Abstract
The single-stranded circular DNA of Abutilon mosaic geminivirions is complemented to double-stranded DNA by host proteins after infecting cells. This double-stranded DNA serves as a template for replication as well as transcription and is assembled into host nucleosomes, yielding circular viral minichromosomes. Their chromatin structure was analyzed by use of isolated nuclei combining nuclease sensitivity assays with ligation-mediated PCR, evaluating nucleosomal ladders and topoisomer distributions in one- and two-dimensional gels by blot hybridization. Viral minichromosomes were found to exist in at least two defined structures covered with 11 or 12 nucleosomes, leaving open gaps accessible for interactions with other host factors. Nucleosome-free gaps were colocalized with promoter structures and the origin of replication in both components of genomic DNA (DNA A and DNA B). Nucleosomes were positioned over the entire viral DNA in at least two alternative phases with different periodicities. The distribution of topoisomers of monomeric viral circular double-stranded DNA confirmed the presence of variable chromatin structures revealing maximum frequencies of molecules with either 11, 12, or 13 superhelical turns (corresponding to respective numbers of nucleosomes) at maximal frequency at different stages during leaf development of infected plants. The role of variable chromatin structures for gene regulation of geminiviruses is discussed.
Replication and transcription are central processes in all living organisms. In comparison to animals, much less is known about their regulation in plants (38). DNA viruses, such as simian virus 40, have been extremely helpful model systems for understanding regulatory cascades in animals (23). A similar tool for plants was lacking, but geminiviruses are appropriate to fill this gap because they are similar to papovaviruses in several aspects of genetic organization. Geminiviruses differ from papovaviruses in that they are tiny circular single-stranded DNA (ssDNA)-containing viruses with one or two genomic components (34), but replicate via double-stranded DNA (dsDNA) intermediates in nuclei, organize their replicative and transcriptional DNA intermediates in minichromosomes (2, 32), and transcribe their genes bidirectionally. Correspondingly, the genome organization of geminiviruses is very similar to that of papovaviruses (42).
Abutilon mosaic virus (AbMV) of the genus Begomovirus (34) possesses a bipartite genome consisting of DNA A and DNA B (see Fig. 4). The DNAs are different from each other, except for a common region of about 200 nucleotides harboring most of the regulatory elements for replication and transcription. As their genome is very small, they rely on host factors, especially a DNA polymerase, for both replication and transcription (reviewed by Hanley-Bowdoin et al. [16]). The common region contains a palindromic sequence forming a hairpin loop, the origin for a rolling-circle type replication, which functions in replication initiation. Recently, we discovered that an alternative replication route, called recombination-dependent replication, is based on recombination and is widespread among begomoviruses (21, 33). Transcription of all geminiviruses is bidirectional, driven by promoters in a long intergenic region, whereas begomoviruses possess additional promoters in both DNA components regulating complementary sense genes (11, 16) (see Fig. 4).
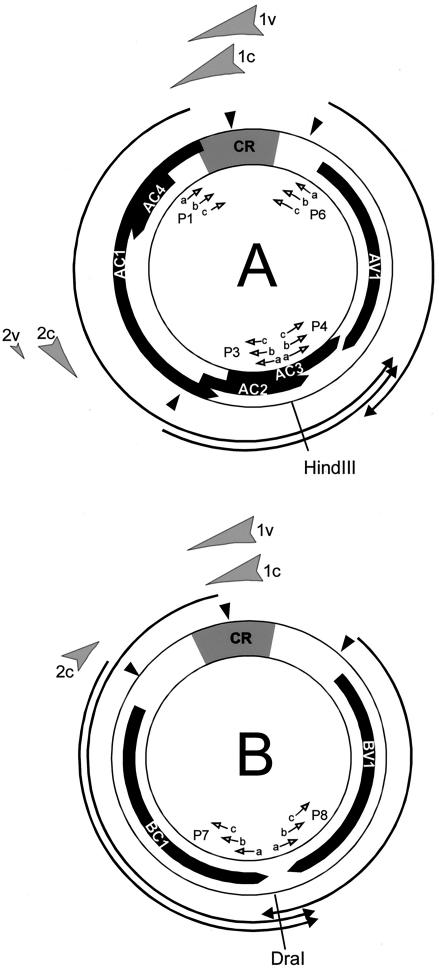
FIG. 4.
Summary of DNase I-hypersensitive sites mapped on DNA A and DNA B. Shaded arrow, DNase I-hypersensitive sites of the c- or v-strand. Solid arrow, open reading frames. Arrows, transcripts (according to Frischmuth et al. [11]). CR, common region. Solid arrowhead, TATA boxes. Open arrows, primers used for LMPCR and hybridization (see Fig. 1).
The genome organization of DNA A of begomoviruses resembles that of papovaviruses (10). Both concentrate regulatory elements for replication and transcription in their intergenic regions, where specific host factors can bind. During multiplication, papovavirus DNA is covered with nucleosomes except for the intergenic region, leaving it open to interact with host transcription and replication factors (6, 15, 53). This chromatin structure modulates gene regulation as it has been subsequently shown for eukaryotic chromatin in general (9, 24, 38, 47, 52).
With respect to the following results, we like to emphasize the role of chromatin structure for geminiviral gene regulation. The regulatory contribution of chromatin modifications has been underestimated in plant virology so far, because only for two families of plant viruses (Pararetroviridae and Geminiviridae) have minichromosomes been detected. AbMV minichromosomes most frequently contain 12 nucleosomes, as shown by electron microscopy (2). Only a minor population was packaged in 13 nucleosomes, which would be sufficient to cover the whole of the DNA circle, estimating that nucleosomal DNAs of about 200 bp each are equally distributed along the 2,600-bp circular DNA. In analogy to papovaviruses, we supposed that a space of about 200 bp should be free of nucleosomes (32). The best guess for a defined location of such a stretch of DNA would have been the intergenic region. However, hybridization analyses with intergenic region-specific probes on nucleosome ladders were not compatible with this simple assumption (32).
The following results resolve these conflicts, demonstrating that at least two alternative viral chromatin structures are present in AbMV-infected nuclei. In order to analyze nucleosome-free gaps, limited nuclease digestion of minichromosomes was combined with ligation-mediated PCR (LMPCR). This sensitive technique was necessary, because AbMV is phloem-limited within its host plants (1, 18, 50), whereby only one nucleus among 200 to 2,000 is infected. Furthermore, gene expression and replication of AbMV might rely on host factors expressed specifically in the phloem. Consequently, we decided to analyze AbMV minichromosomes as close as possible to their in situ environment, e.g., in isolated phloem nuclei.
MATERIALS AND METHODS
Standard techniques for manipulation and analysis of DNA were carried out as described previously (36).
Plants and viruses.
Naturally occurring AbMV-infected Abutilon plants, as they are propagated for ornamental purposes, have been described (49). Clones of DNA A and DNA B were those sequenced previously (13).
Agroinoculation of Nicotiana benthamiana.
Agrobacterium clones containing AbMV DNA A and DNA B (pDE201 and pST201) were introduced into stems of seedlings (for systemic infection) (7) or into leaf disks (for local infection) (21).
Isolation of nuclei.
The method of Watson and Thompson (48) was modified for the preparation of nuclei. The youngest leaves were collected at 8.00 a.m. in buffer 1 (1 M hexylene glycol; 10 mM MOPS; 10 mM MgCl2; 0.01% polyvinylpyrrolidone 40; 5 mM 2-mercaptoethanol; pH 7; 0°C) and homogenized in a Waring blender for 2 min. Debris was removed by filtration through 135-μm nylon gauze. Brij 35 was added to the filtrate under stirring to a final concentration of 0.45%. After three further filtrations (60-, 40-, and 20-μm nylon gauze), nuclei were pelleted (600 rpm for 30 min at 0°C; Heraeus Variofuge) and the sediment was resuspended in 8 ml of buffer 2 (0.5 M hexylene glycol; 10 mM MOPS; 10 mM MgCl2; 0.3% Brij 35; 5 mM 2-mercaptoethanol, 0°C; pH 7) per g (fresh weight) of initial plant material.
Nuclei from 2.5 g of leaf material were loaded onto a step gradient (6 ml of 90%; 8 ml of 60% and 5 ml of 30% Percoll in buffer 2) and centrifuged (1,100 rpm for 30 min at 0°C; Heraeus Variofuge). Nuclei accumulating in 60% Percoll were washed three times with buffer 2 (600 rpm; 0°C; 30 min, Heraeus Variofuge) and resuspended in incubation buffer (50 mM Tris-HCl, pH 8; 0.3 M sucrose; 5 mM MgCl2; 1.5 mM NaCl; 0.1 mM CaCl2; 5 μM 2-mercaptoethanol; 0°C).
DNase I digestion for LMPCR.
We incubated 107 nuclei in a volume of 500 μl with 10 units of DNase I at 37°C for different times as indicated in the figure legends. The reaction was stopped by adding 100 mM EDTA (pH 7,0) and cooling on ice. Proteins were digested with proteinase K (300 μg/ml) in the presence of 1% sopdium dodecyl sulfate for 1 h at 56°C. DNA was sheared by passing the solution through yellow Eppendorf tips 100 times. Following phenol extraction and ethanol precipitation, viral open circular together with a minor amount of linear DNAs were purified from agarose gels. Due to their low amounts in plants, viral open circular and linear DNAs were not visible in ethidium bromide-stained gels. They therefore had to be excised blindly with reference to known marker bands in parallel lanes. With this approach, it was impossible to separate open circular DNA completely from traces of linear DNA, but the resulting fractions were free of supercoiled dsDNA and ssDNA as shown by control hybridizations. DNAs were extracted from the excised gel pieces by electroelution or by centrifugation through blotting paper (51), phenol purified, and ethanol precipitated.
Serving as controls, protein-free nuclear DNAs (proteinase K digested, phenol extracted, and ethanol precipitated as described above) from parallel samples were treated the same way.
DNase I-treated samples were further analyzed by LMPCR (see below).
Micrococcal nuclease digestion for nucleosomal ladders.
Micrococcal nuclease treatments of nuclei were done (30) with modifications. Nuclei (2 × 107 in a volume of 500 μl) were incubated with 1 unit of micrococcal nuclease at 37°C for different times (0.5, 1, 1.5, 2, 3, and 4 min) and deproteinized as described above. Samples were either directly separated in 1% agarose gels or treated with HindIII (for DNA A) or DraI (for DNA B), which cut the respective DNAs at a single position opposite the common region (see Fig. 4). The DNA was blotted to membranes and hybridized with digoxigenin-labeled primers adjacent to the restriction site (see Fig. 4).
LMPCR.
In order to detect nuclease-hypersensitive sites, ligation-mediated PCR (LMPCR) (27) with modifications (14, 19, 35) was applied. In short (Fig. 1), dsDNA is nicked at random, preferentially at one single site per molecule, then melted and annealed with a first sequence-specific primer (Pa) to be elongated by a polymerase as far as the nick site is reached. The resulting blunt end is ligated to a synthetic asymmetric linker prepared by annealing two complementary but unequally long primers (PLong and PShort). A subsequent PCR with a second, nested sequence-specific primer (Pb) amplifies the informative fragment which is detected by blot hybridization after gel electrophoresis with a third, nested sequence-specific primer (Pc) which is labeled. Subtracting the nucleotide length of the synthetic primer (PL), the genomic position of the nick can be calculated from the length of the LMPCR fragment with reference to the 5′ end of primer Pb.
FIG. 1.
Schematic representation of the LMPCR technique. Circles at the ends of lines indicate 5′, arrows indicate 3′ ends, P1 to P6 indicate nested primer sets used for LMPCR and detection, with PL and PS primers generating the asymmetric linker (27). The locations of primers are shown in the genomic map of AbMV in Fig. 4. Numbers refer to the genomic positions of the primers (13).
The templates used in this study were viral open circular and linear DNAs (900 amol). Primers are listed in Fig. 1, and their genomic positions are summarized in Fig. 4. Four nested primer sets (P3, P4, P7, and P8) were used to map larger genomic distances, two (P1 and P6) for high-resolution mapping of the common region in DNA A.
High-pressure liquid chromatography-purified primers were obtained from MWG-Biotech (Ebersberg, Germany). To generate the asymmetric linker, 20 μM each of primers PL and PS were heated for 5 min at 95°C in 250 mM Tris-HCl (pH 7.8) and transferred into a water bath of 70°C, which was gradually cooled down to 4°C over 3 h. Aliquots were stored frozen at −20°C.
Primer annealing and elongation.
We added 5 μl of DNA solutions (860 amol of viral DNA in H2O) to 25 μl of 1.2× reaction mix (48 mM NaCl; 12 mM Tris-HCl, pH 8,9; 0.24 mM each dATP, dCTP, dGTP, and dTTP; MgSO4 in concentrations of 5 mM for P1a, 3.3 mM for P6a, and 2 mM for P3a, P4a, P7a, and P8a; 0.3 pmol of virus-specific primer PXa; and 1 unit of Vent DNA polymerase [New England Biolabs]). The mix was covered with 50 μl of mineral oil. DNA was denatured for 5 min at 95°C, and the primer was subsequently annealed for 30 min at 60°C. The elongation reaction was completed for 10 min at 75°C. Samples were then stored on ice.
Ligation of asymmetric linkers.
For ligation, 30 μl of DNA from the elongation reaction was mixed with 20 μl dilution buffer (110 mM Tris-HCl, pH 7.5; 18 mM MgCl2; 50 mM dithiothreitol) and with 25 μl of ligation solution (10 mM MgCl2; 20 mM dithiothreitol; 3 mM ATP; 100 pmol of asymmetric linker; 4.5 units of T4 DNA ligase [BRL]). The mix was incubated for a minimum of 5 h at 16°C. Yeast tRNA (1 μl; 10 mg/ml), 8.5 μl of 3 M sodium acetate (pH 7.0), and 220 μl of ethanol were added to precipitate DNA for 2 h at −20°C. The DNA was pelleted in an Eppendorf centrifuge, washed with 70% ethanol, dried at room temperature, and then dissolved in 20 μl of H2O.
PCR.
The resulting DNA was diluted to 100 μl of PCR mix (final concentrations: 10 mM KCl; 10 mM (NH4)2SO4; 20 mM Tris-HCl, pH 8.8; 0.1% Triton X-100; 0.24 mM each dATP, dCTP, dGTP, and dTTP; MgSO4 at 4 mM for P6b, 2 mM for P1b, P3b, P4B, P7b, and P8b; 100 pmol of one of the virus-specific primers [Fig. 1, PXb]; 100 pmol of the linker primer [Fig. 1, PL], and 2 units of Vent DNA polymerase). The resulting solution was covered with 100 μl of mineral oil and processed for PCR in a Hybaid thermocycler (5 min at 95°C, 2 min at 65°C, 3 min at 75°C; 19 cycles of 1 min at 95°C, 2 min at 65°C and an extension at 75°C, starting with 3 min for the first cycle, prolonging each consecutive cycle for 5 s, a last cycle under the same conditions but elongation for 10 min). PCR products were extracted with phenol-chloroform and with chloroform, precipitated with ethanol, and resuspended in 5 to 20 μl of H2O. Two thirds of this solution was applied to a single slot of a gel.
Gels and blotting.
For long-range mapping, LMPCR products were separated on a nondenaturing 1.8% agarose gel in Tris-acetate-EDTA (36) and blotted onto nylon membranes (40). The gels were calibrated with appropriate restriction fragments hybridizing with the probe used to detect the LMPCR fragments.
For high-resolution mapping, samples were denatured and run on a denaturing 4% polyacrylamide gel in Tris-borate-EDTA (36) in a direct-blotting device (GATC1500-System; MWG) (3). For calibration, sequencing reactions of the interesting genome regions were run in parallel according to the manufacturer's recommendations.
Hybridization.
Primers were 3′ end labeled with digoxigenin in a terminal transferase reaction adding digoxigenin-dATP and detected according to the manufacturer's instruction (digoxigenin oligonucleotide tailing kit; digoxigenin-detection kit, Boehringer, Mannheim, Germany). Hybridization in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 1% blocking reagent, 0.1% N-lauroylsarcosine, and 0.02% sodium dodecyl sulfate was carried out at 5 to 10°C below the melting temperature of the labeled primers. The last washing step was in 0.1× SSC-0.1% sodium dodecyl sulfate at the same temperature.
Sequencing.
Sequencing according to Maxam and Gilbert (26) was done by use of an oligonucleotide sequencing kit (Merck, Darmstadt, Germany) and processed in parallel for LMPCR, serving as calibration standards. Before ligation, products of the second-strand synthesis were phosphorylated (31).
DNA purification.
Total viral DNA from infected plants was purified as described previously (21). Some samples were further enriched for viral dsDNA by benoylated naphtoylated DEAE-cellulose chromatography (33).
Gel electrophoresis.
Two-dimensional gel electrophoresis with the addition of sodium dodecyl sulfate in the first dimension but chloroquine in the second dimension was performed as described previously (21). A detailed overview of the variety of bands and their identification has been given (33). One-dimensional gel electrophoresis, as documented in the present publication, was performed under similar conditions as used for the second dimension of the two-dimensional gel, applying the samples directly into preformed slots.
Evaluation of fragment sizes and band intensities.
X-ray films, after exposure to chemiluminescent signals, were scanned and digitized with an AGFA transillumination scanner. Bitmap files were analyzed with the program ScanPro (Jandel, Erkrath, Germany) to obtain relative pixel intensities of the lanes in one-dimensional gels and to measure the position of bands (arbitrary pixel length units). Graphs (pixel intensities versus pixel numbers) were processed with Microsoft Excel.
To rescale the x axis for better evaluation, the sizes of DNA fragments were determined either by the program Scanpack (Biometra, Göttingen, Germany) or with the algorithm of Schaffer and Sederoff in the best-fit option of the program SigmaPlot (Jandel, Erkrath, Germany) by comparison with appropriate standard fragments. From the resulting relation between migration distance and molecular weight, each pixel in a gel could be assigned to a certain fragment length or genomic position, in case a reference point was available. Calculations were performed with the transform option of SigmaPlot with reference to appropriate restriction or primer start sites.
RESULTS
Localization of DNase I-hypersensitive sites in AbMV minichromosomes.
Usually, AbMV has been detected in the tissues of infected plants in only one of 2,000 nuclei (18) and at maximum in one of a hundred isolated nuclei (unpublished observation). Therefore, a highly sensitive technique was needed to detect the minor amounts of minichromosomes among total viral DNA in order to analyze their chromatin structure. LMPCR proved to be suitable to reach the necessary level of detection, but its advantage also implies its limitation. Whenever a nick is produced in the plant or during purification, it will be seen after LMPCR. As a consequence, host or viral nucleases do the job before exogenous nucleases can be applied to detect hypersensitive sites. Therefore, the zero control lanes, which used to be empty with comparable techniques, show the first signals in protein-containing as well as in deproteinzized samples (Fig. 2).
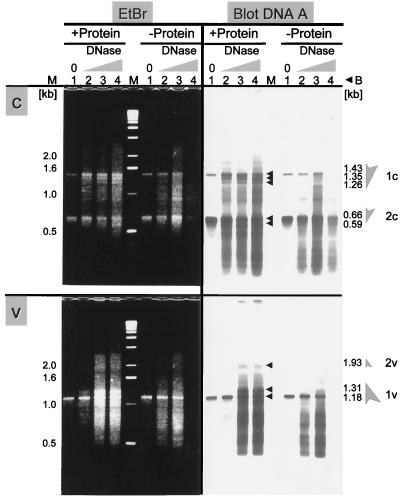
FIG. 2.
Nuclease-hypersensitive sites of c-strand and v-strand for AbMV DNA A. Nuclei (+Protein) and protein-free DNA (−Protein) as a control were incubated with DNase I for different times. +Protein: lane 1, without DNase I, 60 sec; lanes 2 to 4, with DNase I, 20, 40, or 60 sec. −Protein: lane 1, without DNase I, 45 sec; lanes 6 to 8, with DNase I, 15, 30, or 45 sec. (c) The c-strand was analyzed with reference to nucleotide 1336 (LMPCR with primer P3a, b). (v) The v-strand with reference to nucleotide 1297 (LMPCR with primer P4a, b). Left side: Ethidium bromide (EtBr)-stained 1.8% agarose gel. Right side: Blotted DNA after hybridization with primer 3c or 4c. Lane M, size markers (Gibco BRL). Column B indicates the calculated fragment sizes corresponding to the bands marked with arrowheads. 1c and -v and 2c and -v and shaded arrows, hypersensitive sites for comparison with Fig. 4.
Despite intensive efforts with a variety of modifications with CsCl gradient purification (21) or excision of the DNA from gels, we consistently failed to isolate nick-free supercoiled DNA fractions to serve as more appropriate controls. In all our attempts, the resulting samples still contained nicks which produced uninformative smears upon LMPCR (data not shown). In the end we had to accept that host or viral nucleases had nicked the DNA either prior to or during extraction, resulting in an elevated starting level of band intensities. As an indication of hypersensitive sites within the viral chromatin, we judged those bands valid which increased in intensity during digestion with exogenous nucleases in comparison to the deproteinized control samples and of course those bands which appeared completely new (marked with arrowheads in Fig. 2).
For generating the blots shown in Fig. 2, nuclei and, for a control, deproteinized DNAs from an equal aliquot of the same nuclei were incubated with DNase I for different periods of time. Viral DNAs were separated into open circular, linear, supercoiled, and ssDNA forms in an agarose gel. Nicking by DNase I resulted in the conversion of supercoiled DNA into open circular DNA, with prolonged incubation into linear DNA (data not shown). Open circular and linear DNAs were excised together from the gel, verified by hybridizing an aliquot, and used as templates for LMPCR. The two AbMV genome components were analyzed in viral (v-strand) and complementary direction (c-strand). The c-strand of DNA A was analyzed with reference to nucleotide 1336 (5′ end of primer 3b). Two hypersensitive sites were represented in bands from 1.26 to 1.43 kb and from 0.59 to 0.66 kb (Fig. 2c; 1c, 2c) after ethidium bromide staining as well as after hybridization. Corresponding bands were found for the opposite direction (v-strand) with reference to nucleotide 1297 (5′ end of primer 4b). Bands appeared with 1.18 to 1.31 kb and 1.93 kb (Fig. 2v; 1v, 2v). A similar analysis of the c-strand of DNA B also revealed two hypersensitive sites at a distance of 1.23 to 1.32 kb and 0.83 to 0.91 kb with reference to nucleotide 1449, i.e., the 5′ end of primer 7b (Fig. 3c; 1c, 2c). For the v-strand of DNA B, only one hypersensitive site was recognized, 1.08 to 1.2 kb in distance from nucleotide 1205 (Fig. 3; 1v).
FIG. 3.
Nuclease-hypersensitive sites determined for the c-strand and v-strand of AbMV DNA B. The lane pattern is the same as described for Fig. 2. Primers used for LMPCR were P7a, b for the c-strand and P8a, b for the v-strand. Hybridization primers were P7c and P8c, respectively. Most of the DNA in lane 3 in c of (−Protein) was lost during this experiment.
The fragment sizes, each minus 26 bases of the artificial linker (PL) were used to map the genomic positions of the hypersensitive sites as shown in Fig. 4. The hypersensitive sites were mapped to identical genome positions (within the limits of estimation for band sizes), irrespective of whether c-strand or v-strand analysis was performed: nucleotides 2600 to 110 and nucleotides 1900 to 1970 for DNA A, nucleotides 70 to 160 and nucleotides 2250 to 2330 for DNA B. The hypersensitive sequences cover either part of the common region which includes two promoter regions or are located upstream of the transcript start sites of the second complementary transcripts which encode AC2/AC3 and BC1, respectively (Fig. 4; for mapping of the transcription start sites, see Frischmuth et al. [11]). The localization of hypersensitive sites completely fits expectations, because open chromatin structures, which are usually attacked more easily by nucleases, are commonly found associated with regions involved in the regulation of transcription. Unlike animal papovaviruses, however, which harbor only one nucleosome-free gap in their minichromosomes, we conclude that AbMV minichromosomes may have two such regions in agreement with their different transcription strategy.
Nuclease-sensitive sites within the common region.
To analyze the hypersensitive sites in closer detail, primers adjacent to the common region (Fig. 4, primer sets 1a, b, c and 6a, b, c) were used on the same template DNA.
As expected, the most prominent hypersensitive sites were found at the hairpin loop, irrespective of whether the v-strand or c-strand was mapped (Fig. 5). Remarkably, in both cases only the 3′ halves of the hairpin were predominantly attacked by the endogenous and exogenous nucleases (Fig. 6), suggesting that these sequences are especially exposed. In the viral strand, they include the sequence-specific nicking site for the replication-associated protein (Rep/AC1; Fig. 6, encircled A becomes covalently attached to Rep).
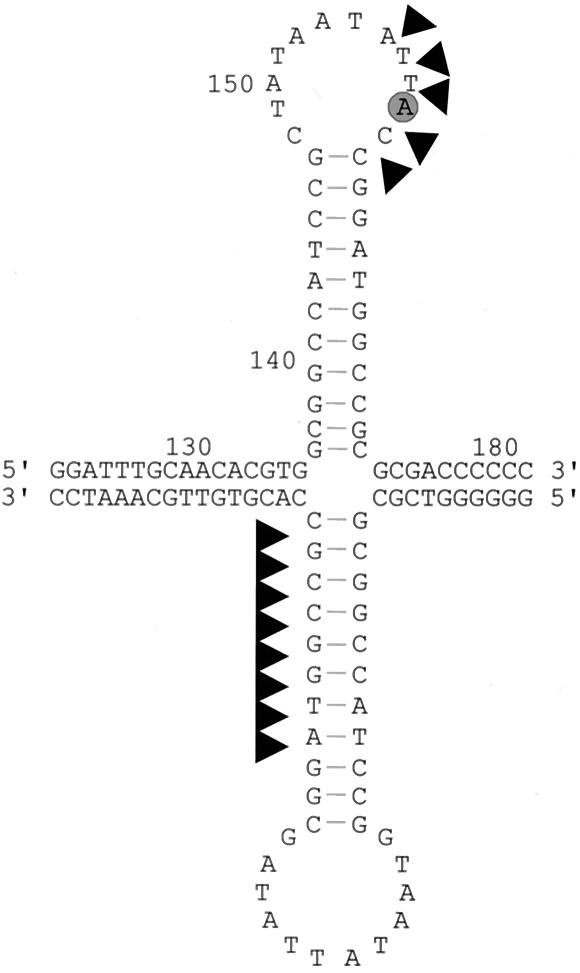
FIG. 5.
Fine mapping of nuclease-hypersensitive sites in the common region of DNA A by LMPCR with primers P1a, b and P6a, b for the c-strand and v-strand, respectively. Template DNA and lanes were the same as described for Fig. 2. LMPCR products were hybridized with primer P1c or P6c. The boxed sequences represent the c-strand or the v-strand of the stem-loop structure as determined by Maxam-Gilbert sequence reactions separated in the same gel (not shown). Arrowheads indicate hypersensitive sites already present before the action of exogenous endonucleases as mapped in Fig. 6, gray bars show regions which are relatively protected from digestion in the presence of protein. Stem-loop-forming sequences are indicated by arrows, and the 5′ A after cleavage by Rep is circled (compare Fig. 6).
FIG. 6.
Map of cleavage sites (arrowheads) found on the sequence of the stem-loop structure (see Fig. 5). The encircled A is the starting point of rolling-circle replication, as reported previously (25, 41).
Nucleosomal phasing of the AbMV minichromosome.
Two hypersensitive sites may indicate at least two different viral chromatin structures. In order to test this hypothesis and to analyze whether nucleosomes are located at fixed positions in the minichromosomes, the arrangement of nucleosomes was analyzed further with a second approach which is less sensitive to nicking activity during purification than LMPCR. Nucleosomal ladders were generated by treating nuclei with micrococcal nuclease, analyzing the products on nondenaturing agarose gels, and, after blotting, by hybridization with labeled primers (Fig. 4 and 7). Aliquots of the samples were additionally cut with restriction enzymes recognizing single sites in DNA A or DNA B (HindIII for DNA A and DraI for DNA B) prior to gel electrophoresis and detection.
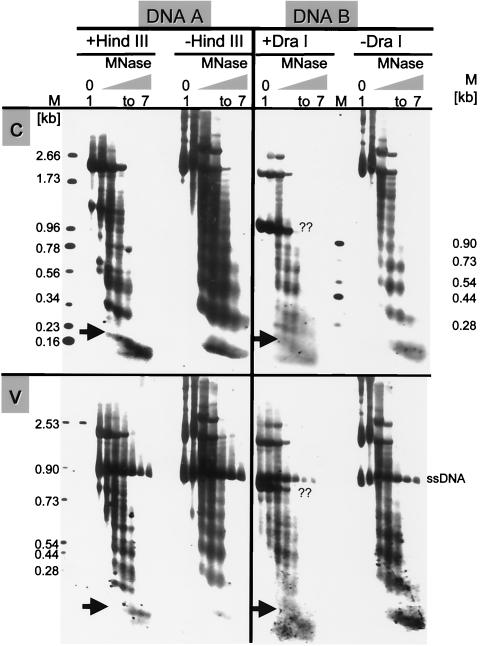
FIG. 7.
Micrococcus nuclease (MNase) digestions revealing the arrangement of nucleosomes in DNA A and B. Nuclei were incubated for different times. Lanes 1, incubation without enzyme for 4 min. Lanes 2 to 7, incubation with enzyme for 0.5, 1, 1.5, 2, 3, or 4 min. One set of aliquots of purified DNAs of every sample was subsequently digested with HindIII (+HindIII) for DNA A or with DraI (+DraI) for DNA B (12 h at 37°C), and the other was left untreated (−HindIII; −DraI). The c-strands (c) and v-strands (v) were hybridized with primer P3a or P4a specific for DNA A or with primer P7c or P8c for DNA B, respectively. The positions of HindIII and DraI sites are indicated in Fig. 4. The sizes of marker bands (M) are shown. Arrows point at the conspicuous swallow-tailed split bands representing mononucleosomes. During the procedure, viral ssDNA was protected by the coat protein. A second prominent band marked with ?? in DNA B might result from subgenomic defective interfering DNA.
If minichromosomes had an arbitrary nucleosomal arrangement, a smear would be expected. If, however, nucleosomes were located at fixed positions, fragments corresponding to multiples of nucleosomal length should appear. Figure 7 shows the results, confirming earlier observations (32) that most of the viral dsDNA can be fragmented into nucleosomal patterns in the absence of restrictions. Under the conditions applied, no such pattern has been observed without addition of exogenous nuclease, indicating that chromatin fragmentation, as it could be expected for apoptotic processes, does not contribute to the AbMV minichromosome patterns considerably.
When viral open circular DNA had been cut additionally by a restriction enzyme, the most conspicuous change was the splitting of the mononucleosome-specific bands into a swallowtail appearance (Fig. 7, arrows), which was most prominent for the c-strand analysis in DNA A as well as DNA B. The rest of the nucleosome-derived ladders exhibited more complex patterns in comparison to unrestricted DNA. To elucidate the regularities underlying these patterns, we scanned the lanes and plotted the respective pixel intensities to a rescaled x axis in order to identify the nucleosome locations in reference to fragment lengths or genomic positions (see Material and Methods).
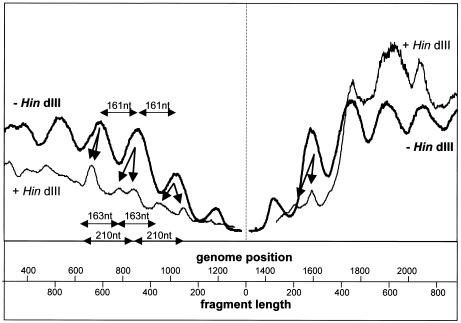
Figure 8 shows that equally spaced nucleosomes were found in the absence of restriction. With HindIII restriction, additional periodicities can be inferred from this plot, at least close to the HindIII site, where resolution is best. As already expected from the split mononucleosome-specific bands, two overlapping patterns, differing not only in phase but also in period (Fig. 8, arrows), were detected for the viral as well as for the complementary strand. With longer DNA digestion products, the patterns merge to the nucleosomal periodicity for the c-strand (genome positions 1700 to 2200 in Fig. 8) due to loss of resolution of the gel.
FIG. 8.
Example of densitometric analysis of micrococcal nuclease digestion experiments, here for lanes 3 for DNA A in Fig. 7 (left part for viral, right part for complementary orientation). The best-fit equation for the relation between the molecular weight and the electrophoretic migration was derived from the marker fragments of Fig. 7, allowing rescaling of the x axis by transformation of migration distances to fragment lengths for untreated and to genome positions for restricted samples with reference to the HindIII site (nucleotide 1317). Harmonic oscillations appeared for untreated (−HindIII) nucleosomal bands with estimated periods of 161 nucleotides This wave was split into at least two oscillations upon restriction (+HindIII; arrows), which are best resolved in the vicinity of the restriction site and merge for more distant genome positions, with estimated periods of 163 and 210 nucleotides.
The second set of experiments with dsDNA products of micrococcal nuclease digestion suggests again that AbMV minichromosomes may exist in at least two conformations with nucleosomes occurring at different fixed positions, with differences in nucleosomal size and phase.
Topoisomer distribution.
Although the second approach is less sensitive to nicking activity, it cannot be completely ruled out that some changes of the isolated chromatin occur during purification. Therefore we employed a third set of experiments which were completely insensitive to nicking because nicked DNA is excluded from the analysis. Moreover, this technique minimizes handling, and special precautions were taken in order to suppress the action of nucleases as well as of topoisomerases during DNA isolation (21). Total DNA enriched for viral DNA was prepared from different leaf materials, separated on one-dimensional or two-dimensional gels, taking advantage of the intercalation of chloroquine to separate different topoisomers (explained in detail previously [33]) and hybridized against viral DNA probes (Fig. 9).
FIG. 9.
Topoisomer distributions of AbMV covalently closed circular DNA obtained by two-dimensional (a to c) or one-dimensional gel(d) electrophoresis with chloroquine as the intercalator. (a to e) Total DNA was enriched for viral molecules and hybridized with full-length DNA A probes as described previously (21), whereby sample c was further purified by benzoylated naphtoylated DEAE-cellulose to enrich for dsDNA (33). Samples from pooled leaves (a and c) and from individual leaves plant (b, d, and e) of a single plant with apex (A) and following leaf numbers (1 to 11) were compared. The following AbMV DNA A- and B-infected plant materials were used: (a) agroinoculated N. benthamiana leaf disks 5 days postinfection; (b, d, and e) naturally infected Abutilon plants, leaf number 11 in b and the indicated leaf numbers in d and e; (c) systemically infected leaves of agroinoculated N. benthamiana. Linear viral DNA (lin), open circular (oc), relaxed covalently closed circular (rccc), supercoiled covalently closed circular (ccc), and single-stranded (ss) DNA forms are indicated. The dotted lines numbered 11 and 13 and the black line numbered 12 in c and e indicate maximally resolved topoisomers (increasing topoisomer density from bottom to top and from right to left). Note the remarkably well preserved spacing of the topoisomers derived from different sources and runs on separate gels in independent experiments (a to c) and the predominant band representing viral DNA with 12 superhelical turns. Only the bands for topoisomers with five or six superhelical turns (?, grey lines in c) had to be interpolated with a polynomial approximation with a second-order equation, resulting in a better fit value for two hypothetical bands (R2 = 0.999) than would be the case for three additional bands (R2 = 0.996) in the gap of unidentifiable bands. Given the ruler for topoisomers obtained from this comparative analysis, it was now possible to determine the linking numbers in covalently closed circular DNA by counting back from topoisomers of higher values (a to c). The densities of every lane of gel d were scanned, and pixel intensities were plotted against pixel distance with Sigma ScanPro software (e), compensating for small differences in migration behavior by searching for the best match of the topoisomers.
In general, the most frequent linking number in a Gaussian distribution of topoisomers reflects the number of nucleosomes, such that one nucleosome causes one superhelical turn (5). Two-dimensional analysis of the supercoiled DNA is of special additional value, because relaxed covalently closed circular DNA is well separated and easily identified (Fig. 9c), providing a first landmark to map the other topoisomers. Moreover, we found two-dimensional analysis extremely reproducible in many experiments with different plants and isolation procedures as far as qualitative band patterns are considered (compare Fig. 9a, b, and c). Band spacing of the consecutive topoisomers is not linear in gels, but migration distances are best fit by a second-order polynomial. Therefore it is always possible to find a unique matching (“resonance”) pattern of bands between different lanes, even in different gels, when the same concentration of chloroquine is used. With various gels and DNA sources, only minimal rescaling is necessary for the second gel dimension to find the best fit (Fig. 9a to c). With this normalization strategy, the most frequent linking number of topoisomers in samples of pooled leaves was found to be 12, with additional high frequencies for 11 and 13. This is in good agreement with electron microscopic data on nucleosomal beads on DNA strings (2), where we found most frequently 12 and, to a smaller extent, 13 beads per minichromosome.
The two-dimensional technique is less convenient for quantitative evaluations of bands because it is not always easy to integrate all spots and tails of the bands accurately and, moreover, it is a laborious technique if several samples have to be compared. However, if at least one sample has been characterized qualitatively with the two-dimensional technique, in the case of Fig. 9b, this can serve as an internal standard for one-dimensional gels to assign the observed bands to certain linking numbers of topoisomers indicating nucleosome numbers (as in Fig. 9d, e). Preparing the viral DNA from single leaves of an individual Abutilon plant in such a way, the infection process was followed at different developmental stages of the host. Abutilon plants are especially suitable, as they do not suffer too much from infection and, in particular, leaf development is not impaired by the presence of the virus.
Figure 9d shows a representative example and exhibits a remarkable shift of the topoisomer patterns during leaf development. Older leaves have accumulated topoisomers of slower mobility. A detailed scanning analysis of the blot (Fig. 9e), which compensates for the differences in DNA migration behaviors in separate lanes, revealed three major peak intensities for most of the samples. These peaks have been identified in reference to sample 11, which had been assigned in two-dimensional gel electrophoresis (compared to Fig. 9b), to represent toposiomers with linking numbers of 11, 12, or 13 at maximum in most of the samples. With aging of leaves, the linking number 13 population increased, whereas in younger leaves lower linking numbers were prevalent. We interpret this phenomenon to be caused by a mixed effect of replication and transcription on viral nucleosome loading and minichromatin condensation.
It is conceivable that single-stranded circular AbMV DNA enters expanding leaves at early stages of development, where it is complemented to circular dsDNA. During this process or just thereafter, dsDNA is packed stepwise into minichromosomes (compare Fig. 9c). At the beginning of assembly, the positions of the nucleosomes might be still less fixed, leading to a scanning profile with less well-defined contours (as in Fig. 9e, A-4). With time, the population of completed minichromosomes should increase and serve as templates for transcription. In general, transcription factors need a less condensed region in the minichromosomes in order to gain access to the DNA. AbMV minichromosomes with 12 nucleosomes could fulfill this prerequisite. Since, however, AbMV uses two separate promoter regions in each DNA, minichromosomes might be opened at either one of the two sites in different molecules (resulting in 12 nucleosomes per minichromosome), or they might be opened simultaneously at both sites in one molecule (resulting in 11 nucleosomes per minichromosome). A clear differentiation between the two alternatives is currently not possible because the two populations overlap and each population produces a Gaussian distribution of topoisomers. With increasing age of leaves, viral transcription might cease completely, explaining the increase of linking number 13 topoisomers as an indication of condensed chromatin packaged into 13 nucleosomes.
DISCUSSION
Chromatin structure is a major determinant of gene regulation (8, 9, 38, 47). Here we present first evidence that this general statement may also hold true for a plant virus. Minichromosomal organization has been reported for one other plant virus, Cauliflower mosaic pararetrovirus (28, 29), but no information about the organization of these minichromosomes is available.
Geminiviruses resemble animal papovaviruses in many aspects (42). This study now adds another similar but also a divergent feature to the comparison. AbMV minichromosomes similarly possess a nucleosome-free space in the intergenic region, but a second minichromosomal region, not found in papovaviruses, is hypersensitive to nucleases, allowing the interaction of viral DNA with host factors at an additional site.
The most prominent DNase-hypersensitive site was assigned to the hairpin loop within the viral origin of replication, showing that this structure is particularly exposed. So far, we have not succeeded in extracting nick-free AbMV DNA from plants, even by very rigorous techniques (39). It therefore remains to be determined which nicks existed before the addition of exogenous nucleases in vivo or which had been produced in early steps of purification when endogenous nucleases had attacked the DNA. The hypersensitive sites detected without the addition of exogenous nucleases have been located in the same regions where the ends of viral heterogeneous high-molecular-weight linear DNA accumulated (21). These molecules have been interpreted as fragments of viral DNA which are generated by interrupted synthesis or by host nucleases and repaired by recombination-dependent replication.
We take this coincidence as an indication that a considerable number of the nicked molecules observed in the absence of exogenous nucleases are produced in vivo by host nucleases, e.g., of the repair machinery, which may continue to operate for some time during nucleus purification. Ultimately, it is less important to decide which nuclease has produced the nicks if we only wish to identify nuclease-hypersensitive sites. Since these sites fit well to the promoter regions and the complementary experimental strategies generated no contradiction to the proposed model of at least two chromatin structures but rather supported it, the results yield evidence to suggest that AbMV minichromosomes have the potential to open two gaps, thereby becoming accessible for transcription factors.
Meanwhile, it is well known (reviewed in reference 16) that the viral strand in the dsDNA intermediate is nicked precisely 5′ to a single defined site (Fig. 6, encircled A) by the viral nick-closing enzyme Rep (AC1, AL1). Geminiviruses replicate in the nucleus of infected cells via a rolling-circle mechanism (37, 43) but may also take advantage of a recombination-dependent route (21, 33). The proposed nicking and ligase activity of AC1 protein (22) was confirmed in vitro, and the introduced nick was localized in the viral strand within the nonanucleotide TAATATT↓AC of the hairpin loop (Fig. 6) for monopartite (17, 25) as well as for bipartite (41) geminiviruses.
Hypersensitivity to DNase I is similar in AbMV DNA A and DNA B. The transcription start sites and the TATA boxes of the complementary transcripts are located at identical positions in the intergenic region of both genome components (11). Additional complementary transcription start sites for open reading frames AC2/AC3 and BC1 have been mapped for AbMV and tomato golden mosaic virus (11, 45). The TATA boxes and initiation sites of these transcripts colocalize with the second hypersensitive sites of DNA A and of DNA B.
Different experimental approaches resulted in converging evidence that AbMV nucleosomes are arranged in phase. In plants, core nucleosomes are wrapped with 146 bp of DNA plus an extra 22-bp binding H1 histone, and holonucleosomes are connected by variable linkers from 0 to 90 bp (38), adding up to from 168 to 258 bp of DNA for the whole subunit, whereby the linker space may vary from species to species as well as in a cell type-specific manner. The estimated values for AbMV nucleosomes fall into this range. Interestingly, at least two and perhaps three different nucleosomal arrangements were found. These might reflect different temporal and/or spatial regulation of the genes. So far, no synchronized infection system for viruses in plants is available to test this hypothesis. It would be challenging to analyze chromosomal phasing under such conditions.
Nucleosome positioning can be directed by proteins or by bendable DNA (23). A bending locus has been demonstrated in the large intergenic region of Wheat dwarf virus, a monopartite mastrevirus (44). Such sequences, however, are absent from the corresponding region of begomoviruses, such as AbMV. For simian virus 40, the position of nucleosomes is directed by the interaction of its early DNA region with proteins, possibly the histone H1 nucleosome (20). In the case of AbMV, it has to be determined which part of the sequence is responsible for nucleosome positioning.
The reported data strongly suggest that the minichromosomes of AbMV are highly ordered entities, which may have important implications for several general fields of interest: for the construction of high-expression gene vectors on the basis of geminiviruses (46), for the optimization of defective interfering DNA serving as a means to induce resistance (12), and for explaining the size selection of viral genomic DNA observed in the absence of coat protein (for a review see reference 4).
Acknowledgments
We thank C. Wege and R. Ghosh for critical reading of the manuscript and W. Preiss for skillful technical assistance.
This research was supported by a grant from the Bundesminister für Forschung und Technologie (BCT 507) and the Deutsche Forschungsgemeinschaft (Je 116-3).
REFERENCES
- 1.Abouzid, A. M., A. Barth, and H. Jeske. 1988. Immunogold labeling of the Abutilon mosaic virus in ultrathin sections of epoxy resin embedded leaf tissue. J. Ultrastruct. Res. 99:39-47. [Google Scholar]
- 2.Abouzid, A. M., T. Frischmuth, and H. Jeske. 1988. A putative replicative form of the Abutilon mosaic virus (gemini group) in a chromatin-like structure. Mol. Gen. Genet. 212:252-258. [Google Scholar]
- 3.Beck, S., and F. M. Pohl. 1984. Sequencing with direct blotting electrophoresis. EMBO J. 3:2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisaro, D. M. 1994. Recombination in geminiviruses: mechanisms for maintaining genome size and generating genomic diversity, p. 39-60. In J. Paszkowski (ed.), Homologous recombination and gene silencing in plants. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 5.Clark, D. J. 1998. Counting nucleosome cores on circular DNA with topoisomerase I, p. 139-152. In H. Gould (ed.), Chromatin—a practical approach. Oxford University Press, Oxford, United Kingdom.
- 6.de Bernardin, W., T. Koller, and J. M. Sogo. 1986. Structure of in-vivo transcribing chromatin as studied in Simian virus 40 minichromosomes. J. Mol. Biol. 191:469-482. [DOI] [PubMed] [Google Scholar]
- 7.Evans, D., and H. Jeske. 1993. Complementation and recombination between mutants of complementary sense genes of DNA A of Abutilon mosaic virus. Virology 197:492-496. [DOI] [PubMed] [Google Scholar]
- 8.Felsenfeld, G. 1992. Chromatin as an essential part of the transcriptional mechanism. Nature 355:219-224. [DOI] [PubMed] [Google Scholar]
- 9.Fransz, P. F., and J. H. de Jong. 2002. Chromatin dynamics in plants. Curr. Opin. Plant Biol. 5:560-567. [DOI] [PubMed] [Google Scholar]
- 10.Fried, M., and C. Prives. 1986. The biology of Simian virus 40 and polyomavirus, p. 1-16. In M. Botchan, T. Grodzicker, and P. A. Sharp (ed.), DNA tumor viruses. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 11.Frischmuth, S., T. Frischmuth, and H. Jeske. 1991. Transcript mapping of Abutilon mosaic virus, a geminivirus. Virology 185:596-604. [DOI] [PubMed] [Google Scholar]
- 12.Frischmuth, T., and J. Stanley. 1993. Strategies for the control of geminivirus disease. Semin. Virol. 4:329-337. [Google Scholar]
- 13.Frischmuth, T., G. Zimmat, and H. Jeske. 1990. The nucleotide sequence of abutilon mosaic virus reveals prokaryotic as well as eukaryotic features. Virology 178:461-468. [DOI] [PubMed] [Google Scholar]
- 14.Garrity, P. A., and B. J. Wold. 1992. Effects of different DNA polymerases in ligation-mediated PCR: Enhanced genomic sequencing and in vivo footprinting. Proc. Natl. Acad. Sci. USA 89:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffith, J. D. 1975. Chromatin structure: deduced from a minichromosome. Science 187:1202-1203. [DOI] [PubMed] [Google Scholar]
- 16.Hanley-Bowdoin, L., S. B. Settlage, B. M. Orozco, S. Nagar, and D. Robertson. 1999. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18:71-106. [PubMed] [Google Scholar]
- 17.Heyraud, F., V. Matzeit, M. Kammann, S. Schaefer, J. Schell, and B. Gronenborn. 1993. Identification of the initiation sequence for viral-strand DNA synthesis of wheat dwarf virus. EMBO J. 12:4445-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horns, T., and H. Jeske. 1991. Localization of Abutilon mosaic virus DNA within leaf tissue by in-situ hybridization. Virology. 181:580-588. [DOI] [PubMed] [Google Scholar]
- 19.Hornstra, I. K., and T. P. Yang. 1993. In vivo footprinting and genomic sequencing by ligation-mediated PCR. Anal. Biochem. 213:179-193. [DOI] [PubMed] [Google Scholar]
- 20.Jeong, S. W., and A. Stein. 1994. DNA sequence affects nucleosome ordering on replicating plasmids in transfected COS-1 cells and in vitro. J. Biol. Chem. 269:2197-2205. [PubMed] [Google Scholar]
- 21.Jeske, H., M. Lütgemeier, and W. Preiss. 2001. Distinct DNA forms indicate rolling circle and recombination-dependent replication of Abutilon mosaic geminivirus. EMBO J. 20:6158-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koonin, E. V., and T. V. Ilyina. 1992. Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA replication initiator proteins. J. Gen. Virol. 73:2763-2766. [DOI] [PubMed] [Google Scholar]
- 23.Kornberg, A., and T. A. Baker. 1992. DNA replication. W. H. Freeman and Company, New York, N.Y.
- 24.Kornberg, R. D., and Y. Lorch. 1992. Chromatin structure and transcription. Annu. Rev. Cell Biol. 8:563-587. [DOI] [PubMed] [Google Scholar]
- 25.Laufs, J., W. Traut, F. Heyraud, V. Matzeit, S. G. Rogers, J. Schell, and B. Gronenborn. 1995. In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc. Natl. Acad. Sci. USA 92:3879-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller, P. R., and B. Wold. 1989. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246:780-786. [DOI] [PubMed] [Google Scholar]
- 28.Olszewski, N., G. Hagen, and T. J. Guilfoyle. 1982. A transcriptionally active, covalently closed minichromosome of cauliflower mosaic virus DNA isolated from infected turnip leaves. Cell 29:395-402. [DOI] [PubMed] [Google Scholar]
- 29.Olszewski, N. E., and T. J. Guilfoyle. 1983. Nuclei purified from cauliflower mosaic virus-infected turnip leaves contain subgenomic, covalently closed circular cauliflower mosaic virus DNAs. Nucleic Acids Res. 11:8900-8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul, A. L., and R. J. Ferl. 1988. Assays for studying chromatin structure, p. 1-11. In S. B. Gelvin and R. A. Schilperoort (ed.), Plant molecular biology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 31.Pfeifer, G. P., and A. D. Riggs. 1991. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells with DNase I and ligation-mediated PCR. Genes Dev. 5:1102-1113. [DOI] [PubMed] [Google Scholar]
- 32.Pilartz, M., and H. Jeske. 1992. Abutilon mosaic geminivirus double-stranded DNA is packed into minichromosomes. Virology 189:800-802. [DOI] [PubMed] [Google Scholar]
- 33.Preiss, W., and H. Jeske. 2003. Multitasking in replication is common among geminiviruses. J. Virol. 77:2972-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rybicki, E. P., R. W. Briddon, J. K. Brown, C. M. Fauquet, D. P. Maxwell, B. D. Harrison, P. G. Markham, D. M. Bisaro, D. Robinson, and J. Stanley. 2000. Family Geminiviridae, p. 285-297. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, et al., Virus taxonomy: classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 35.Saluz, H. P., K. Wiebauer, and A. Wallace. 1991. Studying DNA modifications and DNA-protein interactions in vivo. Trends Genet. 7:207-211. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 37.Saunders, K., A. Lucy, and J. Stanley. 1991. DNA forms of the geminivirus African cassava mosaic virus consistent with a rolling circle mechanism of replication. Nucleic Acids Res. 19:2325-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, J. G., R. S. Hill, and J. P. Baldwin. 1995. Plant chromatin structure and post-translational modifications. Crit. Rev. Plant Sci. 14:299-328. [Google Scholar]
- 39.Song, J. Y., and H. Jeske. 1994. The level of Abutilon mosaic geminivirus in leaf discs and wound callus. J. Phytopathol. 140:45-52. [Google Scholar]
- 40.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 41.Stanley, J. 1995. Analysis of African cassava mosaic virus recombinants suggests strand nicking occurs within the conserved nonanucleotide motif during the initiation of rolling circle DNA replication. Virology 206:707-712. [DOI] [PubMed] [Google Scholar]
- 42.Stanley, J., and J. W. Davies. 1985. Structure and function of the DNA genome of geminiviruses, p. 191-218. In J. Davies (ed.), Molecular plant virology. CRC Press, Boca Raton, Fla.
- 43.Stenger, D. C., G. N. Revington, M. C. Stevenson, and D. M. Bisaro. 1991. Replication release of geminivirus genomes from tandemly repeated copies: evidence for a rolling circle replication of a plant viral DNA. Proc. Natl. Acad. Sci. USA 88:8029-8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suarez-Lopez, P., E. Martinez-Sals, P. Hernandez, and C. Gutierrez. 1995. Bent DNA in the large intergenic region of wheat dwarf geminivirus. Virology 208:303-311. [DOI] [PubMed] [Google Scholar]
- 45.Sunter, G., and D. M. Bisaro. 1989. Transcription map of the B genome component of tomato golden mosaic virus and comparison with A component transcripts. Virology 173:647-655. [DOI] [PubMed] [Google Scholar]
- 46.Timmermans, M. C. P., O. P. Das, and J. Messing. 1994. Geminiviruses and their uses as extrachromosomal replicons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45:79-112. [Google Scholar]
- 47.Verbsky, M. L., and E. J. Richards. 2001. Chromatin remodeling in plants. Curr. Opin. Plant Biol. 4:494-500. [DOI] [PubMed] [Google Scholar]
- 48.Watson, J. C., and W. F. Thompson. 1986. Purification and restriction endonuclease analysis of plant nuclear DNA. Methods Enzymol. 118:57-75. [Google Scholar]
- 49.Wege, C., R. D. Gotthardt, T. Frischmuth, and H. Jeske. 2000. Fulfilling Koch's postulates for Abutilon mosaic virus. Arch. Virol. 145:2217-2225. [DOI] [PubMed] [Google Scholar]
- 50.Wege, C., K. Saunders, J. Stanley, and H. Jeske. 2001. Comparative analysis of tissue tropism of bipartite geminiviruses. J. Phytopathol. 149:359-368. [Google Scholar]
- 51.Weichenhan, D. 1990. Fast recovery of DNA from agarose gels by centrifugation through blotting paper. Trends Genet. 6:172. [DOI] [PubMed] [Google Scholar]
- 52.Wolffe, A. P. 1994. Nucleosome positioning and modification: chromatin structures that potentiate transcription. Trends Biochem. Sci. 19:240-244. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, L., and J. D. Gralla. 1990. In situ nucleoprotein structure involving origin-proximal SV40 DNA control elements. Nucleic Acids Res. 18:1797-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]