Abstract
Hantavirus nucleocapsid (N) protein has been suggested to form homodimers and homotrimers that are further integrated into the nucleocapsid filaments around the viral RNA. Here we report detailed mapping of the regions involved in the homotypic N protein interactions in Tula hantavirus (TULV). Peptide scan screening was used to define the interaction regions, and the mammalian two-hybrid assay was used for the functional analysis of N protein mutants. To study linear regions responsible for N protein interaction(s), we used peptide scanning in which N peptides synthesized on membranes recognize recombinant TULV N protein. The data showed that the N protein bound to membrane-bound peptides comprising amino acids 13 to 30 and 41 to 57 in the N-terminal part and 340 to 379, 391 to 407, and 410 to 419 in the C-terminal part of the molecule. Further mapping of the interaction regions by alanine scanning indicated the importance of basic amino acids along the N protein and especially asparagine-394, histidine-395, and phenyalanine-396 in forming the binding interface. Analysis of truncated mutants in the mammalian two-hybrid assay showed that N-terminal amino acids 1 to 43 are involved in and C-terminal amino acids 393 to 398 (VNHFHL) are absolutely crucial for the homotypic interactions. Furthermore, our data suggested a tail-to-tail and head-to-head binding scheme for the N proteins.
Hantaviruses (genus Hantavirus, family Bunyaviridae) are enveloped viruses with three negative-sense single-stranded RNA genome segments. They encode the RNA-dependent RNA polymerase, two integral membrane glycoproteins (G1 and G2), and nucleocapsid (N) protein. Many rodent species carry hantaviruses, and they can be transmitted via aerosolized excreta to humans, in whom they can cause hemorrhagic fever with renal syndrome or hantavirus pulmonary syndrome (for a review, see reference 13).
The N protein is the major structural component in virions; it accumulates in the cytoplasm of infected cells. The three-dimensional structure of N protein has not been solved yet, but some functionally important regions have been defined. Most importantly, hantavirus N protein has been shown to bind RNA (4, 14, 15). Binding of the N protein to viral RNA is a necessary step during ribonucleoprotein (RNP) formation and viral replication. The RNA binding domain of the Hantaan virus N protein has been mapped between amino acids 175 and 217 (18). Hantavirus N protein has been shown to associate with membranes via electrostatic interactions, and the C-terminal amino acids were responsible for the N protein Golgi localization in the perinuclear area (11).
The N protein is capable of taking part in many protein-protein interactions. An earlier study showed that transportation of newly synthesized RNPs to the plasma membrane, where Black Creek Canal hantavirus is assembled, could be mediated through actin filaments (12). Recently, other cellular proteins have also been reported as binding partners for the N protein, i.e., Daxx and SUMO-1 (5, 8, 10).
We have been interested in homotypic interactions of hantavirus N protein. In our previous paper, direct protein-protein interaction was reported for Puumala virus and Tula virus (TULV) N proteins (6). The interaction was found to be noncovalent and enhanced by divalent cations. Furthermore, our preliminary mammalian two-hybrid data indicated that the interaction region is located within the C-terminal 125 amino acid residues. Homotypic interaction of Sin Nombre virus N protein has been also reported (1). The N-terminal 40 amino acids and C-terminal 72 amino acids were shown to participate in the interaction (1). It seems that N protein can form trimers, both in vitro and in vivo; these trimers can further act as assembly intermediates during RNP formation (1, 6). Further studies indicated the importance of the coiled-coil domains for the trimerization process presumably formed in the N-terminal region of the N protein molecule (2).
In the present study, we used three approaches, peptide scanning, alanine scanning, and the mammalian two-hybrid assay, to map the regions involved in TULV N-N interactions, and we discuss a binding scheme that may be applicable to all hantavirus N proteins.
MATERIALS AND METHODS
Peptide scanning.
The N-terminal 75 and C-terminal 125 amino acids of TULV N protein were synthesized as 15-mer overlapping peptides with one-residue shifts on cellulose filter with the Abimed Autospot robot ASP 222. The filter was blocked overnight at 4°C with 3% bovine serum albumin in TBST (10 mM Tris [pH 7.4], 150 mM NaCl, 0.05% Tween 20) and incubated with baculovirus-expressed TULV N protein (6) at a concentration of 1 μg/ml in TBST for 1 h at room temperature. The peptide-bound N protein was transferred electrophoretically to a nitrocellulose membrane and detected with mouse anti-N monoclonal antibody 3D3 (1 μg/ml, 2 h, room temperature) (9), and with horseradish peroxidase-conjugated mouse antibody (1:1,000) (Dako A/S) (antibodies diluted in 1% skim milk powder-0.05% Tween 20-20mM Tris, pH 8-1 mM EDTA-50 mM NaCl) with enhanced chemiluminescence.
Alanine scanning.
The following peptides were synthesized by changing each amino acid one by one to alanine (amino acids in the TULV N protein sequence are shown in parentheses): (13 to 30) RHEQQIVIARQKLKDVEK; (41 to 58) KSTLQSRRAAVSALEDKL; (394 to 411) NHFHLGDDMDPELRTLAQ; and (407 to 424) RTLAQSLIDQKVKEISNQ. Then 20 μl of in vitro-translated [35S]methionine-labeled TULV N protein (in 5 ml of TBST) was incubated with the membrane for 1 h at room temperature. Bound protein was detected by direct exposure on X-ray film.
In vitro transcription-translation.
N protein was produced from plasmid pcDNA3-N with the TNT coupled reticulocyte lysate system kit according to the manufacturer's instructions (Promega). For radiolabeling, Redivue l-[35S]methionine (>1,000 Ci/mmol) was purchased from Amersham Pharmacia Biotech. The labeled N protein band in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels was visualized by fluorography with Amplify reagent (Amersham Pharmacia Biotech).
Expression of truncated N proteins.
Plasmids encoding the full-length N protein of TULV strain Tula/Moravia/Ma5302V and truncated constructs (N44-429, N1-304, N1-329, N1-379, N1-386, N1-392, N1-398, N1-404, N1-416, N1-421, and N1-425) were created by PCR from a cDNA clone containing the complete sequence of the S segment (17). The PCR amplicons were cloned into the mammalian two-hybrid assay plasmids pM1 (GAL4 DNA-binding domain [DBD]) and/or pVP16 (VP16 activation domain [AD]) (BD Biosciences Clontech) between the BamHI and XbaI restriction sites. All the plasmids were characterized by restriction analysis and sequenced with the ABI Prism dye terminator sequencing kit (Perkin-Elmer).
COS7 cells were cultivated in Eagle's minimal essential medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, penicillin, and streptomycin on six-well culture plates to 70% confluence and transfected with 1 μg of pM1 or pVP16 DNA (expressing different N protein constructs). Transfections were done with 6 μl of FuGene6 reagent for each transfection according to the manufacturer's instructions (Boehringer Mannheim). After 24 h, cell lysates were prepared in 100 μl of 1% Triton X-100-phosphate-buffered saline with an EDTA-free cocktail of protease inhibitors. The proteins were separated on SDS-8% polyacrylamide gels and transferred onto a nitrocellulose membrane. Immunoblotting was done with monoclonal antibodies against GAL4 and VP16 (Santa Cruz Biotechnology, Inc.), as described above.
Mammalian two-hybrid system.
HeLa cells were cultivated in minimal essential medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, penicillin, and streptomycin on 24-well culture plates to 70% confluence and transfected with 0.5 μg of pM1 and pVP16 DNA (expressing different N protein constructs), 0.5 μg of reporter DNA pG5luc (expressing firefly luciferase), and 0.01 μg of control DNA pRL-SV40 (expressing Renilla luciferase) (Promega). Transfections were performed in triplicate with 9 μl of FuGene6 reagent for each transfection according to the manufacturer's instructions (Boehringer Mannheim). After 24 h, the luciferase activities were determined with the dual-luciferase reporter assay system (Promega). Light intensities of samples were measured with a DCR-1 luminometer (Digene Diagnostics Inc.). The linear range of light detection for the luminometer was determined with QuantiLum recombinant luciferase according to the instructions (Promega). Due to inherent variations, the Renilla luciferase values were used to normalize firefly luciferase values by the formula: normalized value of experiment A = [(Renilla luciferase value from N − N/Renilla luciferase value of experiment A) × firefly luciferase value of experiment A. The formula for interaction: %= (normalized value of experiment A/normalized value of N − N) × 100.
RESULTS
Peptide scanning for mapping N protein interaction regions.
Our preliminary data obtained with the mammalian two-hybrid assay suggested that the regions involved in the N-N interaction are located within the last 125 amino acid residues of the TULV N protein (6). For the Sin Nombre virus N protein, it has been shown that the regions involved in the interactions are located within the first 40 and the last 72 amino acid residues (1) (see also Fig. 4). Based on these observations, we carried out peptide scan screening of the 62 N-terminal and 125 C-terminal amino acid residues to map linear binding sites in the N protein. The corresponding sequences were synthesized as 15-mer overlapping peptides with one-residue shifts attached to cellulose membrane. Binding of the recombinant N protein produced in the baculovirus expression system (6) to these peptides was detected by immunoblotting. The spot intensities were supposed to correlate with the strength of interaction between the recombinant N protein and a given peptide. Each spot was then mapped to the N protein primary sequence.
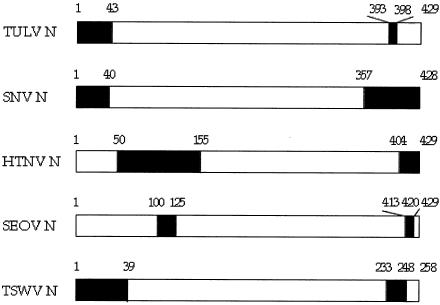
FIG. 4.
Schematic drawing of homotypic interaction regions of different hantavirus and tospovirus TSWV N proteins. The interaction regions (in black) were determined by two-hybrid assays.SNV. SinNombre virus; HTNV, Hantaan virus; SEOV, Seoul virus; TSWV, •• virus.
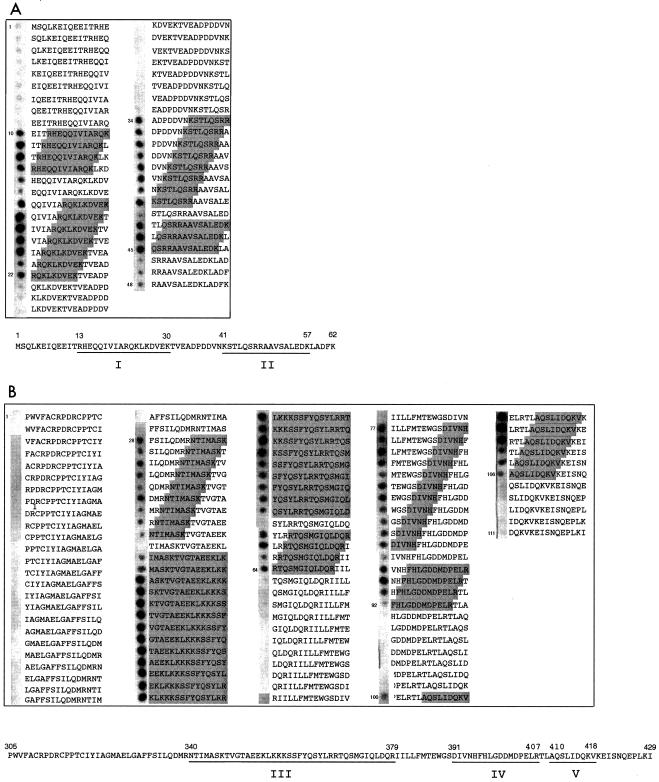
In the N-terminal part of the N protein molecule, several peptides were found to bind the recombinant N protein (Fig. 1a, gray boxes). Two binding regions consisting of adjacent peptides were identified: binding region I was presented by peptides 10 to 22 (amino acids 13 to 30), and binding region II was presented by peptides 34 to 45 (amino acids 41 to 57) (Fig. 1a). In the C terminus, due to several overlapping binding regions, binding region III peptides from 28 to 64 (amino acids 340 to 379) was seen, region IV peptides from 77 to 92 (amino acids 391 to 407), and region V peptides from 100 to 106 (amino acids 410 to 419) were detected as well (Fig. 1b, gray boxes). The reactivity of some peptides in the middle of the binding regions, e.g., peptides 15 and 42 (Fig. 1a) and peptides 37 and 60 (Fig. 1b), was lower or undetectable. This indicates that the binding region may consist of separate subregions or that not all peptides can obtain the conformation required for the interaction. 35S-labeled N protein, produced in a coupled transcription-translation system, showed essentially the same pattern of reactivity as the recombinant N protein (data not shown).
FIG. 1.
Identification of linear regions involved in the TULV N-N interaction. Peptide scanning analysis of the N-terminal 62 amino acid residues (A) and the C-terminal 125 amino acid residues (B). Gray boxes indicate amino acids that are involved in binding. Shading of an entire peptide sequence indicates that it belongs to one of the overlapping binding regions. The amino acid sequences below summarize the peptide spotting results; interaction regions are underlined.
Alanine scanning.
Alanine scanning was used to pinpoint the amino acids that are primarily responsible for the interaction. Regions I, II, IV, and V were included in the alanine scanning analysis. Peptides were synthesized on a membrane similarly as for peptide scanning, but in this case each amino acid was replaced with alanine one by one. In this experiment, in vitro-produced 35S-labeled N protein was used to make detection easier. The labeled N protein was incubated with the membrane, and bound radioactivity was detected (Fig. 2). In region I, residues R13, K24, K26, and K30 seemed to be involved in the binding, as the peptides in which these residues were replaced with alanines showed reduced binding compared to the parental peptide (Fig. 2a). Correspondingly, residues R47 and R48 in region II (Fig. 2b), residues N394, H395, F396 and R407 in region IV (Fig. 2c), and residues K417 and K419 in region V (Fig. 2d) were important for binding. Taken together, the alanine scanning results showed that positively charged amino acid residues (lysine and arginine) are involved in TULV N protein binding. In region IV, polar, uncharged amino acid residues (N394 and H395) at neutral pH and the aromatic F396 were crucial as well.
FIG. 2.
Identification of important amino acid residues by alanine scanning. The alanine scanning assay was performed for the TULV N peptides comprising amino acids 13 to 30 (a), 41 to 58 (b), 394 to 411 (c), and 407 to 424 (d). The parental sequence is boxed. The arrows show the amino acid residues that reduced N protein binding to a given peptide when replaced with alanine.
Mapping of interaction regions with the mammalian two-hybrid assay.
As an alternative approach to more precise mapping of the TULV N-N interaction regions, the mammalian two-hybrid assay was carried out. The full-length N protein (N1-429), the N-terminal deletion mutant N44-429, and C-terminal deletion mutants were coexpressed as GAL4 DNA binding domain and VP16 activation domain fusions (Fig. 3a). To ensure that the lack of interaction in the assay was not due to unsuccessful protein expression, all the constructs expressed as DNA-binding domain and/or activation domain fusions were checked by immunoblotting (data not shown). Measurement of firefly luciferase activity from HeLa cell lysates allowed quantification of the interaction between the two fusion proteins. To verify the specificity of the interactions, each fusion protein was tested together with the nonfused GAL4 DNA-binding domain or VP16 activation domain. None of those combinations was able to activate firefly luciferase activity (data not shown). To normalize the firefly luciferase values for transfection efficiency, Renilla luciferase activity was measured in the same lysates.
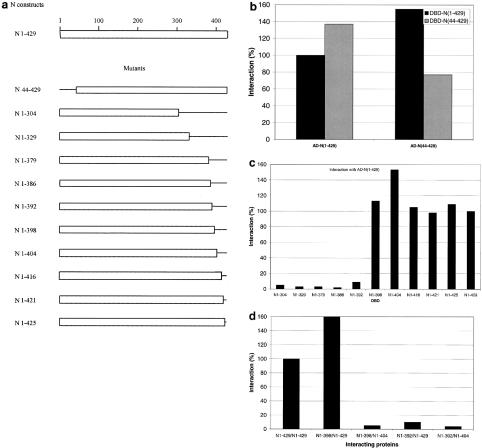
FIG. 3.
Summary of mammalian two-hybrid assay results. (a) Schematic representation of the truncated TULV N proteins used to assay N-N interactions. For the assay, HeLa cells were transfected with DBD-N- and AD-N-expressing plasmids depicted below the columns together with reporter plasmids expressing firefly luciferase and Renilla luciferase. Firefly luciferase and Renilla luciferase values were determined, the firefly luciferase values were normalized to the Renilla luciferase values, and the interaction was calculated (see Materials and Methods). (b) The interactions of N1-429 and N44-429 with different DNA-binding domain and activation domain combinations. (c) The interactions of different C-terminally deleted DBD-N proteins with full-length AD-N protein. (d) The interactions of N proteins with C-terminal deletions in both partners.
First, the involvement of the N-terminal and C-terminal regions in the interaction was studied with constructs N44-429 and N1-304, respectively. Removal of the N-terminal region (DBD-N44-429 or AD-N44-429) did not reduce the interaction (Fig. 3b). An interaction of greater than 100% could appear if truncation of the N protein exposes regions that are hidden in the native protein. However, when 43 amino acid residues were deleted from both constructs, the firefly luciferase activity was reduced to 77% of the control value (Fig. 3b). The same effect was seen in another experiment, i.e., it was reproducible. The results indicated that (i) the N-terminal region is involved in the homotypic interaction and (ii) other regions participate in the interaction, too. On the contrary, the C-terminal deletion mutant N1-304 did not interact with the full-length N protein at all, indicating that the last 125 amino acid residues were crucial for the interaction (Fig. 3c). This region was selected because the last 125 amino acid residues are highly conserved among all hantavirus N proteins. Therefore, further studies were focused on mapping the C-terminal region.
To map the C-terminal interaction region(s) in more detail, other deletion constructs, N1-329, N1-379, N1-386, N1-392, N1-398, N1-404, N1-416, N1-421, and N1-425, were prepared. These constructs were assayed with the full-length N protein AD-N1-429 (Fig. 3c). While N1-329, N1-379, N1-386, and N1-392 did not interact, N1-398, N1-404, N1-416, N1-421, and N1-425 proteins showed full binding activity. These results indicated that the region between amino acid residues 393 and 398 (VNHFHL) is crucial for the interaction and that the last 31 amino acid residues are not directly involved.
Next, the interaction of N1-398, the shortest N protein mutant that interacted with full-length N protein, was tested with N1-404 (Fig. 3d). These two C-terminally truncated proteins were not able to interact with each other, indicating that the interaction between N proteins occurs via C-terminal regions. Thus, this result supports a tail-to-tail binding model. As a negative control, N1-392 was tested as well (Fig. 3d). Furthermore, C-terminal mutants N1-416, N1-421, and N1-425 showed full binding with the N-terminally truncated N protein N44-429, indicating that removal of the 43 N-terminal amino acids does not have any effect on the interaction capacity of the C-terminal part of the molecule unless crucial amino acids are not removed (data not shown). Taken together, the mammalian two-hybrid data revealed two regions involved in the homotypic interaction: a secondary interaction region among amino acids 1 to 43 in the N-terminal part and a primary interaction region involving amino acids 393 to 398 (VNHFHL) in the C-terminal part of the molecule.
Comparison of mammalian two-hybrid results with peptide scanning.
The secondary interaction region (amino acids 1 to 43) included region I and part of region II, which were seen in the peptide scanning assay. The primary interaction region (amino acids 393 to 398, VNHFHL) is included in region IV. Thus, the regions mapped by the mammalian two-hybrid assay partially overlap the regions defined by peptide scanning. The likely reason why different regions were found is that these two methods may deal with different aspects of the interaction. In the peptide scanning assay, N protein bound to linear binding sites presented by the peptides. In the mammalian two-hybrid assay, some linear epitopes can be hidden, and conformational binding sites can be recognized as well (see Discussion).
DISCUSSION
In this study, homotypic interaction regions of TULV hantavirus N protein were mapped with a peptide scanning assay and the mammalian two-hybrid system. The peptide scanning assay can locate linear sequences involved in the interaction; in the mammalian two-hybrid assay, both linear and discontinuous epitopes are detected. The mammalian two-hybrid system has been increasingly used to study the interactions of cellular proteins because mammalian proteins can undergo proper posttranslational modifications and therefore are more likely to gain their native conformation. Interactions of viral proteins e.g., coxsackievirus B3 proteins 2B, 2C, and 2BC, have been studied with this method as well (3).
The results obtained with the mammalian two-hybrid system pointed out that the contribution of the N- and C-terminal regions in the N-N interaction is different. The N-terminal region is involved, but the C-terminal region is absolutely crucial for the interactions. The N-terminal interaction region was mapped within the first 43 amino acid residues, and the C-terminal interaction region was mapped with high precision between amino acids 393 and 398 (VNHFHL). Secondary-structure prediction algorithms showed the presence of α-helices formed by amino acids 374 to 394 and amino acids 404 to 421 (Vibhor Kumar and Peter Engelhardt, personal communication). When the second helix was removed from one of the two interacting N protein molecules (N1-398) in the mammalian two-hybrid assay, the interaction still occurred, but the shorter mutant N1-392 was not able to interact (Fig. 3c). Taken together, these observations suggest that removal of V393 and N394 may disrupt the α-helix and thus destroy the interaction ability of the N protein. Consequently, residues 393 to 398 may not be directly involved in the interaction but, instead, be a part of the scaffolding structure which forms the interface for the interaction.
The fact that these residues are almost completely conserved among different hantavirus species supports the idea that the region is involved in performing some very basic functions common to all hantaviruses. In the model we currently favor, the second helix formed by amino acids 404 to 421 contacts another monomer, and thus the trimer formation which was suggested to be the initial step for hantavirus RNP assembly (1, 6) occurs via these helix “protrusions” from one monomer to another. This model is supported by the data showing that removal of the last α-helix from one of the interacting partners in the mammalian two-hybrid system was not enough to disrupt the interaction, but when helixes were removed from both partners, the interaction was abolished (Fig. 3d).
When amino acid residues important for the N-N interaction were searched by alanine scanning, the data pointed out that basic amino acid residues (K and R) are crucial for binding. This is in agreement with our earlier findings showing that N protein molecules interact via electrostatic forces, e.g., positively charged amino acid residues may facilitate the process (6). Although the possibility that positively charged peptides can mediate nonspecific interactions with the N protein cannot be completely ruled out, it seems less likely than their involvement in a specific action. Notably, amino acid residues N394, H395, and F396 were found to be important for the interaction, indicating the involvement of nonbasic amino acid residues as well. Taken together, the mammalian two-hybrid and alanine scanning results indicated that the same region took part in the interaction, either directly or indirectly.
Our data are in good agreement with the observations made for Sin Nombre virus N protein (1), showing that the C-terminal (amino acids 357 to 428) and the N-terminal (amino acids 1 to 40) regions are involved in the homotypic interaction. Most recently, a study of Hantaan virus, Seoul virus, and Dobrava virus N protein homotypic interactions was published (19). In that paper, the authors used the yeast two-hybrid assay and competitive enzyme-linked immunosorbent assay for mapping the interaction regions. The C-terminal interaction region was mapped within amino acids 404 to 429 for Hantaan virus and amino acids 413 to 420 for Seoul virus. These observations do not exactly match our results but show the involvement of the C-terminal region. For the N-terminal interaction region, Yoshimatsu et al. pointed out amino acids 50 to 155 for Hantaan virus and amino acids 100 to 125 for Seoul virus. This is in contrast to our data on TULV and also to the data of Alfhadli et al. on Sin Nombre virus (1). The reasons for these contradictions remain unclear.
Hantavirus N proteins are very similar both in length (428 to 433 amino acids) and in primary sequence. Some regions, e.g., amino acids 396 to 404, are identical in all hantaviruses, most probably due to some basic functions they perform, e.g., interactions with viral RNA and with other N protein molecules. This suggests similar modes of folding, similar functional domains, and common properties for all hantavirus N proteins.
Our peptide scanning results showed that two regions in the N terminus (amino acids 13 to 30 and 41 to 57) and three regions in the C terminus (amino acids 340 to 379, 391 to 407, and 410 to 419) are involved in the interaction. It seems that an epitope(s) recognized by the peptide scanning assay is mainly that involved in the multimerization step in hantavirus RNP assembly, since purified, recombinant N protein is mainly in the multimeric form (i.e., trimers, dimers, and higher oligomers); monomers are in the minority (6).
The combined data from peptide scanning and the mammalian two-hybrid assay presented here show that the regions involved in multimerization and trimerization are at least partially separated. For the multimerization process, at least two different binding surfaces in the N protein are required, one for the trimerization of monomers, and another for the multimerization of trimers. Similarly, in hepatitis B virus, separate regions involved in core protein dimerization and multimerization were identified by a combination of yeast two-hybrid and peptide scanning assays. The results showed that amino acids 78 to 117 in the core protein form part of the dimer interface, while amino acids 113 to 143 are responsible for dimer multimerization (7).
The involvement of separate binding regions for trimerization and multimerization may explain why N-terminally truncated Seoul virus N proteins (amino acids 155 to 429) interacted in the competitive enzyme-linked immunosorbent assay but failed to do so in the yeast two-hybrid system (19). Since the enzyme-linked immunosorbent assay and two-hybrid system cannot distinguish between N protein trimerization and multimerization, it is impossible to conclude whether this was the case with Seoul virus N protein as well. However, the authors are in the same line by reasoning that the contradictory results of the enzyme-linked immunosorbent assay and the two-hybrid system were caused by the differences in the maturation stage of the nucleocapsid (19). Trimer multimerization during nucleocapsid assembly needs further studies.
Recently, several reports on the interaction regions of hantaviral N proteins have been published (Fig. 4). The consensus appears to be that homotypic interaction regions are located near the N and C termini of the polypeptide chain. The fact that an N-terminal deletion mutant of TULV (N44-429) is still able to interact (Fig. 3b) suggests the head-to-head and tail-to-tail interaction model for the TULV N protein. It seems that Sin Nombre virus N protein utilizes a similar interaction mode (2). Furthermore, the C-terminal regions (amino acids 155 to 429) of the Hantaan virus and Dobrava virus N proteins were sufficient for maintaining the multimers in competitive enzyme-linked immunosorbent assay, suggesting a head-to-head and tail-to-tail binding model (19). In another bunyavirus, the tomato spotted wilt virus (TSWV), the N protein interaction regions are located similarly in the N and C termini (amino acids 1 to 39 and 233 to 248) but opposite their positions in hantavirus N proteins, and the authors postulated a head-to-tail interaction for TSWV virus N proteins (16).
Several studies on hantavirus N protein homotypic interactions have been published in recent years. The interaction regions have been mapped and suggestions about trimerization have been made. Further studies are needed on the N protein multimerization process and on N protein interactions with both viral RNA and RNA polymerase to understand RNP assembly in greater detail.
Acknowledgments
We thank Hilkka Lankinen and Jussi Hepojoki for help with peptide synthesis. Vesa Koistinen, Kirsi Tulimäki, and Leena Kostamovaara are acknowledged for advice and assistance.
This work was supported by EU contracts QLK2-CT-1999-01119 and QLK2-CT-2002-01358, the Academy of Finland, and the Sigrid Jusélius Foundation, Helsinki.
REFERENCES
- 1.Alfadhli, A., Z. Love, B. Arvidson, J. Seeds, J. Willey, and E. Barklis. 2001. Hantavirus nucleocapsid protein oligomerization. J. Virol. 75:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfadhli, A., E. Steel, L. Finlay, H. P. Bachinger, and E. Barklis. 2002. Hantavirus nucleocapsid protein coiled-coil domains. J. Biol. Chem. 277:27103-27108. [DOI] [PubMed] [Google Scholar]
- 3.de Jong, A. S., I. W. Schrama, P. H. Willems, J. M. Galama, W. J. Melchers, and F. J. van Kuppeveld. 2002. Multimerization reactions of coxsackievirus proteins 2B, 2C and 2BC: a mammalian two-hybrid analysis. J. Gen. Virol. 83:783-793. [DOI] [PubMed] [Google Scholar]
- 4.Gott, P., R. Stohwasser, P. Schnitzler, G. Darai, and E. K. Bautz. 1993. RNA binding of recombinant nucleocapsid proteins of hantaviruses. Virology 194:332-337. [DOI] [PubMed] [Google Scholar]
- 5.Kaukinen P., A. Vaheri, and A. Plyusnin. 2003. Non-covalent interaction between nucleocapsid protein of Tula hantavirus and small ubiquitin-related modifier-1, SUMO-1. Virus Res. 92:37-45. [DOI] [PubMed] [Google Scholar]
- 6.Kaukinen, P., V. Koistinen, O. Vapalahti, A. Vaheri, and A. Plyusnin. 2001. Interaction between molecules of hantavirus nucleocapsid protein. J. Gen. Virol. 82:1845-1853. [DOI] [PubMed] [Google Scholar]
- 7.Konig, S., G. Beterams, and M. Nassal. 1998. Mapping of homologous interaction sites in the hepatitis B virus core protein. J. Virol. 72:4997-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, X. D., T. P. Makela, D. Guo, R. Soliymani, V. Koistinen, O. Vapalahti, A. Vaheri, and H. Lankinen. 2002. Hantavirus nucleocapsid protein interacts with the Fas-mediated apoptosis enhancer Daxx. J. Gen. Virol. 83:759-766. [DOI] [PubMed] [Google Scholar]
- 9.Lundkvist, A., O. Vapalahti, A. Plyusnin, K. B. Sjolander, B. Niklasson, and A. Vaheri. 1996. Characterization of Tula virus antigenic determinants defined by monoclonal antibodies raised against baculovirus-expressed nucleocapsid protein. Virus Res. 45:29-44. [DOI] [PubMed] [Google Scholar]
- 10.Maeda, A., B. H. Lee, K. Yoshimatsu, M. Saijo, I. Kurane, J. Arikawa, and S. Morikawa. 2003. The intracellular association of the nucleocapsid protein (NP) of hantaan virus (Hantaan virus) with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9). Virology 305:288-297. [DOI] [PubMed] [Google Scholar]
- 11.Ravkov, E. V., and R. W. Compans. 2001. Hantavirus nucleocapsid protein is expressed as a membrane-associated protein in the perinuclear region. J. Virol. 75:1808-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravkov, E. V., S. T. Nichol, C. J. Peters, and R. W. Compans. 1998. Role of actin microfilaments in Black Creek Canal virus morphogenesis. J. Virol. 72:2865-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severson, W., L. Partin, C. S. Schmaljohn, and C. B. Jonsson. 1999. Characterization of the Hantaan nucleocapsid protein-ribonucleic acid interaction. J. Biol. Chem. 274:33732-33739. [DOI] [PubMed] [Google Scholar]
- 15.Severson, W. E., X. Xu, and C. B. Jonsson. 2001. cis-Acting signals in encapsidation of Hantaan virus S-segment viral genomic RNA by its N protein. J. Virol. 75:2646-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhrig, J. F., T. R. Soellick, C. J. Minke, C. Philipp, J. W. Kellmann, and P. H. Schreier. 1999. Homotypic interaction and multimerization of nucleocapsid protein of tomato spotted wilt tospovirus: identification and characterization of two interacting domains. Proc. Natl. Acad. Sci. USA 96:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vapalahti, O., A. Lundkvist, S. K. Kukkonen, Y. Cheng, M. Gilljam, M. Kanerva, T. Manni, M. Pejcoch, J. Niemimaa, A. Kaikusalo, H. Henttonen, A. Vaheri, and A. Plyusnin. 1996. Isolation and characterization of Tula virus, a distinct serotype in the genus Hantavirus, family Bunyaviridae. J. Gen. Virol. 77:3063-3067. [DOI] [PubMed] [Google Scholar]
- 18.Xu, X., W. Severson, N. Villegas, C. S. Schmaljohn, and C. B. Jonsson. 2002. The RNA binding domain of the hantaan virus N protein maps to a central, conserved region. J. Virol. 76:3301-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshimatsu, K., B. H. Lee, K. Araki, M. Morimatsu, M. Ogino, H. Ebihara, and J. Arikawa. 2003. The multimerization of hantavirus nucleocapsid protein depends on type-specific epitopes. J. Virol. 77:943-952. [DOI] [PMC free article] [PubMed] [Google Scholar]