stability. COS7 cells were transfected with ITAM, ΔC51, ITAM-FY, ITAM-YF, and ITAM-FF constructs. In parallel, cells transfected with ITAM constructs were treated with the proteasome inhibitor ALLN for 16 h. Samples were pulsed with [35S]methionine for 1.5 h and were chased for the indicated times (0, 0.5, or 1 h). Lysates of collected samples were immunoprecipitated with Gal4 antibody and were resolved by SDS-PAGE. Samples were visualized by autoradiography, and protein half-lives (T1/2) were calculated by measuring and comparing band densities.
DISCUSSION
We have determined that the cytoplasmic tail of the hantavirus G1 protein directs its ubiquitination and degradation. Our findings show that NY-1V G1 ITAM tyrosines Y619 and Y632 are required for protein ubiquitination and that deletion or mutagenesis of these tyrosines stabilized expressed proteins. In contrast, Y627, which is not part of the ITAM motif, was present in all constructs (except ΔC51) but had no apparent effect on G1 protein ubiquitination or stability. This indicates a specific role for ITAM tyrosines in the ubiquitination and degradation of the NY-1V G1 protein and the functional importance of the ITAM motif which is conserved in all HPS-associated hantaviruses. These findings demonstrate the importance of specific tyrosine residues within the G1 protein cytoplasmic tail and provide a beginning understanding of conserved regulatory functions of hantavirus proteins within cells.
Hantaviruses have very few proteins by which they can convey a broad array of viral functions, including viral maturation and assembly, viral persistence, and viral regulation of cellular responses (49). Hantaviruses contain four proteins: a polymerase, a nucleocapsid protein, and two virion surface glycoproteins. Outside of the role of the viral polymerase, there are only two substantial cytoplasmic proteins that are available to regulate cell signaling, the N protein and the long cytoplasmic tail of G1. Clearly these hantavirus proteins must perform multiple functions for the virus to establish persistence within its animal host, a task that is accomplished by a large number of DNA virus or retrovirus proteins. Within their hosts, hantaviruses somehow fail to initiate a disease process during infection while avoiding innate and cellular immune responses that should clear the virus from the host. Because hantaviruses infect endothelial cells as well as immune and dendritic cells, there are many points for hantaviruses to regulate viral clearance during infection (33, 41, 61, 65). The fact that each hantavirus is associated with a singular primary host suggests that differences in virally regulated host proteins could limit the success of hantaviruses to selected mammals (6). In contrast to animals, transmission of hantaviruses to humans results in two diseases as well as viral clearance from patients and the failure of viruses to establish persistence (33, 65, 67). This suggests that tactics for cell regulation which are successful in small mammals fail in humans and instead may contribute to pathogenesis.
ITAM sequences have been associated with viral pathogenesis. The ITAM present in the simian immunodeficiency virus (SIV) Nef protein is directly associated with acute pathogenesis caused by the virus (9). Macaques inoculated with ITAM-mutated SIV variants had no clinical symptoms, whereas ITAM-containing SIV caused acute disease. The viruses replicated identically and with the same kinetics, and the Nef ITAM altered the pathogenesis of SIV regardless of the viral background (9). It has yet to be determined whether Nef-carrying ITAMs contribute to pathogenesis through kinase regulation or ubiquitination and degradation of specific cellular proteins. Our findings that ITAMs are present only in hantaviruses associated with HPS suggest that the presence of the viral ITAM could contribute to differences in the pathogenesis of HPS- and HFRS-associated hantaviruses that lead to the high mortality rate of HPS-causing strains.
Cellular ITAM signaling elements are also present in the cytoplasmic tails of T- and B-cell receptors where they direct extracellular ligand binding signals to activate immune cells (45). Viruses which establish persistence regulate immune cell functions by using various mechanisms, including the regulation of cellular ITAM signaling. The Epstein-Barr virus latent membrane protein 2A (LMP2A) is a prominent example of virus-regulated ITAM functions (29, 58). Like immunoreceptors, LMP2A contains an ITAM element in its cytoplasmic tail that interacts with Src and Syk family kinases (13, 29, 51, 58). The LMP2A ITAM is reported to be essential for providing B cells with development and survival signals required for viral success within immune cells (13, 29). LMP2A enhances Lyn and Syk ubiquitination and destabilizes the Lyn Src family kinase (29, 58). Within LMP2A there are at least two domains that direct kinase and ubiquitin pathway interactions, the ITAM which binds Src and Syk family kinases and PPPY motifs that bind WW domains of E3 protein ubiquitin ligases and target proteins to the proteasome (58). Mutation of the LMP2A ITAM eliminates Syk binding and allows for B-cell receptor signaling and normal B-cell maturation, while LMP2A transgenic lines are unable to promote B-cell development (29).
As for LMP2A, the interaction of the hantavirus G1 ITAM with Src and Syk family kinases was recently reported (15). Here we have demonstrated that G1 ITAM tyrosines direct protein ubiquitination and degradation, suggesting that hantaviruses may similarly regulate immune and endothelial cell signaling pathways that direct cellular activation and adherence functions (15). Unlike LMP2A, there are no PY motifs within the ITAM domain of G1 to recruit E3 ligases. Our findings are consistent with the ITAM tyrosines themselves directing the recruitment of G1 to complexes containing E3 ubiquitin ligases. Alternatively, there may be as-yet unidentified motifs within the G1 ITAM domain that recruit E3 ligases.
Cellular adapter proteins have also been shown to down-regulate immunoreceptor-directed ITAM signaling responses via the proteasome (42-44). The cellular adapter proteins, src-like adapter protein-2 (SLAP-2) and Cbl, bind and target Src and Syk family kinases for degradation in the proteasome, down-regulating T- and B-cell receptor activation signals (28, 31, 32, 42-44, 47, 55, 63, 66). Cbl is a cellular protein which recruits E2 conjugases as well as Src and Syk family kinases, thereby attenuating T- and B-cell receptor signaling responses that direct immune cell activation (1, 3, 19, 42-44). SLAP-2 also interacts with Cbl complexes and directs the degradation of Syk family kinases cooperatively with Cbl (28). Cbl knockout mice have hyperresponsive T-cell receptor and B-cell receptor activities, enhanced positive selection of CD4+ thymocytes, and increased surface T-cell receptor levels (31, 32, 42, 47, 66). Cells from Cbl knockout mice have elevated Lck activity and enhanced Lck and Fyn levels in T-cell lines and are defective in ubiquitinating Src family kinases (32, 42). These findings define specific roles for Cbl in regulating T-cell receptor activation responses at multiple levels (3, 32, 42, 44, 66). The ability of the hantavirus G1 protein to bind the Src and Syk kinases and direct protein ubiquitination suggests that HPS-associated hantavirus G1 proteins may also regulate signaling pathways which direct and modulate immune cell activation signals through Cbl- or SLAP-2-like mechanisms.
In addition to immune cell regulation, Cbl has been found to be a central regulator of cell surface receptor activation responses, regulating growth factor receptor and integrin signaling responses in addition to immunoreceptors (8, 11, 22, 26, 27, 52, 56, 57, 59, 64). This is accomplished through the central regulation of Syk and through Cbl's ability to bind, ubiquitinate, and target vascular endothelial growth factor receptor and other growth factor receptors for degradation in the lysosome (18, 25, 30, 37, 42, 56, 62). These findings suggest that G1 complexes with E3 ligases and Src and Syk family kinases could have broad effects on integrin- and growth factor-directed endothelial cell functions which are likely to participate in vascular permeability defects observed in HPS (4, 20, 60). However, there is little information on hantavirus regulation of endothelial or immune cell responses or whether regulation of specific responses contributes to viral pathogenesis or persistence (14, 41, 54, 61, 65).
Since HPS-associated hantaviruses contain ITAMs, these findings do not suggest that ITAMs are required for viral persistence and instead point to a potential role for the ITAM in HPS-specific pathogenic responses. Nonpathogenic, HPS- and HFRS-associated hantaviruses all establish persistence in their hosts, and this suggests a common viral mechanism of persistence. All hantaviruses contain tyrosine 619 in their G1 cytoplasmic tails, and our findings demonstrate that this tyrosine has a dominant effect on protein ubiquitination and degradation. The role of Y619 in the ubiquitination and degradation of G1 proteins from other hantaviruses needs to be addressed in order to determine if this is a common G1 function. However, there are many potential roles for Y619 in G1. One possibility is that Y619 could specify differences in the signaling proteins recruited to the G1 tail for down regulation. Ubiquitination of G1 could also serve additional roles in hantavirus budding, as described for retroviruses (36, 38). Unfortunately, testing many of these questions awaits reverse genetic approaches, which at this point are not established for hantaviruses but appear to be nearly complete (12).
Further differences in cellular regulation between pathogenic and nonpathogenic hantaviruses have recently been reported. Nonpathogenic Prospect Hill virus induced early interferon responses that were not directed by pathogenic NY-1 or HTN viruses (16). Differential cellular interferon responses between pathogenic and nonpathogenic hantaviruses suggest an active role for hantavirus proteins in regulating innate immune responses. In fact, the G1 cytoplasmic tail or the N protein might contribute to additional cell signaling regulatory responses. This could relate to the G1-directed ubiquitination and degradation of other proteins bound to additional domains on G1 or to completely unique interactions with cellular proteins.
The importance of the first YxxL in the G1 cytoplasmic tail and its conservation in all hantaviruses suggests that G1 cytoplasmic tails in general may be targets for ubiquitination and degradation and may play common roles during hantavirus infection. The presence of a G1 ITAM that directs protein ubiquitination and degradation as well as interacts with key immune and endothelial cell kinases provides a potential role for hantavirus signaling interactions in modulating immune and endothelial cell activation responses and as determinants of hantavirus pathogenesis.
Acknowledgments
We thank Tracy Raymond for critical review of the manuscript.
This work was supported by a Merit Award from the Veterans Administration and by NIH grants AI047873 and AI044917.
REFERENCES
- 1.Andoniou, C. E., N. L. Lill, C. B. Thien, M. L. Lupher, Jr., S. Ota, D. D. Bowtell, R. M. Scaife, W. Y. Langdon, and H. Band. 2000. The Cbl proto-oncogene product negatively regulates the Src-family tyrosine kinase Fyn by enhancing its degradation. Mol. Cell. Biol. 20:851-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel, P., C. T. Chien, R. Sternglanz, and S. Fields. 1993. Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14:920-924. [PubMed] [Google Scholar]
- 3.Ben-Neriah, Y. 2002. Regulatory functions of ubiquitination in the immune system. Nat. Immunol. 3:20-26. [DOI] [PubMed] [Google Scholar]
- 4.Byzova, V. T., K. C. Goldman, N. Pampori, A. K. Thomas, A. Bett, J. S. Shattil, and F. E. Plow. 2000. A mechanism for modulation of cellular responses to VEGF: activation of the integrins. Mol. Cell 6:851-860. [PubMed] [Google Scholar]
- 5.Campanero, M. R., and E. K. Flemington. 1997. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:2221-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Childs, J. E., T. G. Ksiazek, C. F. Spiropoulou, J. W. Krebs, S. Morzunov, G. O. Maupin, K. L. Gage, P. E. Rollin, J. Sarisky, R. E. Enscore, et al. 1994. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J. Infect. Dis. 169:1271-1280. [DOI] [PubMed] [Google Scholar]
- 7.Cosgriff, T. M., and R. M. Lewis. 1991. Mechanisms of disease in hemorrhagic fever with renal syndrome. Kidney Int. Suppl. 35:S72-S79. [PubMed] [Google Scholar]
- 8.de Melker, A. A., G. van der Horst, J. Calafat, H. Jansen, and J. Borst. 2001. c-Cbl ubiquitinates the EGF receptor at the plasma membrane and remains receptor associated throughout the endocytic route. J. Cell Sci. 114:2167-2178. [DOI] [PubMed] [Google Scholar]
- 9.Dehghani, H., C. R. Brown, R. Plishka, A. Buckler-White, and V. M. Hirsch. 2002. The ITAM in Nef influences acute pathogenesis of AIDS-inducing simian immunodeficiency viruses SIVsm and SIVagm without altering kinetics or extent of viremia. J. Virol. 76:4379-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devergne, O., E. Hatzivassiliou, K. M. Izumi, K. M. Kaye, M. F. Kleijnen, E. Kieff, and G. Mosialos. 1996. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol. Cell. Biol. 16:7098-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ettenberg, S. A., A. Magnifico, M. Cuello, M. M. Nau, Y. R. Rubinstein, Y. Yarden, A. M. Weissman, and S. Lipkowitz. 2001. Cbl-b-dependent coordinated degradation of the epidermal growth factor receptor signaling complex. J. Biol. Chem. 276:27677-27684. [DOI] [PubMed] [Google Scholar]
- 12.Flick, K., J. W. Hooper, C. S. Schmaljohn, R. F. Pettersson, H. Feldmann, and R. Flick. 2003. Rescue of Hantaan virus minigenomes. Virology 306:219-224. [DOI] [PubMed] [Google Scholar]
- 13.Fruehling, S., and R. Longnecker. 1997. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BCR-mediated signal transduction. Virology 235:241-251. [DOI] [PubMed] [Google Scholar]
- 14.Gavrilovskaya, I. N., T. Peresleni, E. Geimonen, and E. R. Mackow. 2002. Pathogenic hantaviruses selectively inhibit beta3 integrin directed endothelial cell migration. Arch. Virol. 147:1913-1931. [DOI] [PubMed] [Google Scholar]
- 15.Geimonen, E., R. LaMonica, K. Springer, Y. Farooqui, I. N. Gavrilovskaya, and E. R. Mackow. 2003. Hantavirus pulmonary syndrome-associated hantaviruses contain conserved and functional ITAM signaling elements. J. Virol. 77:1638-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geimonen, E., S. Neff, T. Raymond, S. S. Kocer, I. N. Gavrilovskaya, and E. R. Mackow. 2002. Pathogenic and nonpathogenic hantaviruses differentially regulate endothelial cell responses. Proc. Natl. Acad. Sci. USA 99:13837-13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas, A. L., and T. J. Siepmann. 1997. Pathways of ubiquitin conjugation. FASEB J. 11:1257-1268. [DOI] [PubMed] [Google Scholar]
- 18.Hicke, L. 1999. Gettin' down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 9:107-112. [DOI] [PubMed] [Google Scholar]
- 19.Howlett, C. J., and S. M. Robbins. 2002. Membrane-anchored Cbl suppresses Hck protein-tyrosine kinase mediated cellular transformation. Oncogene 21:1707-1716. [DOI] [PubMed] [Google Scholar]
- 20.Inatome, R., S. Yanagi, T. Takano, and H. Yamamura. 2001. A critical role for Syk in endothelial cell proliferation and migration. Biochem. Biophys. Res. Commun. 286:195-199. [DOI] [PubMed] [Google Scholar]
- 21.Kamrud, K. I., and C. S. Schmaljohn. 1994. Expression strategy of the M genome segment of Hantaan virus. Virus Res. 31:109-121. [DOI] [PubMed] [Google Scholar]
- 22.Kassenbrock, C. K., S. Hunter, P. Garl, G. L. Johnson, and S. M. Anderson. 2002. Inhibition of Src family kinases blocks epidermal growth factor (EGF)-induced activation of Akt, phosphorylation of c-Cbl, and ubiquitination of the EGF receptor. J. Biol. Chem. 277:24967-24975. [DOI] [PubMed] [Google Scholar]
- 23.Latour, S., and A. Veillette. 2001. Proximal protein tyrosine kinases in immunoreceptor signaling. Curr. Opin. Immunol. 13:299-306. [DOI] [PubMed] [Google Scholar]
- 24.Lee, H. W. 1982. Hemorrhagic fever with renal syndrome (HFRS). Scand. J. Infect. Dis. Suppl. 36:82-85. [PubMed] [Google Scholar]
- 25.Lee, P. S., Y. Wang, M. G. Dominguez, Y. G. Yeung, M. A. Murphy, D. D. Bowtell, and E. R. Stanley. 1999. The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. EMBO J. 18:3616-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levkowitz, G., H. Waterman, S. A. Ettenberg, M. Katz, A. Y. Tsygankov, I. Alroy, S. Lavi, K. Iwai, Y. Reiss, A. Ciechanover, S. Lipkowitz, and Y. Yarden. 1999. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 4:1029-1040. [DOI] [PubMed] [Google Scholar]
- 27.Longva, K. E., F. D. Blystad, E. Stang, A. M. Larsen, L. E. Johannessen, and I. H. Madshus. 2002. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 156:843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loreto, M. P., D. M. Berry, and C. J. McGlade. 2002. Functional cooperation between c-Cbl and Src-like adaptor protein 2 in the negative regulation of T-cell receptor signaling. Mol. Cell. Biol. 22:4241-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merchant, M., R. G. Caldwell, and R. Longnecker. 2000. The LMP2A ITAM is essential for providing B cells with development and survival signals in vivo. J. Virol. 74:9115-9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monsonego-Ornan, E., R. Adar, E. Rom, and A. Yayon. 2002. FGF receptors ubiquitylation: dependence on tyrosine kinase activity and role in downregulation. FEBS Lett. 528:83-89. [DOI] [PubMed] [Google Scholar]
- 31.Murphy, M. A., R. G. Schnall, D. J. Venter, L. Barnett, I. Bertoncello, C. B. Thien, W. Y. Langdon, and D. D. Bowtell. 1998. Tissue hyperplasia and enhanced T-cell signaling via ZAP-70 in c-Cbl-deficient mice. Mol. Cell. Biol. 18:4872-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naramura, M., H. K. Kole, R. J. Hu, and H. Gu. 1998. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc. Natl. Acad. Sci. USA 95:15547-15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nolte, K. B., R. M. Feddersen, K. Foucar, S. R. Zaki, F. T. Koster, D. Madar, T. L. Merlin, P. J. McFeeley, E. T. Umland, and R. E. Zumwalt. 1995. Hantavirus pulmonary syndrome in the United States: a pathological description of a disease caused by a new agent. Hum. Pathol. 26:110-120. [DOI] [PubMed] [Google Scholar]
- 34.O'Mahoney, J. V., and T. E. Adams. 1994. Optimization of experimental variables influencing reporter gene expression in hepatoma cells following calcium phosphate transfection. DNA Cell Biol. 13:1227-1232. [DOI] [PubMed] [Google Scholar]
- 35.Ota, Y., and L. E. Samelson. 1997. The product of the proto-oncogene c-cbl: a negative regulator of the Syk tyrosine kinase. Science 276:418-420. [DOI] [PubMed] [Google Scholar]
- 36.Ott, D. E., L. V. Coren, R. C. Sowder, Jr., J. Adams, and U. Schubert. 2003. Retroviruses have differing requirements for proteasome function in the budding process. J. Virol. 77:3384-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panigada, M., S. Porcellini, E. Barbier, S. Hoeflinger, P. A. Cazenave, H. Gu, H. Band, H. von Boehmer, and F. Grassi. 2002. Constitutive endocytosis and degradation of the pre-T cell receptor. J. Exp. Med. 195:1585-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pensiero, M., and J. Hay. 1992. The Hantaan virus M-segment glycoproteins G1 and G2 can be expressed independently. J. Virol. 66:1907-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pensiero, M. N., G. B. Jennings, C. S. Schmaljohn, and J. Hay. 1988. Expression of the Hantaan virus M genome segment by using a vaccinia virus recombinant. J. Virol. 62:696-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raftery, M. J., A. A. Kraus, R. Ulrich, D. H. Kruger, and G. Schonrich. 2002. Hantavirus infection of dendritic cells. J. Virol. 76:10724-10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao, N., I. Dodge, and H. Band. 2002. The Cbl family of ubiquitin ligases: critical negative regulators of tyrosine kinase signaling in the immune system. J. Leukoc. Biol. 71:753-763. [PubMed] [Google Scholar]
- 43.Rao, N., A. K. Ghosh, S. Ota, P. Zhou, A. L. Reddi, K. Hakezi, B. K. Druker, J. Wu, and H. Band. 2001. The non-receptor tyrosine kinase Syk is a target of Cbl-mediated ubiquitylation upon B-cell receptor stimulation. EMBO J. 20:7085-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao, N., S. Miyake, A. L. Reddi, P. Douillard, A. K. Ghosh, I. L. Dodge, P. Zhou, N. D. Fernandes, and H. Band. 2002. Negative regulation of Lck by Cbl ubiquitin ligase. Proc. Natl. Acad. Sci. USA 99:3794-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reth, M., and J. Wienands. 1997. Initiation and processing of signals from the B cell antigen receptor. Annu. Rev. Immunol. 15:453-479. [DOI] [PubMed] [Google Scholar]
- 46.Rothe, M., J. Xiong, H. B. Shu, K. Williamson, A. Goddard, and D. V. Goeddel. 1996. I-TRAF is a novel TRAF-interacting protein that regulates TRAF-mediated signal transduction. Proc. Natl. Acad. Sci. USA 93:8241-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudd, C. E., and H. Schneider. 2000. Lymphocyte signaling: Cbl sets the threshold for autoimmunity. Curr. Biol. 10:R344-R347. [DOI] [PubMed] [Google Scholar]
- 48.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 49.Schmaljohn, C. 1996. Bunyaviridae and their replication, p. 1447-1471. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven Press, Philadelphia, Pa.
- 50.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scholle, F., R. Longnecker, and N. Raab-Traub. 2001. Analysis of the phosphorylation status of Epstein-Barr virus LMP2A in epithelial cells. Virology 291:208-214. [DOI] [PubMed] [Google Scholar]
- 52.Taher, T. E., E. P. Tjin, E. A. Beuling, J. Borst, M. Spaargaren, and S. T. Pals. 2002. c-Cbl is involved in Met signaling in B cells and mediates hepatocyte growth factor-induced receptor ubiquitination. J. Immunol. 169:3793-3800. [DOI] [PubMed] [Google Scholar]
- 53.Turner, M., E. Schweighoffer, F. Colucci, J. P. Di Santo, and V. L. Tybulewicz. 2000. Tyrosine kinase SYK: essential functions for immunoreceptor signalling. Immunol. Today 21:148-154. [DOI] [PubMed] [Google Scholar]
- 54.Vapalahti, O., A. Lundkvist, and A. Vaheri. 2001. Human immune response, host genetics, and severity of disease. Curr. Top. Microbiol. Immunol. 256:153-169. [DOI] [PubMed] [Google Scholar]
- 55.Wang, H. Y., Y. Altman, D. Fang, C. Elly, Y. Dai, Y. Shao, and Y. C. Liu. 2001. Cbl promotes ubiquitination of the T cell receptor zeta through an adaptor function of Zap-70. J. Biol. Chem. 276:26004-26011. [DOI] [PubMed] [Google Scholar]
- 56.Wang, Y., H. Miao, S. Li, K. D. Chen, Y. S. Li, S. Yuan, J. Y. Shyy, and S. Chien. 2002. Interplay between integrins and FLK-1 in shear stress-induced signaling. Am. J. Physiol. Cell Physiol. 283:C1540-C1547. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Y., Y. G. Yeung, and E. R. Stanley. 1999. CSF-1 stimulated multiubiquitination of the CSF-1 receptor and of Cbl follows their tyrosine phosphorylation and association with other signaling proteins. J. Cell Biochem. 72:119-134. [PubMed] [Google Scholar]
- 58.Winberg, G., L. Matskova, F. Chen, P. Plant, D. Rotin, G. Gish, R. Ingham, I. Ernberg, and T. Pawson. 2000. Latent membrane protein 2A of Epstein-Barr virus binds WW domain E3 protein-ubiquitin ligases that ubiquitinate B-cell tyrosine kinases. Mol. Cell. Biol. 20:8526-8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woodside, D. G., A. Obergfell, A. Talapatra, D. A. Calderwood, S. J. Shattil, and M. H. Ginsberg. 2002. The N-terminal SH2 domains of Syk and ZAP-70 mediate phosphotyrosine-independent binding to integrin beta cytoplasmic domains. J. Biol. Chem. 277:39401-39408. [DOI] [PubMed] [Google Scholar]
- 60.Yanagi, S., R. Inatome, J. Ding, H. Kitaguchi, V. L. Tybulewicz, and H. Yamamura. 2001. Syk expression in endothelial cells and their morphologic defects in embryonic Syk-deficient mice. Blood 98:2869-2871. [DOI] [PubMed] [Google Scholar]
- 61.Yanagihara, R., and D. J. Silverman. 1990. Experimental infection of human vascular endothelial cells by pathogenic and nonpathogenic hantaviruses. Arch. Virol. 111:281-286. [DOI] [PubMed] [Google Scholar]
- 62.Yokouchi, M., T. Kondo, A. Houghton, M. Bartkiewicz, W. C. Horne, H. Zhang, A. Yoshimura, and R. Baron. 1999. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J. Biol. Chem. 274:31707-31712. [DOI] [PubMed] [Google Scholar]
- 63.Yokouchi, M., T. Kondo, A. Sanjay, A. Houghton, A. Yoshimura, S. Komiya, H. Zhang, and R. Baron. 2001. Src-catalyzed phosphorylation of c-Cbl leads to the interdependent ubiquitination of both proteins. J. Biol. Chem. 276:35185-35193. [DOI] [PubMed] [Google Scholar]
- 64.Yokouchi, M., T. Wakioka, H. Sakamoto, H. Yasukawa, S. Ohtsuka, A. Sasaki, M. Ohtsubo, M. Valius, A. Inoue, S. Komiya, and A. Yoshimura. 1999. APS, an adaptor protein containing PH and SH2 domains, is associated with the PDGF receptor and c-Cbl and inhibits PDGF-induced mitogenesis. Oncogene 18:759-767. [DOI] [PubMed] [Google Scholar]
- 65.Zaki, S., P. Greer, L. Coffield, C. Goldsmith, K. Nolte, K. Foucar, R. Feddersen, R. Zumwalt, G. Miller, P. Rollin, T. Ksiazek, S. Nichol, and C. Peters. 1995. Hantavirus pulmonary syndrome: pathogenesis of an emerging infectious disease. Am. J. Pathol. 146:552-579. [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, J., T. Bardos, D. Li, I. Gal, C. Vermes, J. Xu, K. Mikecz, A. Finnegan, S. Lipkowitz, and T. T. Glant. 2002. Cutting edge: regulation of T cell activation threshold by CD28 costimulation through targeting Cbl-b for ubiquitination. J. Immunol. 169:2236-2240. [DOI] [PubMed] [Google Scholar]
- 67.Terajima, M., J. D. Hendershot, H. Kariwa, F. T. Koster, B. Hjelle, D. Goade, M. C. DeFronzo, and F. A. Ennis. 1999. High levels of viremia in patients with the Hantavirus pulmonary syndrome. J. Infect. Dis. 180:2030-2034. [DOI] [PubMed] [Google Scholar]