Abstract
Porcine circovirus type 2 (PCV2) is the primary causative agent of postweaning multisystemic wasting syndrome (PMWS), whereas the ubiquitous porcine circovirus type 1 (PCV1) is nonpathogenic for pigs. We report here the construction and characterization of two chimeric infectious DNA clones of PCV1 and PCV2. The chimeric PCV1-2 clone contains the PCV2 capsid gene cloned in the backbone of the nonpathogenic PCV1 genome. A reciprocal chimeric PCV2-1 DNA clone was also constructed by replacing the PCV2 capsid gene with that of PCV1 in the backbone of the PCV2 genome. The PCV1, PCV2, and chimeric PCV1-2 and PCV2-1 DNA clones were all shown to be infectious in PK-15 cells, and their growth characteristics in vitro were determined and compared. To evaluate the immunogenicity and pathogenicity of the chimeric infectious DNA clones, 40 specific-pathogen-free (SPF) pigs were randomly assigned into five groups of eight pigs each. Group 1 pigs received phosphate-buffered saline as the negative control. Group 2 pigs were each injected in the superficial inguinal lymph nodes with 200 μg of the PCV1 infectious DNA clone. Group 3 pigs were each similarly injected with 200 μg of the PCV2 infectious DNA clone, group 4 pigs were each injected with 200 μg of the chimeric PCV1-2 infectious DNA clone, and group 5 pigs were each injected with 200 μg of the reciprocal chimeric PCV2-1 infectious DNA clone. As expected, seroconversion to antibodies to the PCV2 capsid antigen was detected in group 3 and group 4 pigs. Group 2 and 5 pigs all seroconverted to PCV1 antibody. Gross and microscopic lesions in various tissues of animals inoculated with the PCV2 infectious DNA clone were significantly more severe than those found in pigs inoculated with PCV1, chimeric PCV1-2, and reciprocal chimeric PCV2-1 infectious DNA clones. These data indicated that the chimeric PCV1-2 virus with the immunogenic ORF2 capsid gene of pathogenic PCV2 cloned into the nonpathogenic PCV1 genomic backbone induces a specific antibody response to the pathogenic PCV2 capsid antigen but is attenuated in pigs. Future studies are warranted to evaluate the usefulness of the chimeric PCV1-2 infectious DNA clone as a genetically engineered live-attenuated vaccine against PCV2 infection and PMWS.
Porcine circovirus (PCV) was first discovered as a noncytopathic contaminant of the porcine kidney cell culture PK-15 (62, 66). PCV is a small icosahedral nonenveloped virus with a single-stranded circular DNA genome of about 1.76 kb. The PCV genome contains at least two potentially functional open reading frames (ORFs): ORF1 (930 bp) encodes the Rep protein involved in viral replication and ORF2 (690 bp) encodes the immunogenic capsid protein (15, 23, 39, 49). PCV belongs to the Circoviridae family along with other animal circoviruses such as Psittacine beak and feather disease virus, Chicken anemia virus (13), and Columbid circovirus (40). There are also three plant circoviruses, banana bunchy top virus, coconut foliar decay virus, and subterranean clover stunt virus (13, 42). Recently, three novel human circoviruses have been discovered, including transfusion-transmitted virus (TTV), SEN virus, and TTV-like minivirus (14, 44, 45, 51, 61, 67, 70). Although antibodies to PCV have been found in many animal species including humans, mice, cattle, and pigs (1, 17, 18, 30, 43, 50, 64, 65), little is known regarding the pathogenesis of PCV in these animal species (55, 65). The PK-15-derived PCV did not produce clinical disease in experimentally inoculated pigs, and thus the virus was considered to be nonpathogenic (3, 63) and was designated PCV1.
Postweaning multisystemic wasting syndrome (PMWS) is an emerging disease in pigs first described in 1991 (27). PMWS primarily affects pigs between 5 and 18 weeks of age. Clinical PMWS signs include progressive weight loss, dyspnea, tachypnea, anemia, diarrhea, and jaundice. The mortality rate may vary from 1 to 2% and up to 40% in some complicated cases in the United Kingdom (47). Characteristics of PMWS include microscopic lesions, granulomatous interstitial pneumonia, lymphadenopathy, hepatitis, and nephritis (4, 8, 11, 27). PMWS has now been recognized in pigs in Canada, the United States (5, 19, 26, 30, 34, 38, 46), most European countries (5, 11, 18, 32, 36, 39, 56, 60, 68), and some countries in Asia (16, 52). PMWS potentially has a serious economic impact on the swine industry worldwide.
The primary causative agent of PMWS is a pathogenic strain of PCV designated PCV2 (2, 5, 9, 11, 19, 21, 22, 23, 43, 46). The complete genomic sequence of PMWS-associated PCV2 has been determined (23, 26, 41). Sequence analyses revealed that PMWS-associated PCV2 shares about 75% nucleotide sequence identity with the nonpathogenic PCV1. The pathogenic PCV2 shares a very similar genomic organization with the nonpathogenic PCV1. The ORF2 gene of both PCV1 and PCV2 encodes the major immunogenic capsid protein (15, 48, 49). Initial attempts to reproduce clinical PMWS in conventional pigs by PCV2 inoculation were unsuccessful (12, 22, 33). Recently, clinical PMWS was reproduced in cesarean-derived-colostrum-deprived pigs and in specific-pathogen-free (SPF) pigs inoculated with PCV2 alone (28, 37). Clinical PMWS was also reproduced in conventional pigs coinfected with PCV2 and either porcine parvovirus (PPV) or porcine reproductive and respiratory syndrome virus (PRRSV) (57; T. Opriessnig, M. Fenaux, S. Yu, R. B. Evans, D. Cavanaugh, J. M. Gallup, F. J. Pallares, E. L. Thacker, K. M. Lagger, X. J. Meng, and P. G. Halbur, submitted for publication). In addition, PMWS was reproduced in PCV2-inoculated gnotobiotic pigs when their immune system was activated by keyhole limpet hemocyanin in incomplete Freund's adjuvant (35; A. Bøtner, S. Ladekjaer-Mikkelsen, J. Nielsen, S. Krakowka, J. Ellis, F. McNeilly, G. Allan, and Y. Storgard, Proc. Conf. ssDNA Viruses Plants Birds Pigs Primates, p. 132, 2002). Two recent field studies by Allan et al. (7) and Kyriakis et al. (36) tested the effect of immunomodulation by Mycoplasma hyopneumoniae vaccine on the development of PMWS in herds in which the disease is endemic and showed a significant decrease in PMWS cases in unvaccinated groups compared to the vaccinated animals. However, a controlled laboratory study by Opriessnig et al. (53) was able to induce significantly longer viremia and more severe lymphoid lesions but was not able to reproduce clinical PMWS in M. hyopneumoniae- and Actinobacillus pleuropneumoniae-vaccinated and PCV2-inoculated SPF piglets. Taken together, PCV2 is generally considered to be the primary, but not the sole, causative agent of PMWS.
It was previously shown that the cloned genomic DNA of PCV2 is infectious when directly injected into the liver and lymph nodes of pigs (22) and that the infectious PCV2 DNA clone produced pathological lesions characteristic of PMWS in SPF pigs (22). Here in this study, we report for the first time that chimeric DNA clones of PCV1 and PCV2 are infectious when transfected into PK-15 cells and express respective capsid antigen and are also infectious when injected directly into the superficial inguinal lymph nodes of SPF piglets. The immunogenicity and pathogenicity of the chimeric infectious DNA clones were characterized in pigs.
MATERIALS AND METHODS
Cell and virus.
The PK-15 cell line used in this study (ATCC CCL-33) was free of PCV1 contamination, as generated by endpoint dilutions (22). The PCV1 virus used in the study originated from the contaminated ATCC PK-15 cell line (63, 66). The PCV2 virus used in the study was originally isolated from a spleen tissue sample of a pig with naturally occurring PMWS (22, 23). The PCV2 infectious DNA clone was described previously (22).
Construction of the nonpathogenic PCV1 infectious DNA clone.
The procedure used for the construction of the PCV1 infectious DNA clone is very similar to that for PCV2 (22). Briefly, the primers KPNPCV1.U and KPNPCV1.L (Table 1) were designed based on the published sequence of PCV1 (42) to amplify the complete PCV1 genome, as a 1,758-bp PCR product, with an overlapping region containing the unique KpnI restriction enzyme site. The PCV1 DNA was extracted from PK-15 cells (ATCC CCL-33) persistently infected with PCV1, by using the QIAmp DNA minikit (Qiagen, Inc., Valencia, Calif.). The extracted DNA was amplified by PCR with Amplitaq Gold polymerase (Perkin-Elmer, Norwalk, Conn.). The PCR cycles consisted of an initial step of 95°C for 10 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 48°C for 1 min, and extension at 72°C for 2 min, and a final extension at 72°C for 7 min. The PCR product of the expected size was separated by gel electrophoresis, purified using a Geneclean kit (Bio 101, Inc., La Jolla, Calif.), digested by the KpnI restriction enzyme, and cloned into pBluescript SK (pSK) vector (Stratagene, La Jolla, Calif.). Escherichia coli DH5α competent cells were used for transformation. Recombinant plasmids containing the full-length PCV1 genome were isolated with a Qiagen plasmid minikit and were verified by restriction enzyme digestion. The full-length PCV1 genome was excised from the pSK vector by KpnI digestion and dimerized as described previously for the PCV2 infectious DNA clone (22) to produce the PCV1 infectious DNA clone. Based on previous data (22), the dimerized tandem DNA clone is more efficient in transfecting cells and producing infectious virions than the clone containing a single copy of the viral genome.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Primer sequence | Application |
|---|---|---|
| KPNPCV1.U. | >a 5′-TTTGGTACCCGAAGGCCGATT-3′ | PCV1 DNA clone construction |
| KPNPCV1.L. | < 5′-ATTGGTACCTCCGTGGATTGTTCT-3′ | PCV1 DNA clone construction |
| HpaI-2 | < 5′-GAAGTTAACCCTAAATGAATAAAAATAAAAACCATTACG-3′ | PCV1-2 DNA clone construction |
| NarI-3 | > 5′-GGTGGCGCCTCCTTGGATACGTCATCCTATAAAAGTG-3′ | PCV1-2 DNA clone construction |
| PsiI-5 | > 5′-AGGTTATAAGTGGGGGGTCTTTAAGATTAA-3′ | PCV1-2 DNA clone construction |
| AclI-6 | < 5′-GGAAACGTTACCGCAGAAGAAGACACC-3′ | PCV1-2 DNA clone construction |
| BglII-ORF2 | > 5′-ACTATAGATCTTTATTCATTTAGAGGGTCTTTCAG-3′ | PCV2-1 DNA clone construction |
| SphI-ORF2 | < 5′-TACGGGCATGCATGACGTGGCCAAGGAGG-3′ | PCV2-1 DNA clone construction |
| BglII-PCV2 | < 5′-AGACGAGATCTATGAATAATAAAAACCATTACGAAG-3′ | PCV2-1 DNA clone construction |
| SphI-PCV2 | > 5′-CGTAAGCATGCAGCTGAAAACGAAAGAAGTG-3′ | PCV2-1 DNA clone construction |
| MCV1 | > 5′-GCTGAACTTTTGAAAGTGAGCGGG-3′ | PCV1 and PCV2 detection |
| MCV2 | < 5′-TCACACAGTCTCAGTAGATCATCCCA-3′ | PCV1 and PCV2 detection |
| Orf.PCV1 | < 5′-CCAACTTTGTAACCCCCTCCA-3′ | PCV1 and PCV2-1 detection |
| Gen.PCV1 | > 5′-GTGGACCCACCCTGTGCC-3′ | PCV1 and PCV1-2 detection |
| nested.Orf.PCV1 | < 5′-CCAGCTGTGGCTCCATTTAA-3′ | PCV1 and PCV2-1 detection |
| nested.Gen.PCV1 | > 5′-TTCCCATATAAAATAAATTACTGAGTCTT-3′ | PCV1 and PCV1-2 detection |
| Orf.PCV2 | < 5′-CAGTCAGAACGCCCTCCTG-3′ | PCV2 and PCV1-2 detection |
| Gen.PCV2 | > 5′-CCTAGAAACAAGTGGTGGGATG-3′ | PCV2 and PCV2-1 detection |
| nested.Orf.PCV2 | < 5′-TTGTAACAAAGGCCACAGC-3′ | PCV2 and PCV1-2 detection |
| nested.Gen.PCV2 | > 5′-GTGTGATCGATATCCATTGACTG-3′ | PCV2 and PCV2-1 detection |
Primer direction.
Construction of a chimeric PCV1-2 infectious DNA clone.
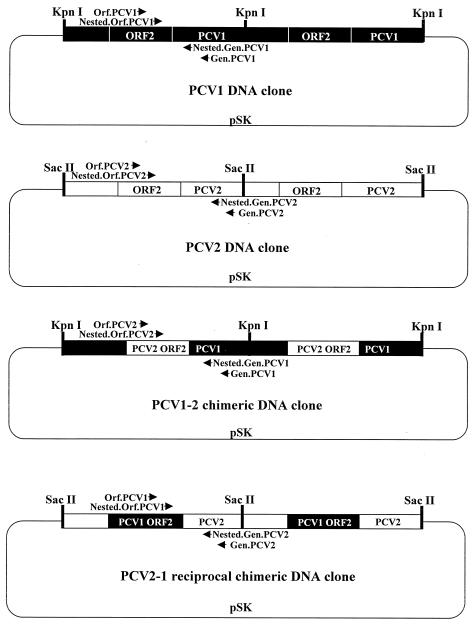
The ORF2 gene of both PCV1 and PCV2 encodes the immunogenic viral capsid protein (49). To construct a chimeric PCV1-2 DNA clone, the ORF2 capsid gene of PCV1 was replaced with that of the pathogenic PCV2 in the genome backbone of PCV1 (Fig. 1). Briefly, two pairs of PCR primers were designed: the first pair, PsiI-5 and AclI-6, amplified the PCV2 ORF2 gene, a fragment of 693 bp, and introduced flanking PsiI and AclI restriction enzyme sites by point mutations. The PCR for the amplification of PCV2 ORF2 consisted of an initial step at 95°C for 9 min, followed by 38 cycles of denaturation at 95°C for 1 min, annealing at 48°C for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 7 min. The second PCR primer pair, HpaI-2 and NarI-3, amplified the pSK vector and the PCV1 genome without the PCV1 ORF2 (pSK-PCV1 ΔORF2), a fragment of 4,023 bp, by using the PCV1 infectious DNA clone as the PCR template, and introduced flanking restriction enzyme sites HpaI and NarI by point mutations. The PCR consisted of an initial step at 95°C for 9 min, followed by 38 cycles of denaturation at 95°C for 1 min, annealing at 50°C for 1 min, extension at 72°C for 3.5 min, and a final extension at 72°C for 7 min. The pSK-PCV1 ΔORF2 PCR product was digested by HpaI and NarI to produce a sticky end and a blunt end complementary to the PCV2 ORF2 PCR product digested by AclI and PsiI restriction enzymes. Once the two PCR products were digested and ligated, all the PCR-introduced point mutations used to facilitate cloning steps were removed in the resulting chimeric DNA clone. E. coli DH5α competent cells were transformed. The recombinant plasmids containing the chimeric PCV1-2 DNA clone were isolated and were verified by restriction enzyme digestion and partial DNA sequencing. The full-length chimeric PCV1-2 genome was excised from the recombinant plasmid by KpnI digestion and dimerized (22) to produce the PCV1-2 chimeric infectious DNA clone (Fig. 1).
FIG. 1.
Organization of the infectious DNA clones. The PCV2 DNA clone was constructed by ligating two full-length linear PCV2 genomes in tandem into pSK as described previously (22). The PCV1 DNA clone was constructed by ligating two full-length linear PCV1 genomes in tandem into pSK. The chimeric PCV1-2 DNA clone was constructed by replacing the ORF2 capsid gene of PCV1 with that of PCV2 in the nonpathogenic PCV1 genomic backbone in pSK. The reciprocal chimeric PCV2-1 DNA clone was constructed by replacing the ORF2 capsid gene of the pathogenic PCV2 with that of the nonpathogenic PCV1 in the PCV2 genomic backbone in pSK. Both chimeric clones were dimers in pSK. The arrows represent the relative locations of the PCR primers used for the detection of PCV1, PCV2, PCV1-2, and PCV2-1 viremia in inoculated animals.
Construction of a reciprocal chimeric PCV2-1 infectious DNA clone.
To construct a reciprocal PCV2-1 chimeric DNA clone, the ORF2 capsid gene of PCV2 was replaced by that of the nonpathogenic PCV1 in the genome backbone of the pathogenic PCV2 (Fig. 1). Briefly, two PCR primer pairs were designed: the first pair, BglII-ORF2 and SphI-ORF2, amplified the PCV1 ORF2 gene, a fragment of 690 bp, and introduced flanking BglII and SphI restriction enzyme sites by point mutations. The second PCR primer pair, BglII-PCV2 and SphI-PCV2, amplified the pSK vector and the PCV2 genome without the ORF2 gene (pSK-PCV2 ΔORF2), a fragment of 4,030 bp, by using the PCV2 infectious DNA clone as the PCR template, and introduced flanking restriction enzymes sites BglII and SphI by point mutations. The pSK-PCV2 ΔORF2 product and the PCV1 ORF2 PCR product were digested by BglII and SphI restriction enzymes to produce complementary sticky and blunt ends and subsequently ligated together. After transformation into E. coli cells, the authentic recombinant plasmids were isolated and confirmed by enzyme digestion and partial DNA sequencing. The full-length reciprocal chimeric PCV2-1 genome was excised from the recombinant plasmid by SacII digestion and dimerized (22) to produce the reciprocal chimeric PCV2-1 infectious DNA clone.
In vitro transfection of PK-15 cells with PCV1, PCV2, chimeric PCV1-2, and reciprocal chimeric PCV2-1 DNA clones.
The infectivity of the PCV2 DNA clone has already been demonstrated both in vitro and in vivo (22). To test the infectivity of the PCV1 and two chimeric clones in vitro, PK-15 cells free of PCV1 contamination (22) were grown in eight-well LabTek chamber slides (Nalge Nunc International, Roskilde, Denmark). When the PK-15 cells reached about 80% confluency, cells were transfected with PCV1, PCV2 (22), PCV1-2, and PCV2-1 DNA clones by using the Lipofectamine Plus reagent according to the protocols supplied by the manufacturer (Life Technologies, Inc.). Mock-transfected cells with pSK+ vector alone were included as controls. Three days after transfection, the cells were fixed with a solution containing 80% acetone and 20% methanol at 4°C for 20 min. Evidence of viral capsid protein expression was confirmed in cells transfected with the PCV1 and PCV2-1 DNA clones by indirect immunofluorescence assay (IFA) with monoclonal antibody to the PCV1 ORF2 capsid gene (1). Briefly, the fixed cells were washed with phosphate-buffered saline (PBS) and incubated with a diluted PCV1 monoclonal antibody at 37°C for 1 h. The cells were then washed three times with PBS buffer and incubated with fluorescein isothiocyanate (FITC)-labeled goat anti-mouse immunoglobulin G (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.) at 37°C for 45 min. After being washed three times with PBS buffer, the slides were mounted with Fluoromount-G, coverslipped, and examined under a fluorescence microscope. The infectivity of cells transfected with the PCV2 and the chimeric PCV1-2 DNA clones was confirmed by IFA with antibody specific for PCV2, as previously described (22).
Transfection and subsequent serial passages of PCV1, PCV2, PCV1-2 and PCV2-1 infectious DNA clones in PK-15 cells.
In order to compare transfection efficiencies of each PCV DNA clone in vitro and to determine the viability of subsequent progeny viruses, four synchronized PK-15 cell cultures in T-25 flasks were transfected each with 44 μg of the PCV1, PCV2, PCV1-2, or PCV2-1 DNA clone. The transfected cell cultures were grown to confluency and then serially passaged five times. At each passage, 1 ml of the suspended cells was harvested, frozen, and thawed three times. The virus titers at each passage were determined according to the Kärber method. Briefly, serial 10-fold dilutions of the samples were used to inoculate PK-15 cells grown on eight-well LabTek chamber slides. After 3 days of incubation, the infected PK-15 cells were fixed, and the virus titers were determined by IFA with PCV1- or PCV2-specific antibodies according to the same IFA protocols described above.
One-step growth curves of PCV1, PCV2, PCV1-2, and PCV2-1 viruses in PK-15 cells.
PCV1-free PK-15 cells were grown on four 12-well plates. Each plate was infected with PCV1, PCV2, PCV1-2, and PCV2-1 live virus, respectively, at a multiplicity of infection (MOI) of 0.1 per well. After 1 h of incubation, the viral inoculum was removed and the cell monolayer was washed five times each with 2 ml of PBS buffer. Maintenance medium (2% bovine calf serum and 1× antibiotics) was subsequently added to each well, and the infected cells were continuously incubated at 37°C. Every 12 h, the medium and cells from one well of each inoculation group were harvested and frozen down at −80°C until virus titration. The PCV1, PCV2, PCV1-2, and PCV2-1 virus infectious titers at different time points were determined by IFA specific for detecting PCV1 or PCV2 as described above by the Kärber method.
Experimental inoculation of pigs with PCV1, PCV2, chimeric PCV1-2, and reciprocal chimeric PCV2-1 DNA clones.
Forty SPF pigs, 3 to 4 weeks of age, were randomly assigned into five rooms of eight animals each. Pregnant sows, from which the piglets used in this study were derived, were negative for antibodies to PCV, PRRSV, PPV, or swine hepatitis E virus (22, 31). In addition, preinoculation serum samples were tested for all piglets by PCR for the presence of PCV1 and PCV2 nucleic acids to confirm that the pigs used in the study were not naturally infected by either virus (54). The PCV1, PCV2, PCV1-2, and PCV2-1 infectious DNA clones were all inoculated by direct injection of the cloned plasmid DNA into the superficial inguinal lymph nodes of pigs as described previously (22). Pigs in group 1 received PBS buffer as a negative control. Group 2 animals were each injected with 200 μg of the infectious PCV1 DNA clone. Group 3 pigs were each injected with 200 μg of the infectious PCV2 DNA clone. Group 4 animals were each injected with 200 μg of the chimeric PCV1-2 infectious DNA clone. Group 5 pigs were each injected with 200 μg of the reciprocal chimeric PCV2-1 infectious DNA clone. All animals were monitored daily for clinical signs of disease. Serum samples were collected from each animal at −2, 7, 14, 21, 28, 35, 42, and 49 days postinoculation (dpi). At 21 dpi, four randomly selected animals from each group were necropsied. The remaining four animals in each group were necropsied at 49 dpi.
Clinical evaluation.
Pigs were weighed on 0 dpi and at the time of necropsies. Rectal temperatures and clinical respiratory scores, ranging from 0 to 6 (0 = normal; 6 = severe) (24), were recorded every other day from 0 to 49 dpi. Clinical observations, including evidence of central nervous system disease, liver disease (icterus), musculoskeletal disease, and changes in body condition, were also recorded daily. All clinical evaluations were performed by a team of two people.
Gross pathology and histopathology.
Four pigs from each group were necropsied at 21 and 49 dpi, respectively (only three pigs from group 4 were necropsied at 21 dpi because one pig died shortly after inoculation). The necropsy team was blinded to the infection status of the pigs at necropsy. Complete necropsies were performed on all pigs. An estimated percentage of the lung with grossly visible pneumonia was recorded for each pig based on a previously described scoring system (24). Other lesions such as enlargement of lymph nodes (ranging from 0 for normal to 3 for three times normal size) were scored separately. Sections for histopathologic examination were taken from nasal turbinate, lungs (seven sections) (24), heart, brain, lymph nodes (tracheobronchial, iliac, mesenteric, subiliac, and superficial inguinal), tonsil, thymus, liver, gall bladder, spleen, joints, small intestine, colon, pancreas, and kidney. The tissues were examined in a blinded fashion and given a subjective score for severity of lung, lymph node, and liver lesions (24). Lung scores ranged from 0 (normal) to 3 (severe lymphohistiocytic interstitial pneumonia). Liver scores ranged from 0 (normal) to 3 (severe lymphohistiocytic hepatitis). Lymph node scores were for an estimated amount of lymphoid depletion of follicles ranging from 0 (normal or no lymphoid depletion) to 3 (severe lymphoid depletion and histiocytic replacement of follicles).
Serology.
Blood was collected from all pigs at −2, 7, 14, 21, 28, 35, 42, and 49 dpi. Serum antibodies to PRRSV were assayed using Herd Check PRRSV enzyme-linked immunosorbent assay (ELISA) (IDEXX Laboratories, Westbrook, Mass.). Serum antibodies to PPV were detected by a hemagglutination inhibition assay (31). Serum antibodies to PCV2 were detected by a modified indirect ELISA based on the recombinant ORF2 capsid protein of PCV2 (48). Serum antibodies to PCV1 were detected by an IFA. Briefly, PK-15 cells infected with PCV1 were grown on eight-well LabTek chamber slides. When the infected PK-15 cells reached about 95 to 100% confluency, the cells were fixed with a solution containing 80% acetone and 20% methanol at 4°C for 20 min. The fixed cells were washed once with PBS buffer. One hundred microliters of 1:10-diluted pig serum sample in PBS was added to the chambers and incubated for 1 h at 37°C. The cells were then washed three times with PBS and incubated for 45 min at 37°C with FITC-labeled goat anti-swine secondary antibody. The slides were subsequently washed three times with PBS, mounted with Fluoromount-G, coverslipped, and examined under a fluorescence microscope. For the positive control, PCV1-infected cells were incubated with a diluted PCV1-specific monoclonal antibody, followed by an incubation with FITC-labeled goat anti-mouse immunoglobulin G (Kirkegaard & Perry Laboratories). For the negative control, PCV1-infected cells were incubated with 1:10-diluted negative swine serum free of PCV1 or PCV2 antibody, followed by an incubation with FITC-labeled goat anti-swine immunoglobulin G (Kirkegaard & Perry Laboratories).
PCR.
To detect PCV1, PCV2, chimeric PCV1-2, and reciprocal chimeric PCV2-1 viremia in sera from inoculated pigs, serum samples collected at different days postinfection were tested by PCR (Fig. 1). Briefly, viral DNA was extracted from 100 μl of each serum sample by using DNAzol reagent according to the manufacturer's protocol (Molecular Research Center, Cincinnati, Ohio). The extracted DNA was resuspended in DNase-, RNase-, and proteinase-free water. To amplify clone-specific genomic sequences of PCV1, PCV2, chimeric PCV1-2, and reciprocal chimeric PCV2-1, two sets of nested PCR primer pairs were designed (Table 1). The first set of nested primers was designed based on published PCV1 sequences. Primers Gen.PCV1 and Orf.PCV1 amplified a 400-bp fragment in the presence of the PCV1 genome. The nested primers, nested.Gen.PCV1 and nested.Orf.PCV1, amplified a 220-bp fragment.
To detect PCV2 viremia, PCV2 primer pair Gen.PCV2 and Orf.PCV2 amplified a 900-bp fragment in the presence of PCV2 in the first round of PCR. Primers nested.Gen.PCV2 and nested.Orf.PCV2 amplified a 600-bp fragment in the nested PCR.
To detect chimeric PCV1-2 viremia, the first round of PCR employed the PCV1-specific primer Gen.PCV1 and the PCV2 ORF2-specific primer Orf.PCV2 to amplify a chimeric PCV1-2 fragment of 580 bp. For the nested PCR, the PCV1-specific primer nested.Gen.PCV1 and the PCV2 ORF2-specific primer nested.Orf.PCV2 were used to amplify a chimeric PCV1-2 fragment of 370 bp.
To detect reciprocal chimeric PCV2-1 viremia, the first round of PCR employed the PCV2-specific primer Gen.PCV2 and the PCV1 ORF2-specific primer Orf.PCV1 to amplify a chimeric PCV2-1 fragment of 700 bp. For the nested PCR, the PCV2-specific primer nested.Gen.PCV2 and the PCV1 ORF2-specific primer nested.Orf.PCV1 were used to amplify a 460-bp chimeric PCV2-1 fragment.
All PCR parameters were essentially the same, consisting of 38 cycles of denaturation at 94°C for 1 min, annealing at 45°C for 1 min, and extension at 72°C for 1.5 min. The serum samples from negative-control pigs were tested by a PCR-restriction fragment length polymorphism diagnostic assay, which can detect and differentiate both PCV1 and PCV2 as described previously (22).
Sequencing.
PCR products from selected animals in each inoculation group, except for the PCV2-1 group, were sequenced to confirm the origin of the infecting virus by comparing the sequences recovered from infected pigs to the sequences of the respective infectious DNA clones with MacVector software (Oxford Molecular Ltd., Beaverton, Oreg.).
Immunohistochemistry (IHC).
IHC detection of PCV2-specific antigen was performed on lymph nodes collected during necropsies at 21 and 49 dpi. A rabbit polyclonal antiserum to PCV2 was used for the IHC, according to the procedures described previously (58). The amount of PCV2 antigen distributed in the lymph nodes was scored in a blinded fashion by assigning a score of 0 for no signal to 3 for a strong positive signal. The mean scores of inoculation groups were determined and statistically analyzed. No IHC detection was performed on PCV1- and PCV2-1-inoculated pig lymph nodes due to an insufficient amount of the gift PCV1 monoclonal antibody.
RESULTS
The PCV1 clone and PCV1-2 and PCV2-1 chimeric clones are infectious when transfected into PK-15 cells and express expected viral capsid antigen.
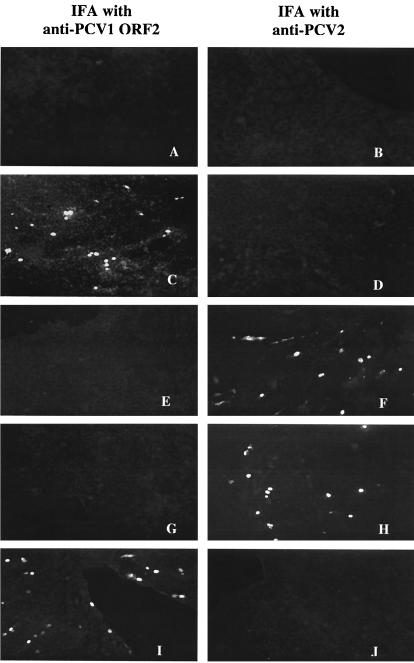
Two identical copies of the complete PCV1 genome were ligated in tandem into the pSK vector to produce the PCV1 DNA clone (Fig. 1). The infectivity of the PCV1 DNA clone was determined by in vitro transfection of PK-15 cells. IFA with monoclonal antibody to the PCV1 ORF2 gene product confirmed that the PCV1 DNA clone was infectious and that about 10 to 20% of the transfected PK-15 cells were positive for PCV1 capsid antigen in the nucleus of transfected cells (Fig. 2C).
FIG. 2.
The PCV1, PCV2, chimeric PCV1-2, and reciprocal chimeric PCV2-1 DNA clones were infectious and expressed respective viral antigens when transfected in vitro in PK-15 cells. The left panels (A, C, E, G, and I) were stained with monoclonal antibody to the PCV1 ORF2. The right panels (B, D, F, H, and J) were stained with antibody to PCV2. (A and B) Mock-transfected PK-15 cells. (C and D) PK-15 cells transfected with the PCV1 DNA clone. (E and F) PK-15 cells transfected with the PCV2 DNA clone. (G and H) PK-15 cells transfected with the chimeric PCV1-2 DNA clone. (I and J) PK-15 cells transfected with the reciprocal chimeric PCV2-1 DNA clone.
The infectious PCV2 DNA clone was constructed by ligating two tandem copies of PCV2 genome into the pSK+ vector (22) (Fig. 1). The PCV2 DNA clone was shown to be infectious in vitro and in vivo in a previous study (22) (Fig. 2F).
The chimeric PCV1-2 DNA clone had the ORF2 capsid gene of PCV1 replaced by that of the pathogenic PCV2 in the backbone of the nonpathogenic PCV1 genome. The infectivity of the PCV1-2 chimeric DNA clone was determined by in vitro transfection of PK-15 cells. Therefore, the chimeric PCV1-2 DNA clone should, if infectious in vitro, have replicated and produced the ORF2 capsid antigen of PCV2. IFA with antibodies to PCV2 confirmed that the PCV1-2 DNA clone was infectious and that about 10 to 20% of the transfected PK-15 cells were positive for PCV2 antigen within the nucleus of infected cells (Fig. 2H).
The reciprocal chimeric PCV2-1 DNA clone had the ORF2 capsid gene of PCV2 replaced by that of the nonpathogenic PCV1 in the backbone of the pathogenic PCV2 genome. Therefore, the reciprocal chimeric PCV2-1 DNA clone should have replicated and expressed PCV1 ORF2 capsid antigen in transfected PK-15 cells, if it was infectious. IFA with PCV1 ORF2-specific monoclonal antibody showed that the PCV2-1 chimeric DNA clone was infectious and that about 10 to 20% of the transfected PK-15 cells expressed PCV1 ORF2 antigen in the nucleus of transfected cells (Fig. 2I).
In vitro characterization of PCV1, PCV2, PCV1-2, and PCV2-1 infectious DNA clones.
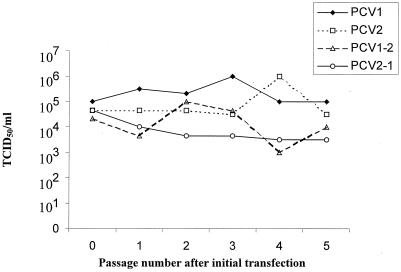
To confirm the viability of the progeny viruses after initial transfection and to compare the in vitro replication levels of the four viruses, four synchronized PK-15 cell cultures were transfected with 44 μg each of PCV1, PCV2, PCV1-2, or PCV2-1 infectious DNA clones. The infectious titers of the initial transfected cell cultures for all four infectious DNA clones ranged from 104.33 to 105 50% tissue culture infective doses (TCID50)/ml (Fig. 3), indicating that PCV1, PCV2, PCV1-2, and PCV2-1 infectious DNA clones all gave rise to similar numbers of progeny viruses after initial transfection. The infectious virus titers fluctuated within 1 log during five serial passages (Fig. 3).
FIG. 3.
In vitro transfection of PK-15 cells with PCV1, PCV2, PCV1-2, and PCV2-1 DNA clones followed by five serial passages of the four viruses in PK-15 cells. Synchronized PK-15 cells were transfected each with 44 μg of the PCV1, PCV2, PCV1-2, or PCV2-1 DNA clone. The PCV1, PCV2, PCV1-2, and PCV2-1 clones had similar infectious viral titers after the initial transfection. The infectious titers were determined at each passage according to the Käber method.
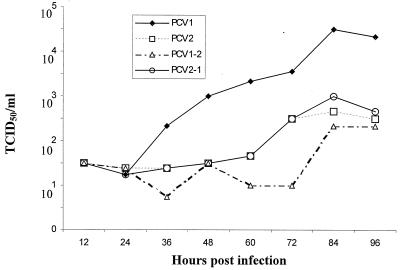
To further determine the growth characteristics of the four viruses, a one-step growth curve was performed simultaneously for each virus. Samples were collected at 12-h intervals from cells infected with either PCV1, PCV2, PCV1-2, or PCV2-1 virus stock at an MOI of 0.1. The infectious titers for each virus at different time points were determined by IFA (Fig. 4). The initial titers after infection at 12 h postinoculation were about 101.5 TCID50/ml for all four viruses. The infectious titers increased from 12 to 96 h for all four viruses. By 96 h postinfection, PCV1 had a titer of 104.33 TCID50/ml, while PCV2, PCV1-2, and PCV2-1 infectious titers ranged from 102.66 to 102.33 TCID50/ml.
FIG. 4.
One-step growth curve of PCV1, PCV2, PCV1-2, and PCV2-1 viruses. Synchronized PK-15 cell cultures were each infected with PCV1, PCV2, PCV1-2, or PCV2-1 virus, all at an MOI of 0.1. All four viruses had a titer of about 101.5 TCID50/ml at 12 h postinoculation. PCV1 replicated more efficiently in vitro than did PCV2, PCV1-2, and PCV2-1 viruses.
Immunogenicity of PCV1, PCV2, chimeric PCV1-2, and PCV2-1 infectious DNA clones in pigs.
Serum samples collected from all control and inoculated animals at −2, 7, 14, 21, 28, 35, 42, and 49 dpi were assayed for PCV1, PCV2, PCV1-2, and PCV2-1 viremia by PCR detection of clone-specific DNA sequences; for anti-PCV1 antibody by IFA; and for anti-PCV2 ORF2 antibody by ELISA. Prior to inoculation at −2 dpi, animals from all five groups tested negative by PCR for both PCV1 and PCV2 nucleic acids.
Negative-control animals were negative for both PCV1 and PCV2 viremia throughout the study (Table 2). Five pigs in the uninoculated control group had detectable PCV2 maternal antibody at −2 dpi, and two pigs had detectable PCV1 maternal antibodies at 7 dpi (Table 3). The maternal antibodies to both PCV1 and PCV2 in these piglets waned by 21 dpi. No seroconversion to antibodies of either PCV1 or PCV2 was detected in any of the eight uninoculated control pigs throughout the study.
TABLE 2.
Detection of viremia by nested PCR in sera of inoculated and control pigs
| Group | Inoculuma | No. of pigs positive/no. testedb
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. at dpt:
|
Total | |||||||||
| −2 | 7 | 14 | 21 | 28 | 35 | 42 | 49 | |||
| 1 | PBS | 0/8 | 0/8 | 0/8 | 0/8 | 0/4 | 0/4 | 0/4 | 0/4 | 0/8 |
| 2 | PCV1 DNA | 0/8 | 1/8 | 1/8 | 2/8 | 0/4 | 2/4 | 0/4 | 0/4 | 5/8 |
| 3 | PCV2 DNA | 0/8 | 3/8 | 6/8 | 7/8 | 1/4 | 2/4 | 2/4 | 0/4 | 8/8 |
| 4 | PCV1-2 DNA | 0/8 | 0/7 | 1/7 | 2/7 | 2/4 | 2/4 | 2/4 | 0/4 | 4/7 |
| 5 | PCV2-1 DNA | 0/8 | 0/8 | 0/8 | 0/8 | 0/4 | 0/4 | 0/4 | 0/4 | 0/8 |
PBS used as negative control. Cloned genomic PCV or chimeric PCV DNA in the pSK plasmid.
Eight pigs in each group; one pig died in group 4 shortly after inoculation.
TABLE 3.
Seroconversion to PCV2 antibodies in pigs inoculated with PCV2 or chimeric PCV1-2 infectious DNA clones and seroconversion to PCV1 antibodies in pigs inoculated with PCV1 or reciprocal chimeric PCV2-1 infectious DNA clones
| Group | Inoculuma | Antibody for which testedb | No. of pigs positive/no. tested at dpic:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| −2 | 7 | 14 | 21 | 28 | 35 | 42 | 49 | |||
| 1 | PBS | PCV1 | NAd | 2/8 | 2/8 | 1/8 | 0/4 | 0/4 | 0/4 | 0/4 |
| PCV2 | 5/8 | 5/8 | 2/8 | 0/8 | 0/4 | 0/4 | 0/4 | 0/4 | ||
| 2 | PCV1 DNA | PCV1 ORF2 | NA | 3/8 | 2/8 | 8/8 | 4/4 | 4/4 | 4/4 | 4/4 |
| 3 | PCV2 DNA | PCV2 | 3/8 | 2/8 | 0/8 | 0/8 | 0/4 | 3/4 | 4/4 | 4/4 |
| 4 | PCV1-2 DNA | PCV2 | 2/8 | 1/7 | 1/7 | 0/7 | 1/4 | 1/4 | 3/4 | 4/4 |
| 5 | PCV2-1 DNA | PCV1 ORF2 | NA | 3/8 | 3/8 | 7/8 | 3/4 | 3/4 | 4/4 | 4/4 |
PBS used as a negative control. The inocula were infectious DNA clones in the pSK plasmid.
PCV1 antibody to ORF2 was measured with an IFA specific for PCV1 ORF2 antigen. PCV2 antibody was measured with an ELISA.
One pig in group 4 died shortly after inoculation.
NA, not applicable.
In the PCV1-inoculated group, viremia was first detected in an inoculated pig at 7 dpi (Table 2) and was last detected at 35 dpi. Five out of eight animals inoculated with the PCV1 infectious DNA clone were positive for PCV1 viremia. The average length of continuous PCV1 viremia was 0.625 weeks. By 21 dpi, all animals in the PCV1-inoculated group had seroconverted to PCV1 antibodies and remained positive for anti-PCV1 antibodies until the end of the study at 49 dpi.
It was previously shown that the PCV2 DNA clone is infectious in pigs (22). In the PCV2 DNA clone-inoculated group, PCV2 viremia was first detected at 7 dpi (Table 2). By 21 dpi, all PCV2-inoculated group 3 animals were positive for PCV2 viremia. The average length of PCV2 viremia was 2.12 weeks. Two pigs in the PCV2-inoculated group had detectable low levels of maternal PCV2 antibodies (optical density at 405 nm [OD405], <0.4) at 7 dpi (Table 3), and the maternal antibodies in these piglets waned by 14 dpi. Seroconversion to antibodies of PCV2, assayed by a PCV2-specific ELISA, was first detected at 35 dpi. By 42 dpi, all pigs inoculated with the PCV2 infectious DNA clone had seroconverted to antibodies of PCV2.
In group 4 pigs inoculated with the PCV1-2 chimeric infectious DNA clone, viremia specific for the chimeric PCV1-2 virus was first detected at 14 dpi (Table 2). Four out of seven inoculated animals became viremic for PCV1-2 between 14 and 42 dpi. The average length of chimeric PCV1-2 viremia was 1 week. One pig had detectable low levels of maternal PCV2 antibodies at 7 and 14 dpi (OD405, <0.4), but the maternal antibody waned by 21 dpi (Table 3). Seroconversion to PCV2 ORF2-specific antibody first occurred at 28 dpi. By 49 dpi, all pigs inoculated with chimeric PCV1-2 infectious DNA clone had seroconverted to PCV2 ORF2-specific antibody.
In pigs inoculated with the reciprocal chimeric PCV2-1 DNA clone, viral DNA specific for PCV2-1 chimeric virus was not detected in serum samples (Table 2). However, by 21 dpi, all animals in group 5 seroconverted to anti-PCV1 antibody.
PCR products amplified from selected pigs in groups 2, 3, and 4 were sequenced and confirmed to be the authentic respective infectious DNA clones used in the inoculation for each group (data not shown). No PCR product from group 5 animals injected with the PCV2-1 reciprocal chimeric clone was sequenced since we were unable to detect viremia.
Pathogenicity of PCV1, PCV2, chimeric PCV1-2, and reciprocal chimeric PCV2-1 infectious DNA clones in pigs. (i) Clinical evaluation.
As expected, none of the control or inoculated pigs showed signs of PMWS (wasting, pneumonia, or icterus). There were no differences in weight gain or mean rectal temperatures among any of the groups (data not shown). One of the pigs from PCV1-2-inoculated group 4 was found dead on the morning following inoculation. No pathogenic agents were detected, lesions consistent with infectious disease were lacking, and it was concluded that the death was not associated with the inoculation procedure or the chimeric PCV1-2 virus.
(ii) Gross lesions.
Pigs in the four inoculated groups 2 to 5 had various degrees of gross lesions limited to the lymph nodes (Table 4). Lymph nodes of animals from the uninoculated control group 1 were normal at both 21 and 49 dpi (Table 4). In PCV1-inoculated group 2 pigs, lymph nodes were grossly normal at 21 dpi; however, mild to moderate enlargement of all lymph nodes was detected at 49 dpi. All PCV2-inoculated group 3 pigs had enlarged tan lymph nodes two to five times the normal size, at both 21 and 49 dpi. Lymph nodes from chimeric PCV1-2-inoculated animals were mildly to moderately enlarged and tan at both 21 and 49 dpi in five out of seven pigs (Table 4). In group 5 pigs, inoculated with the PCV2-1 clone, one out of eight animals had mildly enlarged and tan lymph nodes at 21 dpi (Table 4). The average scores of gross lesions of the lymph nodes in pigs inoculated with the chimeric PCV1-2 clone were not statistically different from those in groups 1, 2, and 5 but were significantly different (P < 0.05) from those of the pathogenic PCV2-inoculated group 3 pigs at 21 dpi. Average lymph node gross lesion scores at 49 dpi from the PCV1-, PCV2-, and PCV1-2-inoculated animals were not significantly different from each other but were all statistically different (P < 0.05) from the average gross lesion scores of group 1 and 5 pigs (Table 4).
TABLE 4.
Gross lymph node lesions in control and inoculated pigs
| Group | Inoculuma | No. of pigs with enlarged lymph nodes/no. necropsied at each dpi-(mean score for estimated enlargement)b
|
|
|---|---|---|---|
| 21 | 49 | ||
| 1 | PBS | 0/4 (0.0)I | 0/4 (0.0)I |
| 2 | PCV1 DNA | 0/4 (0.0)I | 4/4 (1.5)II |
| 3 | PCV2 DNA | 4/4 (2.5)II | 4/4 (2.25)II |
| 4 | PCV1-2 DNA | 2/3 (0.66)I | 3/4 (1.25)II |
| 5 | PCV2-1 DNA | 1/4 (0.25)I | 0/4 (0.0)I |
PBS used as a negative control. The inocula were infectious DNA clones in the pSK plasmid.
Four pigs from each group were necropsied at 21 dpi and the remaining pigs were necropsied at 49 dpi. Different superscripts (I and II) within each column indicate significantly different mean value scores between groups.
(iii) Microscopic lesions.
No microscopic lesions were detected in either uninoculated control group 1 pigs or PCV1-inoculated group 2 pigs at any day postinfection (Table 5). Microscopic lung lesions characterized as mild peribronchiolar lymphoplasmacytic and histiocytic bronchointerstitial pneumonia (Table 5) were observed in one out of the eight PCV2-inoculated pigs. In the PCV1-2- and PCV2-1-inoculated animals, no microscopic lesions were observed in the lungs. No lesions were observed in the thymuses of any inoculated pigs (Table 5). Mild multifocal lymphoplasmacytic myocarditis was observed in two of eight pigs in the PCV2-inoculated group (Table 5). Heart tissues from PCV1-2- and PCV2-1-inoculated animals were free of microscopic lesions. Mild multifocal lymphoplasmacytic interstitial nephritis was observed in four out of eight pigs in the PCV2-inoculated group, in two out of seven PCV1-2-inoculated pigs, and in one out of eight PCV2-1-inoculated pigs (Table 5). Mild to moderate lymphoid depletion and histiocytic replacement of follicles were observed in the tonsil in five out of eight pigs, in the spleen in three out of eight pigs, and in the lymph nodes in eight out of eight pigs in the PCV2-inoculated group. In the chimeric PCV1-2-inoculated animals, mild lymphoid depletion and histiocytic replacement of follicles were observed in the lymph nodes of two out of seven pigs but were not detected in either the spleen or tonsils. No lymphoid depletion and histiocytic replacement of follicles were observed in the lymph nodes, spleen, or tonsils of the reciprocal chimeric PCV2-1-inoculated animals (Table 5). Mild to moderate lymphoplasmacytic hepatitis was observed in seven out of the eight PCV2-inoculated pigs. Mild lymphoplasmacytic hepatitis was observed in two out of the seven chimeric PCV1-2-inoculated pigs. No lymphoplasmacytic hepatitis was observed in reciprocal chimeric PCV2-1-inoculated pigs (Table 5). Lesions in other tissues were unremarkable, except for one animal inoculated with PCV2 that showed mild microscopic lesions in the brain.
TABLE 5.
Distribution of histopathological lesions in different tissues and organs from control and inoculated pigs
| Group | Inoculuma | dpib | No. of pigs positive/no. tested
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lungc | Liverc | Lymph nodesc | Spleen | Thymus | Ileum | Brain | Heart | Kidney | Tonsil | |||
| 1 | PBS | 21 | 0/4 (0.0)I | 0/4 (0.0)I | 0/4 (0.0)I | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 |
| 49 | 0/4 (0.0)I | 0/4 (0.0)I | 0/4 (0.0)I | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | ||
| 2 | PCV1 DNA | 21 | 0/4 (0.0)I | 0/4 (0.0)I | 0/4 (0.0)I | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 |
| 49 | 0/4 (0.0)I | 0/4 (0.0)I | 0/4 (0.0)I | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | ||
| 3 | PCV2 DNA | 21 | 0/4 (0.0)I | 4/4 (1.5)II | 4/4 (1.75)II | 3/4 | 0/4 | 0/4 | 1/4 | 1/4 | 2/4 | 3/4 |
| 49 | 1/4 (0.25)I | 3/4 (0.75)II | 4/4 (1.0)II | 0/4 | 0/4 | 0/4 | 0/4 | 1/4 | 2/4 | 2/4 | ||
| 4 | PCV1-2 DNA | 21 | 0/3 (0.0)I | 1/3 (0.33)I | 1/3 (0.33)I | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| 49 | 0/4 (0.0)I | 1/4 (0.25)I,II | 1/4 (0.25)I | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 2/4 | 0/4 | ||
| 5 | PCV2-1 DNA | 21 | 0/4 (0.0)I | 0/4 (0.0)I | 0/4 (0.0)I | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 |
| 49 | 0/4 (0.0)I | 0/4 (0.0)I | 0/4 (0.0)I | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 1/4 | 0/4 | ||
PBS was used as a negative control. The inocula were infectious DNA clones in the pSK plasmid.
Four pigs from each group were necropsied at 21 dpi, and the remaining pigs were necropsied at 49 dpi.
Values in parentheses are mean histological scores (0, normal; 1, mild; 2, moderate; 3, severe) for interstitial pneumonia for lung, hepatitis for liver, and lymphoid depletion for lymph nodes. Different superscripts (I and II) within each column indicate significantly different mean value scores between groups.
Microscopic lesions in the lung, liver, and lymph nodes were scored according to a published scoring system (24). Mean scores of lesions in lymph nodes in pigs from the chimeric PCV1-2-inoculated group 4 were similar to those from groups 1, 2, and 5 but were significantly different (P < 0.05) from those of the pathogenic PCV2-inoculated group 3 pigs, at both 21 and 49 dpi (Table 5). Mean microscopic liver lesion scores from the chimeric PCV1-2-inoculated group at 21 dpi were significantly different (P < 0.05) from those of PCV2-inoculated group 3 animals but were similar to those of group 1, 2, and 5 pigs at 21 dpi. At 49 dpi, the mean microscopic liver scores from group 4 chimeric PCV1-2-inoculated pigs were not statistically different from those of group 1, 2, 3, and 5 pigs.
(iv) Detection of PCV antigen.
IHC staining of PCV2-specific antigen was performed on lymph nodes of all pigs necropsied at 21 and 49 dpi. As expected, lymph nodes from the uninoculated control and PCV1- and PCV2-1-inoculated pigs were negative for PCV2 antigen. PCV2 antigen was detected in lymphoid tissues of four out of four animals in the PCV2-inoculated group at 21 dpi and in three out of four animals at 49 dpi. PCV2 antigen was also detected in lymphoid tissue of one out of four pigs from the chimeric PCV1-2-inoculated group at 49 dpi, but no PCV2 antigen was detected in lymphoid tissues of PCV1-2-inoculated animals at 21 dpi (Table 6). At 21 dpi, the mean scores for the estimated level of PCV2 antigen in PCV1-2-inoculated animals were significantly different (P < 0.05) from those for PCV2-inoculated pigs but were not significantly different from those for the control group 1. At 49 dpi, the mean IHC score of the PCV1-2-inoculated animals was not significantly different from that of the PCV2-inoculated group 3 or the control group 1.
TABLE 6.
Immunohistochemical detection of PCV2 antigen in lymph nodes of control and PCV2- and PCV1-2-inoculated pigs
| Group | Inoculuma | No. of pigs positive/no. tested at dpib:
|
|
|---|---|---|---|
| 21 | 49 | ||
| 1 | PBS | 0/4 (0 ± 0.00)I | 0/4 (0 ± 0.00)I |
| 3 | PCV2 DNA | 4/4 (2 ± 0.67)II | 3/4 (0.75 ± 0.25)II |
| 4 | PCV1-2 DNA | 0/3 (0 ± 0.00)I | 1/4 (0.25 ± 0.25)I,II |
PBS was used as a negative control. The inocula were infectious DNA clones in the pSK plasmid.
Four pigs from each group were necropsied at 21 dpi, and the remaining pigs were necropsied at 49 dpi. Values in parentheses are the mean estimated levels of PCV2 antigen in lymphoid tissue (ranging from 0, no antigen detected, to 3, high levels of antigen). Different superscripts within each column (I and II) indicate significantly different mean value scores between groups.
DISCUSSION
PMWS has become a major global swine disease. It has become a serious economic problem for the swine industry, and there is an urgent need to develop a vaccine against PCV2, the primary causative agent of PMWS. We report here, for the first time, the construction and characterization of a chimeric PCV1-2 infectious DNA clone and demonstrate that the chimeric PCV1-2 infectious clone induces an immune response specific to the pathogenic PCV2 immunogenic ORF2 capsid protein while it is attenuated in SPF pigs. Animals inoculated via intralymphoid injections with the chimeric PCV1-2 infectious DNA clone developed a mild infection resembling that of PCV1-inoculated animals while seroconverting to antibody of the ORF2 capsid protein of the pathogenic PCV2. The average length of viremia observed in PCV1- and chimeric PCV1-2-inoculated animals was shorter, 0.625 and 1 week, respectively, than that in pathogenic PCV2-inoculated animals, which was about 2.12 weeks. The lack of detectable chimeric PCV1-2 viremia in sera or viral antigen in lymphoid tissues in some of the inoculated animals did not affect seroconversion to antibody of PCV2 ORF2 capsid protein, as all pigs in PCV1-2-inoculated group 4 pigs had seroconverted. The results suggested that, even though the chimeric PCV1-2 viremia and viral antigen were undetectable in some inoculated animals, chimeric PCV1-2 virus was still able to induce an antibody response to PCV2 ORF2 capsid protein. Similarly, none of the PCV2-1-inoculated pigs had detectable viremia, but they all seroconverted to anti-PCV1 capsid antibody by 49 dpi. The lack of viremia or viral antigen in some PCV1-2-inoculated pigs and the lack of viremia in PCV2-1-inoculated pigs could be due to a lower replication level of the PCV1-2 and PCV2-1 chimeric viruses in pigs. The one-step growth curve results indicated that PCV1-2 and PCV2-1 chimeric viruses had similar in vitro growth patterns, comparable to that of the wild-type PCV2. Thus, PCV2-1 and PCV1-2 are viable infectious DNA clones capable of producing infectious virions in vitro. We found that PCV1 replicated more efficiently than PCV2, PCV1-2, and PCV2-1 in PK-15 cells. This is likely due to the fact that the PCV1 isolate used in this study originated from the PK-15 cell line and thus may have already adapted to replicate in the PK-15 cells. After the initial transfection which produced similar infectious virions for all four viruses, the serially passaged PCV1, PCV2, PCV1-2, and PCV2-1 viruses replicated well in PK-15 cells, although the two chimeric viruses replicated slightly less efficiently in vitro.
The presence of low levels of maternal antibodies in some pigs did not seem to have any confounding influences on the onset of viremia or seroconversion. Since only a few animals in each experimental group had low levels of maternal antibodies at the time of inoculation, we were unable to make a definitive assessment as to whether low levels of maternal antibodies could prevent infection. However, a recent study by Harms et al. (29) showed that a PCV2 maternal antibody titer of an OD405 of 0.6 or above is required to prevent PCV2 replication. The highest maternal antibody titer detected in this study was an OD405 of 0.4 in only one pig, and all other pigs with maternal antibodies had very low titers (OD405 of <0.4) at the time of inoculation.
It is known that PCV2 replicates in the lymph nodes, lungs, and liver and that one of the major pathogenic effects is the impairment of the immune system by degradation of the lymphoid structures (10, 11, 20, 27, 33, 68). Gross lesions affecting the lymph nodes in both the nonpathogenic PCV1- and the chimeric PCV1-2-inoculated pigs were mild and limited to only a few pigs, whereas the pathogenic PCV2-inoculated pigs all had moderate to severe enlargement and discoloration of lymph nodes (Table 4). The gross lymph node lesions found in PCV1-inoculated pigs at 49 dpi are likely nonspecific, which is sometimes seen in pigs, and are not induced by PCV1, since no microscopic lesions in the lymph nodes were found in these same animals at 49 dpi. Statistical analysis revealed that the scores of the gross lesions in the lymph nodes of the chimeric PCV1-2-inoculated animals were similar to those in nonpathogenic PCV1-inoculated pigs. At 21 dpi, PCV2-inoculated pigs had gross lesions that were significantly (P < 0.05) more severe than those of the PCV1- and of the chimeric PCV1-2-inoculated pigs. Microscopically, at both 21 and 49 dpi, the chimeric PCV1-2-inoculated animals had significantly (P < 0.05) fewer and less severe microscopic lesions than did the PCV2-inoculated animals. The microscopic lesion scores in lymph nodes of the chimeric PCV1-2-inoculated pigs were similar to those of the nonpathogenic PCV1-inoculated pigs, the reciprocal chimeric PCV2-1-inoculated pigs, and the uninoculated control animals. IHC detection of PCV2 antigen within the lymph nodes from inoculated animals showed that the mean score of PCV2 antigen in PCV2-inoculated animals was significantly more intense (P < 0.05) than that from PCV1-2-inoculated animals, suggesting that during PCV1-2 replication in vivo there is a reduced level of antigen presence within the lymphoid tissues, the main target tissue of pathogenic PCV2. Mild to moderate lesions were found in multiple tissues of pathogenic PCV2-inoculated animals including lungs, liver, lymphoid tissues, spleen, brain, heart, kidneys, and tonsil tissue. However, in the chimeric PCV1-2-inoculated animals, mild to moderate microscopic lesions were limited to only liver, lymph nodes, and kidney tissues (Table 5). The microscopic liver lesion score at 21 dpi in chimeric PCV1-2-inoculated pigs was significantly less severe (P < 0.05) than that of PCV2-inoculated animals. However, at 49 dpi the microscopic liver lesion scores in both PCV2- and PCV1-2-inoculated animals were not significantly different (P > 0.05). It is important, however, that the microscopic liver lesion score of chimeric PCV1-2-inoculated animals at 49 dpi was also not significantly different (P > 0.05) from the scores of PCV1- and PCV2-1-inoculated pigs and of the negative-control pigs (Table 5). Recent studies showed that the pathological lesions in PCV2-infected pigs generally peak at about 21 to 28 dpi (53; Opriessnig et al., submitted), and this could explain why there was no significant difference in liver lesion scores at 49 dpi.
A previous study showed that the infectious PCV2 DNA clone induced pathological lesions characteristic of PMWS but did not induce clinical PMWS (22). It is generally believed that PCV2 is the primary, but not the sole, pathogenic agent responsible for the onset of clinical PMWS. In most experimental models to date, clinical PMWS was reproduced only in SPF pigs coinfected with PCV2 and either PPV or PRRSV (57; Opriessnig et al., submitted) or in PCV2-inoculated gnotobiotic pigs when their immune system was activated by keyhole limpet hemocyanin in incomplete Freund's adjuvant (35) or administration of bacterins (6). In cases of a PRRSV-PCV2 coinfection, the PMWS characteristic pathological signs such as lymphoid depletion, granulomatous inflammation, and necrotizing hepatitis were induced by PCV2 and not by PRRSV (28). Field studies continue to indicate a reduction of PMWS cases when piglets in herds in which PMWS is endemic and with PPV circulation are vaccinated against PPV (25). However, under laboratory conditions, upon challenge with PCV2, PPV vaccination of SPF pigs failed to decrease the occurrence of PMWS lesions compared to that for unvaccinated animals (Opriessnig et al., submitted). Therefore, vaccinations against PPV and possibly PRRSV, and thus limitation of the infections caused by them, may not prevent the onset of PMWS in PCV2-infected pigs, and a vaccine against PCV2 and PMWS is needed.
The availability of the PCV2, PCV1, chimeric PCV1-2, and reciprocal chimeric PCV2-1 infectious DNA clones enables us to study the structural and functional relationships of PCV genes. In this study, we swapped the immunogenic ORF2 capsid gene between the pathogenic PCV2 and the nonpathogenic PCV1 and showed that the chimeric PCV1-2 infectious DNA clone replicated and expressed the immunogenic ORF2 capsid antigen of PCV2 in vitro and in vivo and induced a specific antibody response to PCV2 ORF2 but retained the nonpathogenic nature of PCV1. Therefore, the chimeric PCV1-2 infectious DNA clone developed in this study could serve as a useful vaccine candidate against PCV2 infection and PMWS. The chimeric PCV1-2 infectious DNA clone has the ability to induce a strong immune response against PCV2 while inducing only a limited infection with mild pathological lesions, low levels of viremia, and low or nondetectable levels of viral antigen in lymphoid tissues, similar to infection by the nonpathogenic PCV1. The average ELISA OD405 values at 49 dpi were 0.511 (with 0.2 as the cutoff) for the remaining four seropositive pigs in the chimeric PCV1-2 group, 0.807 for the remaining four seropositive pigs in the PCV2 group, and 0.046 for the remaining four seronegative pigs in the negative-control group. Therefore, the chimeric PCV1-2 infectious DNA clone induced a relatively high anti-PCV2 antibody response comparable to that for PCV2. Future studies are warranted to determine if the PCV2 ORF2-specific antibody response induced by the chimeric PCV1-2 infectious DNA clone can protect pigs against challenge with wild-type pathogenic PCV2. The relatively easy storage and stability of cloned DNA and the economy of large-scale chimeric PCV1-2 DNA clone production should provide an attractive means of delivering a live infectious viral DNA vaccine to pigs. However, the intralymphoid route of inoculation used in this study is not practical for vaccine delivery, and thus, future studies will determine if pigs can be infected by alternate routes such as intramuscular injection of the chimeric PCV1-2 infectious DNA clone. The intramuscular and intradermal routes of inoculation have been successful in other studies with viral infectious DNA clones (59, 69).
Acknowledgments
We thank Gordon Allan of Belfast, United Kingdom, for providing the PCV1 monoclonal antibody; Mike Gill and Jay Srinivasappa of Fort Dodge Animal Health, Inc., and S. M. Boyle, L. A. Eng, R. Duncan, J. Sible, and T. Toth for their support; and Jillian Fenaux and Denis Guenette for their editorial assistance.
This study was supported by a grant from Fort Dodge Animal Health, Inc., Fort Dodge, Iowa.
REFERENCES
- 1.Allan, G. M., D. P. Mackie, J. McNair, B. M. Adair, and M. S. McNulty. 1994. Production, preliminary characterization and applications of monoclonal antibodies to porcine circovirus. Vet. Immunol. Immunopathol. 43:357-371. [DOI] [PubMed] [Google Scholar]
- 2.Allan, G. M., B. Meehan, D. Todd, S. Kennedy, F. McNeilly, J. Ellis, E. G. Clark, J. Harding, E. Espuna, A. Botner, and C. Charreyre. 1998. Novel porcine circoviruses from pigs with wasting disease syndromes. Vet. Rec. 142:467-468. [PubMed] [Google Scholar]
- 3.Allan, G. M., F. McNeilly, J. P. Cassidy, G. A. Reilly, B. Adair, W. A. Ellis, and M. S. McNulty. 1995. Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet. Microbiol. 44:49-64. [DOI] [PubMed] [Google Scholar]
- 4.Allan, G. M., F. McNeilly, J. Ellis, S. Krakowka, B. Meehan, I. McNair, I. Walker, and S. Kennedy. 2000. Experimental infection of colostrum deprived piglets with porcine circovirus 2 (PCV2) and porcine reproductive and respiratory syndrome virus (PRRSV) potentiates PCV2 replication. Arch. Virol. 145:2421-2429. [DOI] [PubMed] [Google Scholar]
- 5.Allan, G. M., F. McNeilly, S. Kennedy, B. Daft, E. G. Clarke, J. A. Ellis, D. M. Haines, B. M. Meehan, and B. M. Adair. 1998. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Investig. 10:3-10. [DOI] [PubMed] [Google Scholar]
- 6.Allan, G. M., F. McNeilly, S. Kennedy, B. Meehan, J. Ellis, and S. Krakowka. 2000. Immunostimulation, PCV-2 and PMWS. Vet. Rec. 147:170-171. [PubMed] [Google Scholar]
- 7.Allan, G. M., F. McNeilly, I. McNair, M. O'Conner, B. M. Meehan, D. Gilpin, J. A. Ellis, H. Townsend, C. Lasagna, G. Boriosi, and S. Krakowka. 2001. Neonatal vaccination for Mycoplasma hyopneumoniae and postweaning multisystemic wasting syndrome: a field trial. Pig J. 48:34-41. [Google Scholar]
- 8.Allan, G. M., F. McNeilly, B. M. Meehan, J. A. Ellis, T. J. Conner, I. McNair, S. Krakowka, and S. Kennedy. 2000. A sequential study of experimental infection of pigs with porcine circovirus and porcine parvovirus: immunostaining of cryostat sections and virus isolation. J. Vet. Med. B Infect. Dis. Vet. Public Health 47:81-94. [DOI] [PubMed] [Google Scholar]
- 9.Allan, G. M., F. McNeilly, B. M. Meehan, S. Kennedy, D. P. Mackie, J. A. Ellis, E. G. Clark, E. Espuna, N. Saubi, P. Riera, A. Botner, and C. E. Charreyre. 1999. Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet. Microbiol. 66:115-123. [DOI] [PubMed] [Google Scholar]
- 10.Allan, G. M., S. Kennedy, F. McNeilly, J. C. Foster, J. A. Ellis, S. J. Krakowka, B. M. Meehan, and B. M. Adair. 1999. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J. Comp. Pathol. 121:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Allan, G. M., and J. A. Ellis. 2000. Porcine circoviruses: a review. J. Vet. Diagn. Investig. 12:3-14. [DOI] [PubMed] [Google Scholar]
- 12.Balasch, M., J. Segales, C. Rosell, M. Domingo, A. Mankertz, A. Urniza, and J. Plana-Duran. 1999. Experimental inoculation of conventional pigs with tissue homogenates from pigs with post-weaning multisystemic wasting syndrome. J. Comp. Pathol. 121:139-148. [DOI] [PubMed] [Google Scholar]
- 13.Bassami, M. R., D. Berryman, G. E. Wilcox, and S. R. Raidal. 1998. Psittacine beak and feather disease virus nucleotide sequence analysis and its relationship to porcine circovirus, plant circoviruses, and chicken anaemia virus. Virology 249:453-459. [DOI] [PubMed] [Google Scholar]
- 14.Biagini, P., P. Gallian, H. Attoui, M. Touinssi, J.-F. Cantaloube, P. de Micco, and X. de Lamballerie. 2001. Genetic analysis of full-length genomes and subgenomic sequences of TT virus-like mini virus human isolates. J. Gen. Virol. 82:379-383. [DOI] [PubMed] [Google Scholar]
- 15.Cheung, A. K. 2003. Transcriptional analysis of porcine circovirus. J. Virol. 305:168-180. [DOI] [PubMed] [Google Scholar]
- 16.Choi, C., C. Chae, and E. G. Clark. 2000. Porcine postweaning multisystemic wasting syndrome in Korean pig: detection of porcine circovirus 2 infection by immunohistochemistry and polymerase chain reaction. J. Vet. Diagn. Investig. 12:151-153. [DOI] [PubMed] [Google Scholar]
- 17.Dulac, G. C., and A. Afshar. 1989. Porcine circovirus antigens in PK-15 cell line (ATCC CCL-33) and evidence of antibodies to circovirus in Canadian pigs. Can. J. Vet. Res. 53:431-433. [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards, S., and J. J. Sands. 1994. Evidence of circovirus infection in British pigs. Vet. Rec. 134:680-681. [DOI] [PubMed] [Google Scholar]
- 19.Ellis, J., L. Hassard, E. Clark, J. Harding, G. Allan, P. Willson, J. Strakappe, K. Martin, F. McNeilly, B. Meehan, D. Todd, and D. Haines. 1998. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can. Vet. J. 39:44-51. [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis, J., S. Krakowka, M. Lairmore, D. Haines, A. Bratanich, E. Clark, G. Allan, C. Konoby, L. Hassard, B. Meehan, K. Martin, J. Harding, S. Kennedy, and F. McNeilly. 1999. Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J. Vet. Diagn. Investig. 11:3-14. [DOI] [PubMed] [Google Scholar]
- 21.Ellis, J. A., A. Bratanich, E. G. Clark, G. Allan, B. Meehan, D. M. Haines, J. Harding, K. H. West, S. Krakowka, C. Konoby, L. Hassard, K. Martin, and F. McNeilly. 2000. Coinfection by porcine circoviruses and porcine parvovirus in pigs with naturally acquired postweaning multisystemic wasting syndrome. J. Vet. Diagn. Investig. 12:21-27. [DOI] [PubMed] [Google Scholar]
- 22.Fenaux, M., P. G. Halbur, G. Haqshenas, R. Royer, P. Thomas, P. Nawagitgul, M. Gill, T. E. Toth, and X. J. Meng. 2002. Cloned genomic DNA of type 2 porcine circovirus is infectious when injected directly into the liver and lymph nodes of pigs: characterization of clinical disease, virus distribution, and pathologic lesions. J. Virol. 76:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenaux, M., P. G. Halbur, M. Gill, T. E. Toth, and X. J. Meng. 2000. Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2. J. Clin. Microbiol. 38:2494-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halbur, P. G., P. S. Paul, M. L. Frey, J. Landgraf, K. Eernisse, X. J. Meng, M. A. Lum, J. J. Andrews, and J. A. Rathje. 1995. Comparison of the pathogenicity of two U. S. porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet. Pathol. 32:648-660. [DOI] [PubMed] [Google Scholar]
- 25.Halbur, P. G. 2001. PMWS improved models and findings of clinical significance. Proc. Swine Dis. Conf. Swine Practitioners 9:162-166. [Google Scholar]
- 26.Hamel, A. L., L. L. Lin, and G. P. Nayar. 1998. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J. Virol. 72:5262-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harding, J. C., and E. G. Clark. 1997. Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS). Swine Health Prod. 5:201-203. [Google Scholar]
- 28.Harms, P. A., S. D. Sorden, P. G. Halbur, S. R. Bolin, K. M. Lager, I. Morozov, and P. S. Paul. 2001. Experimental reproduction of severe disease in CD/CD pigs concurrently infected with type 2 porcine circovirus and porcine reproductive and respiratory syndrome virus. Vet. Pathol. 38:528-539. [DOI] [PubMed] [Google Scholar]
- 29.Harms, P. A., S. D. Sorden, P. G. Halbur, P. Nawagitgul, K. Lagger, S. R. Bolin, and P. S. Paul. 2002. Role of maternal immunity to PCV2 and PRRSV co-infection in the pathogenesis of PMWS. Proc. Am. Assoc. Swine Vet. 33:307-311. [Google Scholar]
- 30.Hines, R. K., and P. D. Lukert. 1995. Porcine circovirus: a serological survey of swine in the United States. Swine Health Prod. 3:71-73. [Google Scholar]
- 31.Joo, H. S., C. R. Donaldson-Wood, and R. H. Johnson. 1976. A standardized haemagglutination inhibition test for porcine parvovirus antibody. Aust. Vet. J. 52:422-424. [DOI] [PubMed] [Google Scholar]
- 32.Kennedy, S., G. Allan, F. McNeilly, B. M. Adair, A. Hughes, and P. Spillane. 1998. Porcine circovirus infection in Northern Ireland. Vet. Rec. 142:495-496. [PubMed] [Google Scholar]
- 33.Kennedy, S., D. Moffett, F. McNeilly, B. Meehan, J. Ellis, S. Krakowka, and G. M. Allan. 2000. Reproduction of lesions of postweaning multisystemic wasting syndrome by infection of conventional pigs with porcine circovirus type 2 alone or in combination with porcine parvovirus. J. Comp. Pathol. 122:9-24. [DOI] [PubMed] [Google Scholar]
- 34.Kiupel, M., G. W. Stevenson, S. K. Mittal, E. G. Clark, and D. M. Haines. 1998. Circovirus-like viral associated disease in weaned pigs in Indiana. Vet. Pathol. 35:303-307. [DOI] [PubMed] [Google Scholar]
- 35.Krakowka, S., J. A. Ellis, F. McNeilly, S. Ringler, D. M. Rings, and G. Allen. 2001. Activation of the immune system is the pivotal event in the production of wasting disease in pigs infected with porcine circovirus-2 (PCV-2). Vet. Pathol. 38:31-42. [DOI] [PubMed] [Google Scholar]
- 36.Kyriakis, S. C., K. Saoulidis, S. Lekkas, C. C. Miliotis, P. A. Papoutsis, and S. Kennedy. 2002. The effects of immuno-modulation on the clinical and pathological expression of postweaning multisystemic wasting syndrome. J. Comp. Pathol. 126:38-46. [DOI] [PubMed] [Google Scholar]
- 37.Ladekjaer-Mikkelsen, A. S., J. Nielsen, T. Stadejek, T. Storgaard, S. Krakowka, J. Ellis, F. McNeilly, G. Allan, and A. Botner. 2002. Reproduction of postweaning multisystemic wasting syndrome (PMWS) in immunostimulated and non-immunostimulated 3-week-old piglets experimentally infected with porcine circovirus type 2 (PCV2). Vet. Microbiol. 89:97-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larochelle, R., M. Morin, M. Antaya, and R. Magar. 1999. Identification and incidence of porcine circovirus in routine field cases in Quebec as determined by PCR. Vet. Rec. 145:140-142. [DOI] [PubMed] [Google Scholar]
- 39.Mankertz, A., M. Domingo, J. M. Folch, P. LeCann, A. Jestin, J. Segales, B. Chmielewicz, J. Plana-Duran, and D. Soike. 2000. Characterisation of PCV-2 isolates from Spain, Germany and France. Virus Res. 66:65-77. [DOI] [PubMed] [Google Scholar]
- 40.Mankertz, A., K. Hattermann, B. Ehlers, and D. Soike. 2000. Cloning and sequencing of columbid circovirus (CoCV), a new circovirus from pigeons. Arch. Virol. 145:2469-2479. [DOI] [PubMed] [Google Scholar]
- 41.Mankertz, J., H. J. Buhk, G. Blaess, and A. Mankertz. 1998. Transcription analysis of porcine circovirus (PCV). Virus Genes 16:267-276. [DOI] [PubMed] [Google Scholar]
- 42.Meehan, B. M., J. L. Creelan, M. S. McNulty, and D. Todd. 1997. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J. Gen. Virol. 78:221-227. [DOI] [PubMed] [Google Scholar]
- 43.Meehan, B. M., F. McNeilly, D. Todd, S. Kennedy, V. A. Jewhurst, J. A. Ellis, L. E. Hassard, E. G. Clark, D. M. Haines, and G. M. Allan. 1998. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J. Gen. Virol. 79:2171-2179. [DOI] [PubMed] [Google Scholar]
- 44.Miyata, H., H. Tsunoda, A. Kazi, A. Yamada, M. A. Khan, J. Murakami, T. Kamahora, K. Shiraki, and S. Hino. 1999. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J. Virol. 73:3582-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moen, E. M., L. Huang, and B. Grinde. 2001. Molecular epidemiology of TTV-like mini virus in Norway. Arch. Virol. 147:181-185. [DOI] [PubMed] [Google Scholar]
- 46.Morozov, I., T. Sirinarumitr, S. D. Sorden, P. G. Halbur, M. K. Morgan, K. J. Yoon, and P. S. Paul. 1998. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J. Clin. Microbiol. 36:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muirhead, M. 2002. Sources of information on PMWS/PDNS. Vet. Rec. 150:456. [PubMed] [Google Scholar]
- 48.Nawagitgul, P., P. A. Harms, I. Morozov, B. J. Thacker, S. D. Sorden, C. Lekcharoensuk, and P. S. Paul. 2002. Modified indirect porcine circovirus (PCV) type 2-based and recombinant capsid protein (ORF2)-based enzyme-linked immunosorbent assays for detection of antibodies to PCV. Clin. Diagn. Lab. Immunol. 9:33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nawagitgul, P., I. Morozov, S. R. Bolin, P. A. Harms, S. D. Sorden, and P. S. Paul. 2000. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 81:2281-2287. [DOI] [PubMed] [Google Scholar]
- 50.Nayar, G. P., A. L. Hamel, L. Lin, C. Sachvie, E. Grudeski, and G. Spearman. 1999. Evidence for circovirus in cattle with respiratory disease and from aborted bovine fetuses. Can. Vet. J. 40:277-278. [PMC free article] [PubMed] [Google Scholar]
- 51.Nishizawa, T., H. Okamoto, K. Konishi, H. Yoshizawa, Y. Miyakawa, and M. Mayumi. 1997. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 241:92-97. [DOI] [PubMed] [Google Scholar]
- 52.Onuki, A., K. Abe, K. Togashi, K. Kawashima, A. Taneichi, and H. Tsunemitsu. 1999. Detection of porcine circovirus from lesions of a pig with wasting disease in Japan. J. Vet. Med. Sci. 61:1119-1123. [DOI] [PubMed] [Google Scholar]
- 53.Opriessnig, T., S. Yu, J. M. Gallup, R. B. Evans, M. Fenaux, F. J. Pallares, E. L. Thacker, C. W. Brockus, M. R. Ackerman, P. Thomas, X. J. Meng, and P. G. Halbur. Effects of vaccination with selective bacterins on conventional pigs infected with wild type 2 porcine circovirus. Vet. Pathol., in press. [DOI] [PubMed]
- 54.Pogranichnyy, R. M., K.-J. Yoon, P. A. Harms, S. L. Swenson, J. J. Zimmerman, and S. D. Sorden. 2000. Characterization of immune response of young pigs to porcine circovirus type 2 infection. Viral Immunol. 13:143-153. [DOI] [PubMed] [Google Scholar]
- 55.Quintana, J., M. Balasch, J. Segales, M. Calsamiglia, G. M. Rodriguez-Arrioja, J. Plana-Duran, and M. Domingo. 2002. Experimental inoculation of porcine circovirus type 1 (PCV1) and type 2 (PCV2) in rabbits and mice. Vet. Res. 33:229-237. [DOI] [PubMed] [Google Scholar]
- 56.Rosell, C., J. Segales, J. A. Ramos-Vara, J. M. Folch, G. M. Rodriguez-Arrioja, C. O. Duran, M. Balasch, J. Plana-Duran, and M. Domingo. 2000. Identification of porcine circovirus in tissues of pigs with porcine dermatitis and nephropathy syndrome. Vet. Rec. 146:40-43. [DOI] [PubMed] [Google Scholar]
- 57.Rovira, A., M. Blasch, J. Segales, L. Garcia, J. Plana-Duran, C. Rosell, H. Ellerbrok, A. Mankertz, and M. Domingo. 2002. Experimental inoculation of conventional pigs with porcine reproductive and respiratory syndrome virus and porcine circovirus 2. J. Virol. 76:3232-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sorden, S. D., P. A. Harms, P. Nawagitgul, D. Cavanaugh, and P. S. Paul. 1999. Development of a polyclonal-antibody-based immunohistochemical method for the detection of type 2 porcine circovirus in formalin-fixed, paraffin-embedded tissue. J. Vet. Diagn. Investig. 11:528-530. [DOI] [PubMed] [Google Scholar]
- 59.Sparger, E. E., H. Louie, A. M. Ziomeck, and P. A. Luciw. 1997. Infection of cats by injection with DNA of feline immunodeficiency virus molecular clone. Virology 238:157-160. [DOI] [PubMed] [Google Scholar]
- 60.Spillane, P., S. Kennedy, B. Meehan, and G. Allan. 1998. Porcine circovirus infection in the Republic of Ireland. Vet. Rec. 143:511-512. [PubMed] [Google Scholar]
- 61.Takahashi, K., Y. Iwasa, M. Hijikata, and S. Mishiro. 2000. Identification of a new human DNA virus (TTV-like mini virus, TLMV) intermediately related to TT virus and chicken anemia virus. Arch. Virol. 145:979-993. [DOI] [PubMed] [Google Scholar]
- 62.Tischer, I., H. Gelderblom, W. Vettermann, and M. A. Koch. 1982. A very small porcine virus with circular single-stranded DNA. Nature 295:64-66. [DOI] [PubMed] [Google Scholar]
- 63.Tischer, I., W. Mields, D. Wolff, M. Vagt, and W. Griem. 1986. Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 91:271-276. [DOI] [PubMed] [Google Scholar]
- 64.Tischer, I., L. Bode, D. Peters, S. Pociuli, and B. Germann. 1995. Distribution of antibodies to porcine circovirus in swine populations of different breeding farms. Arch. Virol. 140:737-743. [DOI] [PubMed] [Google Scholar]
- 65.Tischer, I., L. Bode, J. Apodaca, H. Timm, D. Peters, R. Rasch, S. Pociuli, and E. Gerike. 1995. Presence of antibodies reacting with porcine circovirus in sera of humans, mice, and cattle. Arch. Virol. 140:1427-1439. [DOI] [PubMed] [Google Scholar]
- 66.Tischer, I., R. Rasch, and G. Tochtermann. 1974. Characterization of papovavirus and picornavirus-like particles in permanent pig kidney cell lines. Zentbl. Bakteriol. Hyg. A 226:153-167. [PubMed] [Google Scholar]
- 67.Umemura, T., A. E. Yeo, A. Sottini, D. Moratto, Y. Tanaka, R. Y. Wang, J. W. Shih, P. Donahue, D. Primi, and H. J. Alter. 2001. SEN virus infection and its relationship to transfusion-associated hepatitis. Hepatology 33:1303-1311. [DOI] [PubMed] [Google Scholar]
- 68.Wellenberg, G. J., S. Pesh, F. W. Berndsen, P. J. Steverink, W. Hunneman, T. J. Van der Vorst, N. H. Peperkamp, V. F. Ohlinger, R. Schippers, J. T. Van Oirschot, and M. F. de Jong. 2000. Isolation and characterization of porcine circovirus type 2 from pigs showing signs of post-weaning multisystemic wasting syndrome in the Netherlands. Vet. Q. 22:167-172. [DOI] [PubMed] [Google Scholar]
- 69.Willems, L., D. Portetelle, P. Kerkhofs, G. Chen, A. Burney, M. Mammerickx, and R. Kettmann. 1992. In vivo transfection of bovine leukemia provirus into sheep. Virology 189:775-777. [DOI] [PubMed] [Google Scholar]
- 70.Wilson, L. E., T. Umemeru, J. Astemborski, S. C. Ray, H. J. Alter, S. A. Strathdee, D. Vlahov, and D. L. Thomas. 2001. Dynamics of SEN virus infection among injection drug users. J. Infect. Dis. 184:1315-1319. [DOI] [PubMed] [Google Scholar]