Abstract
Infection of cells with adeno-associated virus (AAV) type 2 (AAV-2) is mediated by binding to heparan sulfate proteoglycan and can be competed by heparin. Mutational analysis of AAV-2 capsid proteins showed that a group of basic amino acids (arginines 484, 487, 585, and 588 and lysine 532) contribute to heparin and HeLa cell binding. These amino acids are positioned in three clusters at the threefold spike region of the AAV-2 capsid. According to the recently resolved atomic structure for AAV-2, arginines 484 and 487 and lysine 532 on one site and arginines 585 and 588 on the other site belong to different capsid protein subunits. These data suggest that the formation of the heparin-binding motifs depends on the correct assembly of VP trimers or even of capsids. In contrast, arginine 475, which also strongly reduces heparin binding as well as viral infectivity upon mutation to alanine, is located inside the capsid structure at the border of adjacent VP subunits and most likely influences heparin binding indirectly by disturbing correct subunit assembly. Computer simulation of heparin docking to the AAV-2 capsid suggests that heparin associates with the three basic clusters along a channel-like cavity flanked by the basic amino acids. With few exceptions, mutant infectivities correlated with their heparin- and cell-binding properties. The tissue distribution in mice of recombinant AAV-2 mutated in R484 and R585 indicated markedly reduced infection of the liver, compared to infection with wild-type recombinant AAV, but continued infection of the heart. These results suggest that although heparin binding influences the infectivity of AAV-2, it seems not to be necessary.
Attachment of a virus to a host cell requires a specific interaction of the virus shell with cellular receptor molecules. These permit uptake and thus intracellular processing of the virus so that it can successfully infect the cell. For adeno-associated virus (AAV) type 2 (AAV-2), a member of the parvovirus family containing a nonenveloped icosahedral capsid, heparan sulfate proteoglycan has been shown to act as a primary receptor (62). However, the contribution of an additional receptor(s) has been postulated because AAV-2 binding to cells and recombinant AAV-2 transduction did not quantitatively correlate with the amount of heparan sulfate on the surface of different cell types tested (26, 51). After binding to the cell surface, AAV-2 is thought to engage a secondary receptor which mediates cell entry. To date, αVβ5 integrin and human fibroblast growth factor receptor 1 have been proposed (49, 61), but the involvement of other molecules has also been suggested (50, 52).
Heparan sulfate glucosaminoglycans (HSGAGs) are complex polysaccharides with high structural diversity. They consist of repeating disaccharide units each composed of glucuronic acid or iduronic acid linked to glucosamine (17, 48). The immense structural diversity of HSGAGs arises from the modification of individual disaccharide units within the oligosaccharide. This diversity enables these molecules to interact with a wide variety of proteins, such as growth factors, chemokines, morphogens, enzymes, matrix proteins, lipoproteins, and antimicrobial peptides, which are involved in diverse biological processes, such as morphogenesis, tissue repair, energy balance, host defense, cell adhesion, proliferation, and growth factor signaling (3, 40, 48). Numerous viruses, e.g., herpes simplex virus type 1 (HSV-1) and HSV-2 (70), human immunodeficiency virus (46), respiratory syncytial virus (38), dengue virus (9), pseudorabies virus (64), foot-and-mouth disease virus (33), vaccinia virus (11), Sindbis virus (4, 36), several papillomaviuses (34, 58), cytomegalovirus (12), AAV-2 and AAV-3 (26, 62), and others, bind to HSGAGs. HSGAG chains are assembled while attached to a proteoglycan core protein. So far, three major protein families have been characterized: the membrane-spanning syndecans, the glycosylphosphatidylinositol-linked glypicans, and the basement membrane proteoglycans perlecan and agrin (17). The sequence of the HSGAGs does not correlate with the protein family to which they are bound but rather correlates with the cell type in which the HSGAGs have been synthesized.
HSGAGs interact with proteins mainly through electrostatic interactions of basic amino acids with the negatively charged sulfate and carboxyl groups of HSGAG chains (5, 30). However, hydrogen bond formation (18, 31) and, to a lesser extent, hydrophobic interactions can also contribute to such interactions (1). Heparin-binding domains of heparin-binding proteins have been shown to contain consensus sequence motifs, such as XBBXB and XBBBXXBX, where B is a basic amino acid exposed on one side and X is a neutral or hydrophobic amino acid directed toward the protein interior. Through an analysis of the available experimentally determined heparin-protein complexes, it became obvious that the spatial orientation of basic residues rather than sequence proximity is an important factor in determining heparin-binding affinity (32). Such binding domains are usually located at the protein surface and form a flat pocket with a positive charge (30).
The capsid of AAV-2 is composed of three overlapping capsid proteins (VP1, VP2, and VP3) containing a unique VP1 N terminus, a portion common to VP1 and VP2, and a portion common to VP1, VP2, and VP3 (2, 6, 60, 63). To date, the VP domains involved in binding to heparin have not been studied in detail. Several short basic sequences located in the N-terminal portion of VP1 and VP2 have been proposed as possible candidates for heparin binding (62). However, capsids with mutations in these amino acids (69; unpublished data) and capsids composed only of VP3 molecules (54) showed binding to heparin, suggesting that the heparin-binding domain is located in the portion common to all three VP molecules. Several groups observed that insertions in capsid proteins or clustered charge-to-alanine mutations affected heparin binding (22, 45, 59, 69).
We set out to define precisely, through point mutational analysis, the amino acids of AAV-2 capsid proteins involved in heparin binding. Using information from the recently resolved atomic structure for the AAV-2 capsid (72), we can now describe a heparin-binding motif which is composed of basic amino acids and which is formed by folding into the threefold spike region of the capsid. Infection analysis in vitro and in vivo showed that although heparin binding contributes to AAV-2 infection, it is not a prerequisite.
MATERIALS AND METHODS
Cell cultures.
293T and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mM l-glutamine, and 100 μg each of penicillin and streptomycin/ml at 37°C in 5% CO2.
Transfection and preparation of virus supernatants.
293T cells were plated at 1.5 × 106 cells per 10 ml of Dulbecco's modified Eagle's medium with 10% FCS per 10-cm petri dish 1 day prior to transfection. Transfection was carried out with 12 μg of DNA per dish by the method of Chen and Okayama (8). At 16 to 18 h posttransfection, the medium was replaced with 10 ml of medium containing adenovirus type 5 (multiplicity of infection, 50). The cultures were incubated for 2 days at 37°C in 5% CO2. Cells were harvested in 10 ml of medium and lysed by three rounds of freezing-thawing (−80 and 37°C) to release the produced virus. Cell debris was removed by centrifugation at 400 × g for 10 min. Prior to cell lysis, an aliquot of 300 μl of harvested cells was taken, washed once with phosphate-buffered saline (PBS), and denatured by being heated at 100°C for 5 min in the presence of sodium dodecyl sulfate sample buffer. After sonication, the samples were analyzed by Western blotting.
Determination of viral titers.
Quantitation of AAV-2 capsid titers was carried out by an enzyme-linked immunosorbent assay (ELISA) as described by Grimm et al. (23) with precoated plates supplied by Progen GmbH (Heidelberg, Germany). Infectious and genomic titers were determined as described previously (35). Randomly primed 32P-labeled probes were generated from a rep fragment (XhoI-BamHI) of plasmid pBSΔTR18 (65).
Plasmids and mutagenesis.
Plasmid pTAV2-0 (29) contains the entire AAV-2 genome from pAV-2 (39), including both inverted terminal repeats, cloned into the BamHI site of pBluescript II. Plasmid pJ407, which was used as the template for the site-directed mutagenesis reactions, contains the BamHI-NotI fragment of the AAV-2 genome from pTAV2-0 cloned into pUC131. Mutagenesis was performed by using a Stratagene (Amsterdam, The Netherlands) QuikChange site-directed mutagenesis kit according to the manufacturer's protocol. For each mutant, two complementary PCR primers were designed to contain the sequence of the substitution or deletion, respectively, flanked by 15 to 20 homologous base pairs on each side of the mutation. Mutant plasmids were identified by DNA sequencing. The BsiWI-XcmI (XcmI-EcoNI, respectively) fragment containing the mutation was then subcloned into the pTAV2-0 backbone. The complete fragment was sequenced to check for additional PCR mutations. Deletion mutant 6 was constructed by inserting a double-stranded oligonucleotide (5′-CGTTAACCCAGGCATGGTCTGGGC-3′ and 5′-CCCAGACCATGCCTGGGTTAACGCATG-3′), providing an additional XcmI site, into the SphI site of mutant 29. The resulting plasmid was digested with XcmI and religated.
Western blot analysis.
Western blot analysis and antibodies 303.9 and B1 were described previously (67). Peroxidase-coupled secondary antibodies and an enhanced chemiluminescence kit (NEN, Cologne, Germany) were used according to standard methods.
Cell-binding assay.
Cell binding was measured essentially as described previously (68). Briefly, HeLa cells in 100 μl of medium were plated at 104 cells/well on microtiter plates (Nunc, Wiesbaden, Germany) 1 day prior to infection. Virus supernatants were diluted in precooled medium without FCS to 109 to 1010 capsids/well. The cells were incubated with 50 μl of the dilutions/well in triplicate for 60 min at 4°C. After being washed with 200 μl of cooled PBS/well, the cells were fixed with 100 μl of methanol/well, cooled to −20°C for 15 min, and dried at room temperature overnight. After being washed three times with PBS containing 0.05% Tween 20 (wash buffer), the cells were blocked with 200 μl of PBS containing 0.2% casein and 0.05% Tween 20 (blocking buffer)/well for 2 h at 37°C. The blocking buffer was removed and replaced with blocking buffer containing 100 ng of biotinylated antibody A20/well (23), and the cells were incubated for 90 min at room temperature. Unbound A20 was removed by three washes with wash buffer. The cells then were incubated with 100 μl of streptavidin-coupled peroxidase diluted to a final concentration of 0.8 μg/ml in blocking buffer/well for 1 h at room temperature. After three washes with wash buffer, the cells were incubated with 100 μl of tetramethylbenzidine substrate (Devitron, Castrop-Rauxel, Germany). The reaction was stopped by adding 50 μl of 1 M H2SO4 to each well. The absorption at 450 nm was measured by using an ELISA reader (MWG, Ebersberg, Germany). Cell binding was expressed as a percentage of absorption measured with the AAV-2 wild-type virus stock by using the same capsid number.
Heparin-binding assay.
A 1-ml heparin column (Sigma, Munich, Germany) preequilibrated with 20 ml of PBS containing 1 mM MgCl2 and 2.5 mM KCl (PBS-MK) was loaded with 5 ml of virus supernatants. The column was washed twice with 5 ml of PBS-MK and eluted twice with 5 ml of PBS containing 1 M NaCl. The flowthrough, wash, and eluate fractions were analyzed by using the A20 capsid ELISA as described above. Values were expressed as a percentage of capsids applied to the column.
In vivo analysis of capsid mutants.
To analyze the in vivo distribution of double mutant R484E/R585E, recombinant AAV (rAAV) vectors carrying a luciferase reporter gene were generated. The capsid region carrying both mutations (R484E and R585E) was excised with XcmI-BsiWI from plasmid pJ407(R484E/R585E) and inserted into pBSΔTR18, providing rep and cap genes without inverted terminal repeats (65). Helper plasmid pDG(R484E/R585E) was obtained by excision of the mutated cap gene from modified pBSΔTR18 with SwaI-ClaI and insertion into the corresponding position of pDGΔVP (16). pUFCMV-Luc is a derivative of pUF3 (74) harboring the firefly luciferase sequence from pGL3-basic (Promega, Mannheim, Germany) inserted into the HindIII site and blunted 3′ XhoI site.
rAAV was produced by cotransfection of 293T cells with pUFCMV-Luc and pDG(R484E/585E) or pDG as described by Hauswirth et al. (28). After 2 days, cells were harvested and viruses were purified by using iodixanol gradients (28). Gradient fractions were concentrated by using VIVASPIN columns (Sartorius, Göttingen, Germany), thereby exchanging iodixanol against PBS.
Female immunocompetent NMRI mice (6 to 8 weeks old) were purchased from the German branch of Charles River Laboratories (Wilmington, Mass.). Six mice per group were injected intravenously via the tail vein with concentrated vectors (1011 genomic particles) from two independent preparations. After 3 weeks, mice were sacrificed, and representative organs (heart, lungs, liver, spleen, kidneys, skeletal muscle, and brain) were harvested and frozen in liquid nitrogen. All procedures involving the use and care of animals were performed according to the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication no. 85-23, revised 1996) and the German Animal Protection Code.
For the detection of AAV genomes in tissues, genomic DNA was extracted by using a Qiamp tissue kit (Qiagen, Hilden, Germany). Genomic DNA (800 ng) was used for PCR amplification (40 cycles) of a 677-bp fragment of the luciferase gene with primers 5′-GACGCCAAAAACATAAAGAAAG-3′ and 5′-CCAAAAATAGGATCTCTGGC-3′ under standard conditions. The integrity of the DNA was determined by amplifying a 492-bp region of the murine β-actin gene with primers 5′-ATGTTTGAGACCTTCAACAC-3′ and 5′-AACGTCACACTTCATGATGG-3′. PCR products were analyzed by agarose gel electrophoresis.
For the determination of luciferase reporter activities, we used a luciferase assay kit (Promega) according to the manufacturer's recommendations. Frozen tissue samples were homogenized with reporter lysis buffer and centrifuged for 10 min at 10,000 × g. Light activities were measured with a Luminometer (Lumat LB9501; Berthold, Bad Wildbad, Germany). A standard curve was generated with luciferin (Roche, Mannheim, Germany). Protein content in tissue homogenates was determined with a NanoOrange kit (Molecular Probes, Leiden, The Netherlands). Luciferase activities were expressed as relative light units per milligram of protein.
Graphics and docking simulation.
Pictures were made with Rasmol 2.6 (57) software and Insight2000 (Accelrys Inc.). Starting structures for the protein monomer and the heparin ligand were obtained from the Brookhaven Protein Data Bank (entry 1LP3 for AAV-2 protein [72] and entry 1HPN for the heparin hexamer [44]) and were minimized with the AMBER force field (66). The three-dimensional structure of the AAV-2 capsid was determined (our own software) with the BIOMT information given in the Brookhaven Protein Data Bank file. Potentials and partial charges were derived from the AMBER force field implemented in Insight2000. Partial charges for heparin had to be corrected for the carboxyl and sulfate groups to fit with the net charge of −12 for the heparin hexamer. Docking on the VP trimer was performed with the program AUTODOCK 3.05 (21, 43) with a grid spacing of 0.0375 nm and a rectangular grid box of 4.5 nm.
RESULTS
Search for a heparin-binding motif in the AAV capsid protein sequence.
The search for a heparin-binding motif in the AAV-2 capsid proteins was guided by a number of theoretical considerations and previous experimental findings. A sequence comparison of the AAV-2 capsid proteins with the capsid proteins of AAV-1, AAV-4, AAV-5, and AAV-6, which do not bind heparin, revealed major differences between amino acids 443 and 602, which are all located in loop IV, according to the alignment with canine parvovirus (CPV) (7). Several published insertion and charge-to-alanine mutations affecting heparin binding are also located in loop IV (22, 54, 59, 69). In addition, the epitope of monoclonal antibody C37, which effectively blocks the binding of AAV-2 capsids to HeLa cells, maps to two stretches of amino acids at the beginning and at the end of loop IV (68).
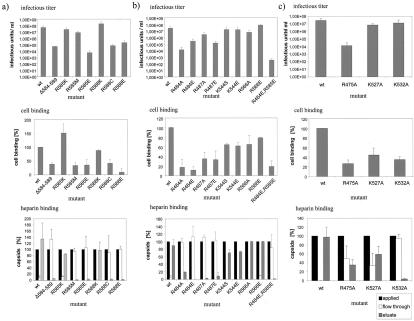
Based on the assumption that AAV-2 and AAV-3 bind to different primary receptors (26, 41), we converted short amino acid stretches in the loop IV region of AAV-2 to those of AAV-3 (Fig. 1, mutants 1 to 5, and Table 1) and analyzed the mutant viruses for capsid, genomic, and infectious titers. We observed a significant reduction in infectivity with mutant 4 and a modest reduction with mutant 5. Analysis of cell binding and heparin binding showed that both were reduced in mutant 4 (comprising arginine 588) but not in neighboring mutant 5. These results made us focus on the region surrounding arginine 588, the basic residue missing in mutant 4. An extended deletion from amino acid positions 587 to 601 (mutant 6), as well as short, partially overlapping deletions from amino acids 582 to 586 (mutant 7), from amino acids 584 to 589 (mutant 8), and from amino acids 587 to 590 (mutant 9), all confirmed that the region including arginines 585 and 588 likely is involved in heparin binding. The mutant viruses showed up to 4-log-unit reductions in infectivity, a loss of heparin binding, and a significant reduction in their ability to bind HeLa cells (Table 1). Further support for the involvement of arginine 588 in heparin binding came from double mutant 29 (N587A and R588C), which also resulted in more than a 1,000-fold reduction in infectivity, reduced cell binding, and a loss of heparin binding (Table 1). For all of the mutants analyzed in this report, no significant effect on genome packaging was detected (Table 1). From these results, we hypothesized that the loss of the basic charge of arginines 585 and 588 located in loop IV resulted in reduced cell binding and infectivity.
FIG. 1.
Amino acids of AAV-2 VPs subjected to site-directed mutagenesis. Blockwise conversions of VP sequences (VP1 numbering) of AAV-2 to those of AAV-3 (mutants 1, 2, 3, 4, and 5) are highlighted by grey rectangles. Single mutated amino acids are highlighted by grey circles. Mutants are numbered as shown in Table 1.
TABLE 1.
Mutant analysis
| Mutant | Mutation(s) (amino acid positions) | Infectivitya
|
Packagingb
|
Binding
|
|||
|---|---|---|---|---|---|---|---|
| No. of capsids/no. of infectious particles | Phenotype | Capsid/genome ratio | Phenotype | Cellc | Heparind | ||
| 1 | DIRD → SMSL (469-472) | 103 | WT | 21 | WT | ND | ND |
| 2 | SEK → TTA (547-549) | 103 | WT | 1 | WT | ND | ND |
| 3 | VDI → AEL (552-554) | 103 | WT | 26 | WT | ND | ND |
| 4 | RQA → TAP (588-590) | 2 × 106 | LI | 42 | WT | ± | − |
| 5 | A; AD → T; GT (591; 593, 594) | 7 × 104 | RI | 5 | WT | + | + |
| 6 | ΔQ589-V600, L601N, NR → AC (587, 588) | >106 | LI | 5 | WT | ND | ND |
| 7 | ΔN582-G586 | 8 × 105 | LI | 2 | WT | − | − |
| 8 | ΔQ584-Q589 | 106 | LI | 1 | WT | − | − |
| 9 | ΔN587-A590 | 3 × 106 | LI | 1 | WT | − | − |
| 10 | R447A | 5 × 103 | WT | 4 | WT | + | + |
| 11 | R459A | 2 × 103 | WT | 4 | WT | ± | + |
| 12 | R475A | 107 | LI | 0.3 | WT | − | ± |
| 13 | R484A | 107 | LI | 5 | WT | − | − |
| 14 | R484E | 106 | LI | 6 | WT | − | − |
| 15 | R487A | 4 × 104 | RI | 7 | WT | − | − |
| 16 | R487E | 3 × 107 | LI | 5 | WT | − | − |
| 17 | R484E, R585E | 6 × 108 | LI | 7 | WT | − | − |
| 18 | K507A | 4 × 103 | WT | 27 | WT | ± | + |
| 19 | K527A | 3 × 104 | RI | 3 | WT | ± | ± |
| 20 | K532A | 2 × 104 | RI | 3 | WT | − | − |
| 21 | K544S | 1.3 × 104 | RI | 1 | WT | ± | + |
| 22 | K544E | 1.1 × 104 | RI | 1 | WT | ± | + |
| 23 | K556A | 7 × 102 | WT | 4 | WT | ± | + |
| 24 | R566A | 6 × 103 | WT | 19 | WT | ± | + |
| 25 | R566E | 3 × 103 | WT | 5 | WT | ± | + |
| 26 | R585E | 1.3 × 107 | LI | 3 | WT | − | − |
| 27 | R585M | 9 × 104 | RI | 3 | WT | − | − |
| 28 | R585K | 2 × 103 | WT | 2 | WT | + | + |
| 29 | NR → AC (587, 588) | 2 × 106 | LI | 5 | WT | ± | − |
| 30 | R588E | 2 × 106 | LI | 3 | WT | − | − |
| 31 | R588C | 106 | LI | 13 | WT | ± | − |
| 32 | R588K | 103 | WT | 3 | WT | + | + |
| 33 | K620A | 104 | WT | 9 | WT | + | + |
Infectious AAV-2 particles were quantified by a dot blot replication assay as described previously (24). The number of assembled capsids was measured by a capsid ELISA (25). Infectivity was expressed as the ratio of the number of capsids to the number of infectious particles. The wild-type (WT) phenotype corresponds to a ratio of ≤104; reduced infectivity (RI) corresponds to ratios of >104 to <106, and low infectivity (LI) corresponds to ratios of ≥106.
Packaging was measured as the ratio of capsids to viral genomes as described previously (23). The WT phenotype indicates a packaging ratio of <50; reduced packaging was indicated by a ratio of >50.
Cell binding was measured as the binding of capsids to HeLa cells by an A20-based ELISA as described in Materials and Methods. It is expressed as the percentage of binding of WT AAV-2: +, more than 80% binding; ±, 40 to 80% binding; −, less than 40% binding. ND, not determined.
Heparin binding was measured as the percentage of viral particles which were applied to a heparin-agarose column and which were recovered after high-salt elution as determined with the A20-based capsid ELISA: +, more than 80% binding; ±, 20 to 80% binding; −, less than 20% binding.
Two distantly located pairs of arginines (R585-R588 and R484-R487) are involved in heparin binding.
In order to substantiate the results obtained so far, we mutated arginines 585 and 588 to the acidic glutamate (E), conservatively to the basic lysine (K), or to neutral methionine or cysteine (M or C). Point mutants were compared to wild-type virus and mutant 8 with respect to infectivity, cell binding, and heparin binding (Fig. 1 and 2a and Table 1). Conversion of arginines 585 and 588 to glutamic acid (R585E and R588E) strongly decreased infectivity, cell binding, and heparin binding, similar to the results obtained with the deletion mutant (mutant 8). In contrast, a conservative exchange of the arginines to lysine (R585K and R588K) had no negative effect on infectivity, cell binding, or heparin binding. The cell binding of R585K was in fact increased. Mutation of arginine 588 to cysteine (R588C) decreased infectivity, cell binding, and heparin binding, whereas mutation of arginine 585 to methionine (R585M) had no significant effect on infectivity, although cell binding was decreased and heparin binding was lost completely. With this exception, heparin binding correlated well with the infectivity of different mutant viruses. Taken together, the results of analyses of these mutants suggest that arginines 585 and 588 participate in the heparin binding of AAV-2 capsids.
FIG. 2.
Influence of mutations of basic amino acids in loop IV on infectivity, cell binding, and heparin binding of AAV-2. Virus stocks were produced by transfection of wild-type (wt) or mutated AAV-2 genomic plasmids into 293T cells and superinfection with adenovirus (multiplicity of infection, 50). (a) Analysis of arginines 585 and 588 for involvement in heparin binding. (b) Analysis of distantly located basic amino acids predicted to possibly contribute to heparin binding. (c) Further loop IV basic amino acids involved in heparin binding. Infectious units were determined as described in Materials and Methods. Capsid titers and genomic titers were not influenced by the mutations (data not shown), implying that they did not affect capsid assembly or genome encapsidation. Virus bound to HeLa cells was measured with monoclonal antibody A20 (68). Heparin binding was assayed by chromatography of virus preparations (freeze-thaw lysates) with heparin-agarose and quantitation of applied, flowthrough, and salt-eluted virus with the A20 capsid ELISA. The amount of applied virus was set to 100%. Error bars indicate standard deviations of at least three independent experiments.
Based on the sequence alignment of AAV-2 VP3 and CPV VP3 (7) and the atomic structure of the CPV capsid, we predicted that arginines 484 and 487, as well as two other basic amino acids, lysine 544 and arginine 566, could be spatially very close to arginines 585 and 588 in the folded state. We therefore mutated arginines 484, 487, and 566 to either alanine or glutamic acid and lysine 544 to serine or glutamic acid. All mutants were analyzed for virus infectivity, packaging, cell binding, and heparin binding (Fig. 1 and 2b and Table 1). Both charge-to-alanine mutations (R484A and R487A) and basic-to-acidic mutations (R484E and R487E) of arginines 484 and 487 (mutants 13, 14, 15, and 16) significantly reduced infectivity, cell binding, and heparin binding. This result strongly suggests that these two arginine residues form a basic cluster together with arginines 585 and 588 to strengthen heparin binding. Double mutant 17 (R484E and R585E) showed a similar loss of cell binding and heparin binding and was even more affected in its infectivity than the single mutants, further supporting our interpretation. Two mutations of lysine 544 (K544S and K544E; mutants 21 and 22) showed no reduction in infectivity and a modest reduction in cell binding. Heparin binding was not significantly reduced, although the recovery of the mutant viruses in the high-salt eluate was slightly reduced. Furthermore, mutations of arginine 566 (R566A and R566E; mutants 24 and 25) were not measurably affected in infectivity and heparin binding but, again, cell binding was slightly reduced. These results support the conclusion that, of the four basic amino acids predicted to be potentially located close to R585 and R588, only R484 and R487 are involved in heparin binding.
Further candidate amino acids in loop IV are involved in heparin binding.
To analyze whether other basic amino acids in loop IV contribute to the basic cluster around arginines 585 and 588 upon folding and assembly, we mutated all other not-yet-analyzed arginines and lysines between amino acid positions 446 and 620 (Fig. 1). Most residues showed no change in the parameters analyzed (Table 1) (mutants 10, 11, 12, 18, 19, 20, 23, and 33), except for mutations of arginine 475 (mutant 12) and of lysines 532 and 527 (mutants 20 and 19, respectively) (Fig. 2c). Conversion of arginine 475 to alanine (R475A; mutant 12) dramatically reduced the infectivity of the mutant virus as well as cell binding, whereas heparin binding was only partially lost. The opposite was observed for mutation of lysine 532 (K532A; mutant 20): while heparin binding was strongly reduced and cell binding was modestly reduced, infectivity remained nearly unaffected. Mutation of lysine 527 (K527A; mutant 19) to alanine produced similar characterisics, except that heparin binding was only moderately reduced. Based on these results, we concluded that R475 and K532 could be part of the heparin-binding motif together with R588, R585, R487, and R484.
Tissue tropism of AAV-2 heparin-binding mutants in vivo.
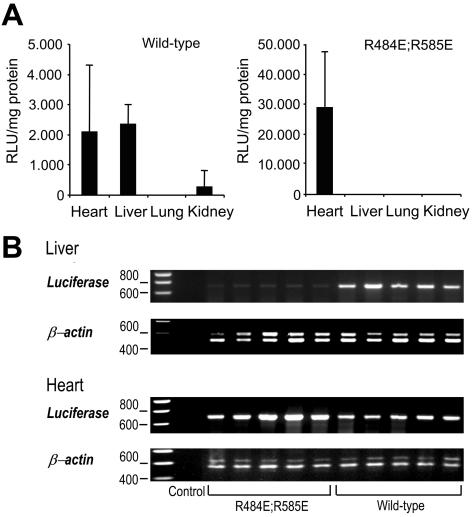
To test whether mutations in the heparin-binding motif also affect the in vivo tropism of the virus, we injected rAAV with either wild-type or heparin-binding mutant 17 (R484E and R585E) capsids into the tail vein of mice and analyzed vector genome distribution and reporter gene expression in different tissues. The reporter gene activity of rAAV packaged into the wild-type capsid could be detected in the liver and heart muscle after injection of 1011 vector genomes (Fig. 3A). Luciferase activity in the lungs and kidneys was close to background levels and could not be detected in the other tissues analyzed. The reporter gene activity of the double mutant was not detectable in the liver; however, it was surprisingly high in the heart sample, suggesting heparan sulfate-independent transduction of heart tissue in vivo. The difference in reporter gene expression between wild-type and double-mutant vectors was confirmed by PCR analysis of vector genomes in the heart and liver in several independently harvested samples (Fig. 3B). In contrast, vector genomes delivered by wild-type or mutant capsids could be detected by PCR in the lungs and kidneys in similar amounts (data not shown).
FIG. 3.
In vivo distribution of wild-type rAAV and rAAV mutant R484E/R585E in mouse tissues. (A) Luciferase activities in different organs after intravenous injection of 1011 wild-type capsids or rAAV mutated in R484E and R585E 3 weeks postinfecion. Luciferase activities are depicted in relative light units (RLU) per milligram of protein. Error bars indicate standard deviations. (B) Detection of luciferase gene transfer by PCR in selected organs 3 weeks after intravenous infusion of 1011 vector genomes of rAAV mutant R484E/R585E and wild-type rAAV. Amplification of β-actin was used as a control. The 677-bp luciferase band is barely detectable in liver tissues of mice injected with double mutant R484E/R585E but is clearly visible in mice injected with wild-type rAAV. PCR amplification of tissue samples from heart, lungs (data not shown), and kidneys (data not shown) did not reveal any differences in luciferase gene transduction.
DISCUSSION
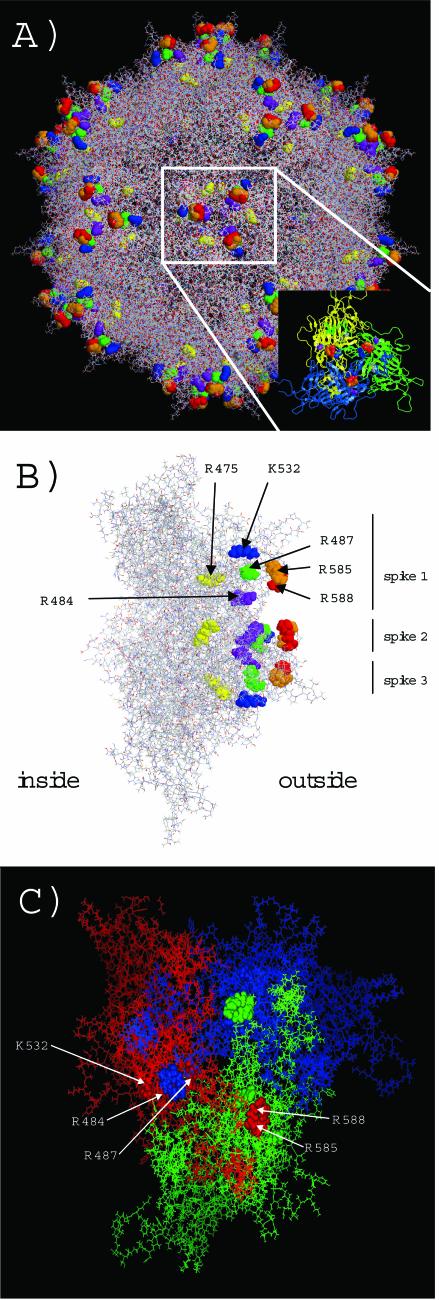
Our experimental analysis of amino acids potentially involved in heparin binding was performed prior to the atomic structure of AAV-2 capsids being known. The availability of the X-ray structure (72) has allowed us now to analyze our data at the molecular level. The basic residues R484, R487, K532, R585, and R588 all cluster in three basic patches at the threefold rotation axis of the AAV-2 capsid structure (Fig. 4A). A side view of the VP trimer reveals that these basic residues surround the surface of a channel with arginines 484 and 588 located at the border (Fig. 4B). According to this view, the binding motif is not located at the tips of the threefold spikes but at the inwardly oriented shoulders. Arginines 585 and 588 are even partially hidden by asparagine 587 if one looks from outside straight onto the threefold spike (data not shown). This finding suggests that the acidic heparin molecules interact with the basic amino acids along this channel predominantly by electrostatic interactions. Arginine 475, on the other hand, is buried within the capsid protein subunits and cannot have direct contact with heparin at the virus surface. It is therefore most likely that mutation of arginine 475 indirectly influences heparin binding and the infectivity of the virus. Indeed, arginine 475 is located at the border of two adjacent capsid proteins and might distort subunit geometry when mutated. This interpretation is further supported by the fact that basic residues K532, R484, and R487 of one subunit surround R585 and R588 of another subunit, suggesting that formation of the basic heparin-binding motif is a consequence of correct subunit interaction and assembly (Fig. 4C). After this article was submitted, a report in which amino acids in the AAV-2 capsid that contribute to heparan sulfate proteoglycan binding were identified was published (47). The identified amino acids there are essentially identical to those of the motif reported in this article.
FIG. 4.
Localization of amino acids involved in heparin binding on the AAV-2 capsid surface. (A) The amino acids involved in heparin binding—R585 (orange), R588 (red), R484 (purple), R487 (green),K532 (blue), and R475 (yellow)—are grouped in three clusters at the threefold spike region of the capsid. The inset shows a VP trimer which forms the threefold spikes. (B) A side view of a VP trimer shows the surface exposure of the basic clusters at the inner shoulder of the threefold spikes and along a channel-like structure between the threefold spikes. R475 is buried in the capsid wall (at the border of two subunits [not shown]) and likely affects heparin binding indirectly when it is mutated to alanine. The unmarked residues are derived from the second and third spikes. (C) Two different subunits of a VP trimer (indicated in red, blue, and green) contribute to the formation of a basic heparin-binding cluster. R585 and R588 of the red, green, or blue subunit (shown as orbitals, with the red subunit labeled) are surrounded by K532, R484, and R487 of another subunit (orbitals of the red subunit which surround R585 and R588 of the blue subunit are indicated by arrows).
Unexpectedly, R585 and R588 of AAV-2 are not conserved in the closely related AAV-3 which also binds to heparin (26, 53). These positions are instead occupied by serine and threonine, respectively. The polar nature of these amino acids could partially substitute for the charged arginines in AAV-2. In addition, they are also able to contribute to heparin binding by hydrogen bond formation. The homologues of arginines 484 and 487 and of lysine 532 are conserved in AAV-3. Together with the observation that AAV-2 and AAV-3 do not compete for binding to HeLa cells (26, 41), this finding may indicate that both viruses bind to different HSGAGs which are both related to and mimicked by heparin but may require slightly different binding sites.
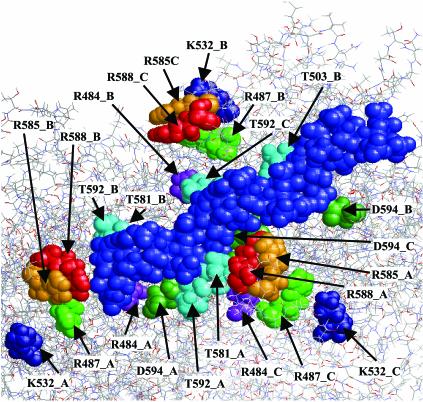
In a computer-simulated heparin-docking experiment involving the AAV-2 capsid surface, the heparin molecule (a heparin hexamer) aligned with the three basic clusters along the channels between the three spikes (Fig. 5). This experiment predicts close contacts (below 3 Å) of several threonines and aspartic acids 269 and 594 with the heparin molecule. While threonines could contribute to heparin binding via hydrogen bond stabilization, the two aspartic acids 269 and 594 may be required for charge neutralization of the basic clusters in the absence of bound heparin molecules. Since this charge neutralization is no longer needed after interaction of the capsid with heparin, a conformational switch might occur upon a capsid-heparin interaction to prevent interference of the acidic amino acids with heparin binding. It will be interesting to test this hypothesis.
FIG. 5.
Computer simulation of heparin docking to the AAV-2 capsid surface. The docking simulation was performed as described in Materials and Methods. The light blue orbitals show the heparin hexamer. Amino acids which are in proximity to the heparin molecule and which might permit electrostatic or hydrogen bond interactions are indicated. Letters A, B, and C indicate the three different VP subunits involved in heparin binding.
Mutational analysis of AAV-2 capsid protein amino acids involved in heparin binding showed a strong correlation with cell binding and virus infectivity, supporting the initial observation that heparin binding is involved in the initiation of an AAV-2 infection (62). Nevertheless, several mutants apparently contradict this interpretation: While heparin binding and cell binding of an R585M mutant (mutant 27) and a K532A mutant (mutant 20) were clearly reduced, the infectivity of these mutants was not affected much. Also, for mutant 12 (R475A) and mutant 19 (K527A), there was no quantitative correlation between infectivity and heparin binding. Mutant 12 had a strong effect on infectivity and only a weak influence on heparin binding, whereas mutant 19 had a very weak effect on infectivity but a moderate influence on heparin binding. In this respect, it should be noted that the infectious titer assay used here to determine infectivity measures more than just infection; it also measures replication and capsid formation. As postbinding events have a strong influence on gene transduction (15, 20, 27, 55, 71, 73), a particular mutation could compensate for a defect in cell binding and entry by improved cytoplasmic and nuclear trafficking or even replication and assembly. Therefore, the lack of a tight correlation between replication titers and heparin binding for these mutants does not necessarily contradict the involvement of AAV-2- HSPAG interactions in the infection process. Furthermore, the heparin-binding assay with a heparin-agarose column might not fully reflect binding to the still unknown HSGAG at the cell surface. There was, however, always a correlation between cell binding and heparin binding, suggesting that binding to heparin-agarose is a reliable measure for the interaction of AAV-2 with its cellular HSGAG.
Alternatively, however, the lack of correlation of heparin binding and infectivity of some heparin-binding mutants could mean that binding to heparan sulfate proteoglycan facilitates and enhances AAV-2 cell interaction but that this interaction is not necessary for infection (51), similar to the situation with adenovirus types 2 and 5, human immunodeficiency virus type 1, and HSV-1 (10, 13, 14, 19, 42, 46, 56, 70). Support for this interpretation comes from the in vivo distribution of wild-type and mutant rAAVs. A double mutation of the heparin-binding motif at R484 and R585 (mutant 17), leading to a loss of heparin binding and to low cell binding on HeLa cells, strongly reduced infection of mouse liver tissue in vivo but had no effect on infection of heart, lung, or kidney tissue. Although in this case the heart would have received a higher amount of virus (due to reduced retention by the liver) than in the wild-type experiment, there must also be a significant permissivity of heart tissue for the mutant virus to explain the observed expression levels. Since heparan sulfate proteoglycans are not internalized upon binding of a ligand when used as receptor molecules, their function in the AAV-2 infection process likely just increases the probability for interaction with a secondary receptor, such as fibroblast growth factor receptor 1 or αVβ5 integrin. Under this assumption, efficient interaction of the double mutant with one of these or an alternative (secondary) receptor could provide the observed infection of heart tissue with the heparin-binding mutant.
This analysis of a heparin-binding motif in the AAV-2 capsid has focused on basic amino acids in loop IV of the three AAV-2 capsid proteins. Several other basic sequence motifs present in the N-terminal region, unique for VP1 and VP2, were shown to play no role in heparin binding (69) (unpublished data). Nevertheless, Shi et al. (59) reported insertion mutants outside the range of of loop IV which inhibited heparin binding. According to the atomic structure of the AAV-2 capsid, either these positions are not surface exposed like amino acids 312 and 520 and most probably reflect an indirect influence on heparin binding or they are located on the surface of the fivefold symmetry element (V323 and K549), which is less exposed on the capsid surface than the threefold spike (37, 72). Nevertheless, the contribution of further nonbasic residues and a definitive description of all residues involved in an AAV-2- heparin interaction would require the X-ray analysis of heparin-bound capsids.
Acknowledgments
We thank P. Poberschin for excellent experimental support. We also acknowledge the initial experimental contributions of Niclas Czeloth and Simone Giese.
C.L. was supported by a grant from the HGF, and O.M. is supported by the Deutsche Forschungsgemeinschaft (MU 1654/2-1).
Footnotes
This article is dedicated to Harald zur Hausen on the occasion of his retirement as head of the German Cancer Research Center with gratitude and appreciation for 20 years of leadership.
REFERENCES
- 1.Bae, J., U. R. Desai, A. Pervin, E. E. Caldwell, J. M. Weiler, and R. J. Linhardt. 1994. Interaction of heparin with synthetic antithrombin III peptide analogues. Biochem. J. 301:121-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becerra, S. P., F. Koczot, P. Fabisch, and J. A. Rose. 1988. Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. J. Virol. 62:2745-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernfield, M., M. Gotte, P. W. Park, O. Reizes, M. L. Fitzgerald, J. Lincecum, and M. Zako. 1999. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68:729-777. [DOI] [PubMed] [Google Scholar]
- 4.Byrnes, A. P., and D. E. Griffin. 1998. Binding of Sindbis virus to cell surface heparan sulfate. J. Virol. 72:7349-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capila, I., and R. J. Linhardt. 2002. Heparin-protein interactions. Angew. Chem. Int. Ed. Engl. 41:391-412. [DOI] [PubMed] [Google Scholar]
- 6.Cassinotti, P., M. Weitz, and J. D. Tratschin. 1988. Organization of the adeno-associated virus (AAV) capsid gene: mapping of a minor spliced mRNA coding for virus capsid protein 1. Virology 167:176-184. [PubMed] [Google Scholar]
- 7.Chapman, M. S., and M. G. Rossmann. 1993. Structure, sequence, and function correlations among parvoviruses. Virology 194:491-508. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. A., and H. Okayama. 1988. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. BioTechniques 6:632-638. [PubMed] [Google Scholar]
- 9.Chen, Y., T. Maguire, R. E. Hileman, J. R. Fromm, J. D. Esko, R. J. Linhardt, and R. M. Marks. 1997. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 3:866-871. [DOI] [PubMed] [Google Scholar]
- 10.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 11.Chung, C. S., J. C. Hsiao, Y. S. Chang, and W. Chang. 1998. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 72:1577-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Compton, T., D. M. Nowlin, and N. R. Cooper. 1993. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology 193:834-841. [DOI] [PubMed] [Google Scholar]
- 13.Dechecchi, M. C., A. Tamanini, A. Bonizzato, and G. Cabrini. 2000. Heparan sulfate glycosaminoglycans are involved in adenovirus type 5 and 2-host cell interactions. Virology 268:382-390. [DOI] [PubMed] [Google Scholar]
- 14.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 15.Duan, D., Y. Yue, Z. Yan, J. Yang, and J. F. Engelhardt. 2000. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J. Clin. Investig. 105:1573-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubielzig, R., J. A. King, S. Weger, A. Kern, and J. A. Kleinschmidt. 1999. Adeno-associated virus type 2 protein interactions: formation of preencapsidation complexes. J. Virol. 73:8989-8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esko, J. D., and S. B. Selleck. 2002. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 71:435-471. [DOI] [PubMed] [Google Scholar]
- 18.Fath, M. A., X. Wu, R. E. Hileman, R. J. Linhardt, M. A. Kashem, R. M. Nelson, C. D. Wright, and W. M. Abraham. 1998. Interaction of secretory leukocyte protease inhibitor with heparin inhibits proteases involved in asthma. J. Biol. Chem. 273:13563-13569. [DOI] [PubMed] [Google Scholar]
- 19.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 20.Girod, A., C. E. Wobus, Z. Zadori, M. Ried, K. Leike, P. Tijssen, J. A. Kleinschmidt, and M. Hallek. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83:973-978. [DOI] [PubMed] [Google Scholar]
- 21.Goodsell, D. S., G. M. Morris, and A. J. Olson. 1996. Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recognit. 9:1-5. [DOI] [PubMed] [Google Scholar]
- 22.Grifman, M., M. Trepel, P. Speece, L. B. Gilbert, W. Arap, R. Pasqualini, and M. D. Weitzman. 2001. Incorporation of tumor-targeting peptides into recombinant adeno-associated virus capsids. Mol. Ther. 3:964-975. [DOI] [PubMed] [Google Scholar]
- 23.Grimm, D., A. Kern, M. Pawlita, F. Ferrari, R. Samulski, and J. Kleinschmidt. 1999. Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 6:1322-1330. [DOI] [PubMed] [Google Scholar]
- 24.Grimm, D., A. Kern, K. Rittner, and J. A. Kleinschmidt. 1998. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 9:2745-2760. [DOI] [PubMed] [Google Scholar]
- 25.Grimm, D., and J. A. Kleinschmidt. 1999. Progress in adeno-associated virus type 2 vector production: promises and prospects for clinical use. Hum. Gene Ther. 10:2445-2450. [DOI] [PubMed] [Google Scholar]
- 26.Handa, A., S. Muramatsu, J. Qiu, H. Mizukami, and K. E. Brown. 2000. Adeno-associated virus (AAV)-3-based vectors transduce haematopoietic cells not susceptible to transduction with AAV-2-based vectors. J. Gen. Virol. 81:2077-2084. [DOI] [PubMed] [Google Scholar]
- 27.Hansen, J., K. Qing, and A. Srivastava. 2001. Adeno-associated virus type 2-mediated gene transfer: altered endocytic processing enhances transduction efficiency in murine fibroblasts. J. Virol. 75:4080-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hauswirth, W. W., A. S. Lewin, S. Zolotukhin, and N. Muzyczka. 2000. Production and purification of recombinant adeno-associated virus. Methods Enzymol. 316:743-761. [DOI] [PubMed] [Google Scholar]
- 29.Heilbronn, R., A. Burkle, S. Stephan, and H. zur Hausen. 1990. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J. Virol. 64:3012-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hileman, R. E., J. R. Fromm, J. M. Weiler, and R. J. Linhardt. 1998. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Bioessays 20:156-167. [DOI] [PubMed] [Google Scholar]
- 31.Hileman, R. E., R. N. Jennings, and R. J. Linhardt. 1998. Thermodynamic analysis of the heparin interaction with a basic cyclic peptide using isothermal titration calorimetry. Biochemistry 37:15231-15237. [DOI] [PubMed] [Google Scholar]
- 32.Huntington, J. A., S. T. Olson, B. Fan, and P. G. Gettins. 1996. Mechanism of heparin activation of antithrombin. Evidence for reactive center loop preinsertion with expulsion upon heparin binding. Biochemistry 35:8495-8503. [DOI] [PubMed] [Google Scholar]
- 33.Jackson, T., F. M. Ellard, R. A. Ghazaleh, S. M. Brookes, W. E. Blakemore, A. H. Corteyn, D. I. Stuart, J. W. Newman, and A. M. King. 1996. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J. Virol. 70:5282-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joyce, J. G., J. S. Tung, C. T. Przysiecki, J. C. Cook, E. D. Lehman, J. A. Sands, K. U. Jansen, and P. M. Keller. 1999. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J. Biol. Chem. 274:5810-5822. [DOI] [PubMed] [Google Scholar]
- 35.King, J. A., R. Dubielzig, D. Grimm, and J. A. Kleinschmidt. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klimstra, W. B., K. D. Ryman, and R. E. Johnston. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72:7357-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kronenberg, S., J. A. Kleinschmidt, and B. Bottcher. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krusat, T., and H. J. Streckert. 1997. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch. Virol. 142:1247-1254. [DOI] [PubMed] [Google Scholar]
- 39.Laughlin, C. A., J. D. Tratschin, H. Coon, and B. J. Carter. 1983. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene 23:65-73. [DOI] [PubMed] [Google Scholar]
- 40.Liu, D., Z. Shriver, Y. Qi, G. Venkataraman, and R. Sasisekharan. 2002. Dynamic regulation of tumor growth and metastasis by heparan sulfate glycosaminoglycans. Semin. Thromb. Hemost. 28:67-78. [DOI] [PubMed] [Google Scholar]
- 41.Mizukami, H., N. S. Young, and K. E. Brown. 1996. Adeno-associated virus type 2 binds to a 150-kilodalton cell membrane glycoprotein. Virology 217:124-130. [DOI] [PubMed] [Google Scholar]
- 42.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 43.Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew, and A. Olson. 1998. Automated docking using a Lamarckian genetic algorithm and empirical binding energy function. J. Comp. Chem. 19:1639-1662. [Google Scholar]
- 44.Mulloy, B., M. J. Forster, C. Jones, and D. B. Davies. 1993. N. m. r. and molecular-modelling studies of the solution conformation of heparin. Biochem. J. 293:849-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicklin, S. A., H. Buening, K. L. Dishart, M. de Alwis, A. Girod, U. Hacker, A. J. Thrasher, R. R. Ali, M. Hallek, and A. H. Baker. 2001. Efficient and selective AAV2-mediated gene transfer directed to human vascular endothelial cells. Mol. Ther. 4:174-181. [DOI] [PubMed] [Google Scholar]
- 46.Ohshiro, Y., T. Murakami, K. Matsuda, K. Nishioka, K. Yoshida, and N. Yamamoto. 1996. Role of cell surface glycosaminoglycans of human T cells in human immunodeficiency virus type-1 (HIV-1) infection. Microbiol. Immunol. 40:827-835. [DOI] [PubMed] [Google Scholar]
- 47.Opie, S. R., K. H. Warrington, Jr., M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77:6995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park, P. W., O. Reizes, and M. Bernfield. 2000. Cell surface heparan sulfate proteoglycans: selective regulators of ligand-receptor encounters. J. Biol. Chem. 275:29923-29926. [DOI] [PubMed] [Google Scholar]
- 49.Qing, K., C. Mah, J. Hansen, S. Zhou, V. Dwarki, and A. Srivastava. 1999. Hum. fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 5:71-77. [DOI] [PubMed] [Google Scholar]
- 50.Qiu, J., and K. E. Brown. 1999. Integrin alphaVbeta5 is not involved in adeno-associated virus type 2 (AAV2) infection. Virology 264:436-440. [DOI] [PubMed] [Google Scholar]
- 51.Qiu, J., A. Handa, M. Kirby, and K. E. Brown. 2000. The interaction of heparin sulfate and adeno-associated virus 2. Virology 269:137-147. [DOI] [PubMed] [Google Scholar]
- 52.Qiu, J., H. Mizukami, and K. E. Brown. 1999. Adeno-associated virus 2 co-receptors? Nat. Med. 5:467-468. [DOI] [PubMed] [Google Scholar]
- 53.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rabinowitz, J. E., W. Xiao, and R. J. Samulski. 1999. Insertional mutagenesis of AAV2 capsid and the production of recombinant virus. Virology 265:274-285. [DOI] [PubMed] [Google Scholar]
- 55.Sanlioglu, S., P. K. Benson, J. Yang, E. M. Atkinson, T. Reynolds, and J. F. Engelhardt. 2000. Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by rac1 and phosphatidylinositol-3 kinase activation. J. Virol. 74:9184-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sattentau, Q. J., A. G. Dalgleish, R. A. Weiss, and P. C. Beverley. 1986. Epitopes of the CD4 antigen and HIV infection. Science 234:1120-1123. [DOI] [PubMed] [Google Scholar]
- 57.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374. [DOI] [PubMed] [Google Scholar]
- 58.Selinka, H. C., T. Giroglou, and M. Sapp. 2002. Analysis of the infectious entry pathway of human papillomavirus type 33 pseudovirions. Virology 299:279-287. [DOI] [PubMed] [Google Scholar]
- 59.Shi, W., G. S. Arnold, and J. S. Bartlett. 2001. Insertional mutagenesis of the adeno-associated virus type 2 (AAV2) capsid gene and generation of AAV2 vectors targeted to alternative cell-surface receptors. Hum. Gene Ther. 12:1697-1711. [DOI] [PubMed] [Google Scholar]
- 60.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Summerford, C., J. S. Bartlett, and R. J. Samulski. 1999. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat. Med. 5:78-82. [DOI] [PubMed] [Google Scholar]
- 62.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trempe, J. P., and B. J. Carter. 1988. Alternate mRNA splicing is required for synthesis of adeno-associated virus VP1 capsid protein. J. Virol. 62:3356-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trybala, E., T. Bergstrom, D. Spillmann, B. Svennerholm, S. J. Flynn, and P. Ryan. 1998. Interaction between pseudorabies virus and heparin/heparan sulfate. Pseudorabies virus mutants differ in their interaction with heparin/heparan sulfate when altered for specific glycoprotein C heparin-binding domain. J. Biol. Chem. 273:5047-5052. [DOI] [PubMed] [Google Scholar]
- 65.Weger, S., A. Wistuba, D. Grimm, and J. A. Kleinschmidt. 1997. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep proteins. J. Virol. 71:8437-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiner, S. J., P. A. Kollman, D. A. Case, U. C. Singh, C. Ghio, G. Alagona, S. Profeta, and P. Weiner. 1986. An all atom forcefield for simulations of proteins and nucleic acids. J. Comp. Chem. 7:230-252. [DOI] [PubMed] [Google Scholar]
- 67.Wistuba, A., S. Weger, A. Kern, and J. A. Kleinschmidt. 1995. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol. 69:5311-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wobus, C. E., B. Hugle-Dorr, A. Girod, G. Petersen, M. Hallek, and J. A. Kleinschmidt. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2- cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.WuDunn, D., and P. G. Spear. 1989. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J. Virol. 63:52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiao, W., K. H. Warrington, Jr., P. Hearing, J. Hughes, and N. Muzyczka. 2002. Adenovirus-facilitated nuclear translocation of adeno-associated virus type 2. J. Virol. 76:11505-11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yan, Z., R. Zak, G. W. Luxton, T. C. Ritchie, U. Bantel-Schaal, and J. F. Engelhardt. 2002. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 76:2043-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zolotukhin, S., M. Potter, W. W. Hauswirth, J. Guy, and N. Muzyczka. 1996. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 70:4646-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]