Abstract
Type 1 wild-vaccine recombinant polioviruses sharing a 367-nucleotide (nt) block of Sabin 1-derived sequence spanning the VP1 and 2A genes circulated widely in China from 1991 to 1993. We surveyed the sequence relationships among 34 wild-vaccine recombinants by comparing six genomic intervals: the conserved 5′-untranslated region (5′-UTR) (nt 186 to 639), the hypervariable portion of the 5′-UTR (nt 640 to 742), the VP4 and partial VP2 genes (nt 743 to 1176), the VP1 gene (nt 2480 to 3385), the 2A gene (nt 3386 to 3832), and the partial 3D gene (nt 6011 to 6544). The 5′-UTR, capsid (VP4-VP2 and VP1), and 2A sequence intervals had similar phylogenies. By contrast, the partial 3D sequences could be distributed into five divergent genetic classes. Most (25 of 34) of the wild-vaccine recombinant isolates showed no evidence of additional recombination beyond the initial wild-Sabin recombination event. Eight isolates from 1992 to 1993, however, appear to be derived from three independent additional recombination events, and one 1993 isolate was derived from two consecutive events. Complete genomic sequences of a representative isolate for each 3D sequence class demonstrated that these exchanges had occurred in the 2B, 2C, and 3D genes. The 3D gene sequences were not closely related to those of the Sabin strains or 53 diverse contemporary wild poliovirus isolates from China, but all were related to the 3D genes of species C enteroviruses. The appearance within approximately 2.5 years of five recombinant classes derived from a single ancestral infection illustrates the rapid emergence of new recombinants among circulating wild polioviruses.
Recombination is integral to poliovirus evolution. The first indication of a role for recombination in the natural evolution of polioviruses was the detection vaccine-related isolates with chimeric sequences excreted by children exposed to the trivalent oral poliovirus vaccine (OPV). These isolates were detected in patients with vaccine-associated paralytic poliomyelitis (6, 9, 12, 16, 18, 21, 22, 25) and in healthy OPV recipients (1, 3, 41). The heterologous sequences of most vaccine-related isolates were derived from the other Sabin OPV strains, with recombinants most frequently found among vaccine-related isolates of serotypes 2 and 3 (3, 11, 22). A small proportion of vaccine-related isolates have capsid sequences derived from the OPV strains and noncapsid sequences derived from other, nonvaccine viruses (10), a property consistently found among circulating vaccine-derived poliovirus isolates (15, 36, 45; B. Thorley, F. Paladin, and H. Shimizu, Abstr. XIIth Int. Congr. Virol., abstr. V-508, 2002). Recombination among the OPV strains is readily detectable because the sequences of the parental vaccine strains are well defined (42).
Recombination also occurs during the circulation of wild polioviruses (5, 23). We recently described natural type 1 wild-vaccine recombinants that circulated widely in China from 1991 to 1993 (23). These recombinants shared a 367-nucleotide (nt) block of sequence derived from Sabin 1 that spanned the VP1-2A gene junction. Phylogenetic analysis suggested that the wild-vaccine recombinants emerged from a mixed type 1 wild Sabin 1 infection in northern China in early 1991. Sequences encompassing the 3′ half of the 2A genes of the wild-vaccine recombinants were derived from a third virus (23), consistent with the active recombination found in the noncapsid region of the indigenous wild polioviruses from China (H.-M. Liu, L.-B. Zhang, O. M. Kew, and M. A. Pallansch, Abstr. 20th Annu. Meet. Am. Soc. Virol., abstr. W34-12, 2001). The generality of recombination in wild polioviruses (21, 35) was supported by the recent description of high heterogeneity in the 3D noncapsid sequences of wild type 1 and type 2 poliovirus isolates from India (5).
The existence of wild-vaccine poliovirus recombinants, derived from a recent common ancestor and having transmissibility properties indistinguishable from those of wild polioviruses, provided the opportunity to monitor the dynamics of poliovirus recombination during the approximately 2.5-year period of their circulation. We found evidence of four additional recombination events, which involved crossovers in the noncapsid 2B, 2C, and 3D genes, following the initial genetic exchange with Sabin 1 in 1991. Complete genomes of the five different classes of wild-vaccine recombinant isolates contained two to four discernible recombination sites relative to the genome of a closely related “nonrecombinant” indigenous type 1 wild poliovirus isolate. One class of recombinant was the product of at least three successive rounds of recombination estimated to have occurred between 1991 and 1993. The donors of the heterologous noncapsid sequences were probably not restricted to the polioviruses indigenous to China but appeared to include other species C enteroviruses related to the polioviruses.
MATERIALS AND METHODS
Poliovirus isolates.
The 34 type 1 wild-vaccine recombinant polioviruses (genotype CHN-R91 [abbreviated R91]) isolated in China from 1991 to 1993 (Table 1) have been described previously (23). A wild poliovirus from genotype CHN-JX89 (abbreviated JX89), Henan/91-3, which showed the highest nucleotide identity with wild-vaccine recombinants in the wild VP1 gene interval, was included as a reference strain. All isolates were obtained from children with acute flaccid paralysis (AFP) as part of the national poliovirus surveillance system in China (46). All of these viruses were isolated in the laboratories of the provincial antiepidemic stations of the People's Republic of China. Viruses were further propagated in RD cell monolayers (human rhabdomyosarcoma cell line; ATCC CCL 136).
TABLE 1.
Type 1 poliovirus isolates sequenced in this study
| Isolate | Laboratory no.a | Location | Date (day mo yr) of:
|
R91 class | Lineagea | |
|---|---|---|---|---|---|---|
| Onset | Sampling | |||||
| Hebei/91-1 | 3648b | Cixian, Hebei | 30 Mar. 1991 | 19 Apr. 1991 | R91-1 | Root |
| Hebei/91-2 | 3645b | Anxin, Hebei | 15 Apr. 1991 | 24 Apr. 1991 | R91-1 | Root |
| Hebei/91-3 | 3646b | Zaoqiang, Hebei | 21 Apr. 1991 | 15 May 1991 | R91-1 | Root |
| Nei Mongol/91-1 | 3640b | Wumeng, Nei Mongol | 21 June 1991 | 2 July 1991 | R91-1 | E |
| Shanxi/91 | 3639b | Datong, Shanxi | 15 July 1991 | 15 July 1991 | R91-1 | F1 |
| Nei Mongol/91-2 | 3641b | Wumeng, Nei Mongol | 10 June 1991 | 18 July 1991 | R91-1 | F1 |
| Henan/91-1 | 3650b | Kaifeng, Henan | 28 Sept. 1991 | 1991 | R91-1 | A1 |
| Henan/91-2 | 3651b | Xinzhen, Henan | 3 Oct. 1991 | 1991 | R91-1 | A1 |
| Henan/91-3 | 3653b,c | Zhenping, Henan | 3 Oct. 1991 | 1991 | JX89 | |
| Henan/91-4 | 3652b | Fugou, Henan | 10 Nov. 1991 | 1991 | R91-1 | A1 |
| Fujian/91-1 | 6331 | Pinghe, Fujian | 1991 | 14 Nov. 1991 | R91-1 | G1 |
| Fujian/91-2 | 6330 | Pinghe, Fujian | 1991 | 12 Dec. 1991 | R91-1 | G1 |
| Henan/91-5 | 3649b | Yuanyang, Henan | 1991 | 1991 | R91-1 | D |
| Sichuan/92 | 5160 | Naxi, Sichuan | 24 Jan. 1992 | 1992 | R91-1 | F2 |
| Fujian/92-1 | 6333 | Jinjiang, Fujian | 5 Nov. 1992 | 9 Nov. 1992 | R91-1 | F4 |
| Fujian/92-2 | 6334 | Jinjiang, Fujian | 10 Dec. 1992 | 17 Dec. 1992 | R91-1 | F4 |
| Guangdong/92-1 | 5158 | Guangdong | 1992 | 1992 | R91-3 | C1 |
| Guangdong/92-2 | 5157 | Guangdong | 1992 | 1992 | R91-3 | C2 |
| Hainan/92-1 | 5153 | Haikou, Hainan | 1992 | 1992 | R91-3 | C1 |
| Hainan/92-2 | 5154 | Dongfang, Hainan | 1992 | 1992 | R91-3 | C2 |
| Hainan/92-3 | 5730 | Hainan | 1992 | 1992 | R91-3 | C1 |
| Yunnan/92 | 6421 | Gengma, Yunnan | 1992 | 1992 | R91-2 | A2 |
| Fujian/93-1 | 6340 | Zhangpu, Fujian | 1 Apr. 1993 | 1993 | R91-1 | G1 |
| Fujian/93-2 | 6337 | Pinghe, Fujian | 12 Apr. 1993 | 1993 | R91-1 | G2 |
| Fujian/93-3 | 6339 | Zhangpu, Fujian | 24 Apr. 1993 | 1993 | R91-1 | G1 |
| Fujian/93-4 | 6338 | Zhangpu, Fujian | 27 Apr. 1993 | 1993 | R91-1 | G1 |
| Fujian/93-5 | 6335 | Ningde, Fujian | 29 Apr. 1993 | 1993 | R91-1 | F4 |
| Fujian/93-6 | 6341 | Fuding, Fujian | 9 July 1993 | 1993 | R91-1 | F4 |
| Fujian/93-7 | 5749 | Fujian | 1993 | 1993 | R91-1 | G1 |
| Fujian/93-8 | 6343 | Zhangpu, Fujian | 23 Oct. 1993 | 1993 | R91-5 | G1 |
| Guangdong/93-1 | 5738 | Guangdong | 1993 | 1993 | R91-1 | B |
| Guangdong/93-2 | 5739 | Guangdong | 1993 | 1993 | R91-1 | B |
| Hainan/93-1 | 5731 | Hainan | 1993 | 1993 | R91-3 | C1 |
| Hainan/93-2 | 5732 | Hainan | 1993 | 1993 | R91-4 | C2 |
| Qinghai/93 | 5742 | Qinghai | 1993 | 1993 | R91-1 | F3 |
Laboratory numbers and lineage designations are from Liu et al. (23).
Isolate contributed by Z.-Y. Fang, Institute of Virology, Chinese Center for Disease Control and Prevention, Beijing, China.
Nonrecombinant wild poliovirus isolate closely related to recombinant group.
Extraction of RNA, RT-PCR, and sequencing.
The methods used for poliovirus viral RNA extraction, in vitro reverse transcription (RT) and PCR amplification, and sequencing of RT-PCR amplicons have been described previously (23). Two RT-PCR primer pairs, L1S (sense; positions 166 to 185, 5′-CACTTCTGTCTCCCCGGTGA-3′) and L2A (antisense; 737 to 756, 5′-ACCTGAGCACCCATTATGAT-3′) and L3S (sense; 621 to 639, 5′-TTGGATTGGCCATCCGGTG-3′) and L4A (antisense; 1177 to 1196, 5′-GTAGTTTCCACCACCACCCT-3′) were used to amplify and sequence the 5′-untranslated region (5′-UTR) and VP4 and partial VP2 genes (VP4-VP2 gene). RT-PCR primer pair L5S (sense; 5756 to 5775, 5′-AGTAAGTACCCCAACATGTA-3′) and L6A (antisense; 6921 to 6940, 5′-TAAATCTATGCCCTTGTAGG-3′) and inner sequencing primer L7A (antisense; 6563 to 6582, 5′-TGAAAAGCAGCATACAGGTT-3′) were used to obtain the partial 3D gene sequences of seven wild-vaccine recombinant isolates (Guangdong/92-1, Guangdong/92-2, Hainan/92-1, Hainan/92-2, Hainan/92-3, Hainan/93-1, and Yunnan/92) and wild isolate Henan/91-3. Primer pair L8S (sense; 5990 to 6009, 5′-GAGATTCAGTGGATGAGACC-3′) and L9A (antisense; 6545 to 6561, 5′-CCAAAGGCCATTCTCAT-3′) was used to amplify and sequence the PCR amplicons for the 3D region for the other 27 isolates. Additional amplification and sequencing primers were used to obtain the complete genome sequences of wild-vaccine recombinants Fujian/93-8, Guangdong/92-2, Hainan/93-2, Hebei/91-2, and Yunnan/92 and of wild poliovirus isolate Henan/91-3. Terminal sequences were determined by using the 5′ and 3′ rapid amplification of cDNA ends system kits (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions.
Sequence analysis.
Sequence data were analyzed with programs in the Wisconsin Sequence Analysis Package, version 10.2 (Accelrys Inc., Madison, Wis.). Phylogenetic trees were constructed from the six different sequence intervals of the 34 recombinant isolates or from the five recombinant complete genomes with the DNA maximum-likelihood method (DNAML) (8) in the PHYLIP software package, version 3.57 (J. Felsenstein, PHYLIP [phylogeny inference package], version 3.5c, Department of Genetics, University of Washington, 1993). The input transition/transversion (Ts/Tv) ratio for DNAML tree construction was estimated from each sequence data set (conserved 5′-UTR, 5.8; variable 5′-UTR, 4.3; VP4-VP2, 12.8; VP1, 8.1; 2A, 7.7; partial 3D, 4.1; complete 3D, 2.8) by the program TREE-PUZZLE, version 5.0 (37), applying the HKY model of nucleotide substitution (13). Phylograms were displayed with the program TreeExplorer, version 2.12 (Koichiro Tamura, Department of Biological Sciences, Tokyo Metropolitan University [http://evolgen.biol.metro-u.ac.jp/TE/TE_man.html]). Radial trees were displayed with the program TreeView (32). Plots of nucleotide similarity between poliovirus isolates were created by use of the program SimPlot (24) to visualize the percentage of nucleotide sequence identity between two poliovirus isolates along the complete genome with a sliding window of 100 nt and moving 50 nt each step. Distances were corrected for superimposed substitutions by the Jukes and Cantor method (14).
Estimation of evolution rate in different genomic intervals.
Rates of fixation of synonymous base substitutions in the VP4-VP2 gene, VP1 gene, 2A gene, and partial 3D gene were estimated as previously described (23), from the sequence differences among 21 wild-vaccine recombinants for which the date of specimen collection or onset of paralysis was available. The rate of evolution at all nucleotide positions was also estimated for the conserved and hypervariable regions of the 5′-UTR and for the VP4-VP2 gene, VP1 gene, 2A gene, and partial 3D gene. The genetic distances at all nucleotide positions were computed according to the Kimura two-parameter model (17) of the Distances program in the Wisconsin Sequence Analysis Package to correct for multiple substitutions at the same site. Genetic distance values from the root sequence (that of Hebei/91-2) were plotted as a function of the date of specimen collection (zero time: 24 April 1991). The evolution rates at all nucleotide positions were estimated by linear regression of genetic distance over the 2.5-year period. Isolate Fujian/93-8 was not included for estimation of the 3D gene evolution rate because an additional recombination site is located in the sequence interval of its 3D gene.
Numbering of nucleotide and amino acid positions.
The coding sequences of all the poliovirus isolates in this study were colinear; however, some of the 5′-UTR sequences differed from one another by a small number of insertions or deletions. To facilitate comparisons, numbering of the nucleotide positions of all isolates followed that described for the Sabin type 1 strain (28). Amino acid positions were indicated by the name of the viral protein and numbered consecutively from residue 1 of each protein. Amino acid substitutions were indicated by the following convention: protein:original residue, position, substituted residue(s). For example, VP4:F46L indicates a phenylalanine-to-isoleucine substitution at amino acid position 46 of VP4.
Nucleotide sequence accession numbers.
The complete genomic sequences of five wild-vaccine recombinant polioviruses (Fujian/93-8, Guangdong/92-2, Hainan/93-2, Hebei/91-2, and Yunnan/92) and the closely related wild poliovirus (Henan/91-3) have been submitted to GenBank under accession numbers AF111981, AF111961, AF111966, AF111953, AF111982, and AF111983, respectively, which replace the previously submitted VP1-2A nucleotide sequences (23). The nucleotide sequences of the other 29 wild-vaccine recombinant poliovirus isolates were submitted to GenBank under accession numbers AF518078 to AF518097 and AY017229 to AY017246 for the 5′-UTR-VP4-VP2 interval and numbers AF518076 to AF518086 and AY017247 to AY017264 for the partial 3D genes.
RESULTS
Comparison of complete genomic sequences of an early wild-vaccine recombinant and a closely related indigenous wild poliovirus.
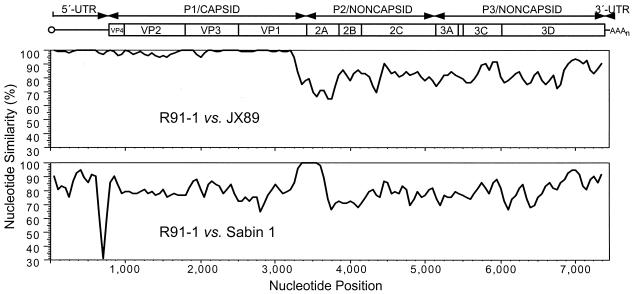
We had previously shown that the early wild-vaccine recombinant isolate Hebei/91-2 had very high nucleotide sequence identity (99.5%) with the indigenous wild poliovirus isolate Henan/91-3 of genotype JX89 in the interval of the VP1 gene (nt 2480 to 3270) derived from wild type 1 poliovirus (23). The sequence identity was much lower (78.2%) across the VP1-2A gene junction (nt 3271 to 3637), where a 367-nt block of sequence derived from Sabin 1 (100% nucleotide identity to Sabin 1) was found in the Hebei/91-2 genome. The sequence identity between the two isolates was even lower (73.8%) in the non-Sabin 1-derived sequences (nt 3638 to 3832) of the last 195 nucleotides of the 2A gene (23). To further investigate the relationship between isolates Hebei/91-2 and Henan/91-3, we compared the complete genomic sequences of both isolates by plotting sequence identity in a sliding window of 100 nt (Fig. 1). Sequences upstream of the Sabin 1 interval (nt 1 to 3270) were very similar to one another (98.8% nucleotide identity overall; 99.2% in the 5′-UTR and 98.7% in the non-Sabin 1 capsid interval) (Table 2). The capsid proteins of isolates Hebei/91-2 and Henan/91-3 differed at only 3 of 842 amino acid positions (VP2:I165N, VP3:Q071R, and VP1:T121A). By contrast, the halves of the genome downstream from the Sabin 1 interval (nt 3638 to 7441) for Hebei/91-2 and Henan/91-3 were markedly dissimilar (84.3% nucleotide identity) (Fig. 1; Table 2). Despite the extensive nucleotide sequence divergence in the noncapsid region, the encoded amino acid sequences were highly conserved (98.2% amino acid identity) (Table 2). Of the 22 total amino acid differences in the noncapsid protein interval not of Sabin 1 origin, 6 mapped to 2A, 4 clustered near the amino terminus of 3Dpol, and the remainder were scattered in 2B, 2C, 3A, 3BVPg, and 3Cpro. The complete genomic sequences of two other type 1 wild poliovirus isolates of genotype JX89 from central and southeastern China (from the provinces of Hubei in 1990 and Fujian in 1991) were very similar to those of Henan/91-3 (Liu et al., Abstr. 20th Annu. Meet. Am. Soc. Virol.), indicating that the 5′-UTR and capsid sequences of the wild-vaccine recombinant Hebei/91-2 were derived from a representative of a major indigenous type 1 wild poliovirus genotype (47).
FIG. 1.
Nucleotide similarity plots comparing the complete genomic sequences of an early poliovirus type 1 wild-vaccine recombinant isolate Hebei/91-2 (recombinant class R91-1) with those of a closely related type 1 wild isolate Henan/91-3 (genotype JX89) (top graph) and with those of the Sabin type 1 OPV strain (bottom graph). The plots are aligned with a schematic of the organization of the poliovirus genome (top); the single open reading frame is flanked by the 5′- and 3′-UTRs.
TABLE 2.
Nucleotide and amino acid sequence identity upstream and downstream of the predicted recombination sites in the complete genome sequences, comparing parent and its recombinant progeny for different groups of wild-vaccine recombinant poliovirus R91
| Parent group (isolate)/progeny group (isolate) | Recombination site(s) | % Identity (5′/3′)
|
|
|---|---|---|---|
| Nucleotide | Amino acid | ||
| JX89 (Henan/91-3)/R91-1 (Hebei/91-2)a | 3263-3270 (VP1) and 3638-3639 (2A) | 98.8/84.3 | 99.6/98.2 |
| R91-1 (Hebei/91-2)/R91-2 (Yunnan/92) | Before 4009 (2B)b | 97.7/84.1 | 99.5/98.1 |
| R91-1 (Hebei/91-2)/R91-3 (Guangdong/92-2) | Before 4843 (2C)b | 97.9/85.8 | 99.5/98.0 |
| R91-1 (Hebei/91-2)/R91-4 (Hainan/93-2) | Before 4843 (2C)b | 97.0/85.1 | 99.4/97.9 |
| R91-3 (Guangdong/92-2)/R91-4 (Hainan/93-2) | Before 4873 (2C)b | 97.8/84.8 | 99.5/97.6 |
| R91-1 (Hebei/91-2)/R91-5 (Fujian/93-8) | Before 6379 (3D)b | 96.9/87.5 | 99.2/99.4 |
Because there is a Sabin 1 sequence interval in VP-2A junction region in wild-vaccine recombinant poliovirus, only the non-Sabin 1 sequences in R91-1 were compared with those of the wild parent.
Only the right boundary of the recombination site was identified.
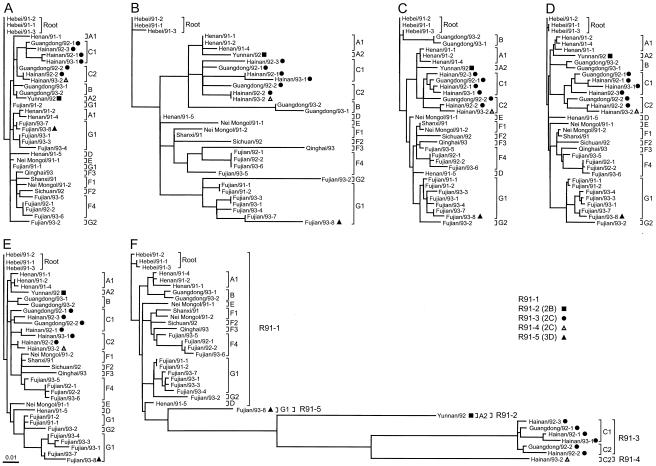
Phylogenetic analysis of the nucleotide sequences from wild-vaccine recombinants in six genomic intervals.
A total of 34 wild-vaccine recombinant viruses (genotype R91) isolated from polio patients in China from 1991 to 1993 were analyzed. In addition to the previous analysis of VP1 and 2A genes (23), sequence comparisons of these 34 isolates were extended to include part of the conserved sequences (nt 186 to 639) and all of the hypervariable sequences (nt 640 to 742) in the 5′-UTR, an interval encoding VP4 and part of VP2 (nt 743 to 1176), and an interval encoding part of 3D (nt 6011 to 6544). Phylogenetic trees were constructed from the sequences in these six genomic intervals with the maximum-likelihood algorithm (8) (Fig. 2). Isolates were assigned to lineages (A1 to G2) based on the sequence relationships previously described for the VP1 and 2A genes (23). Apart from that for the 3D region tree (Fig. 2F), the branch topologies of the five other trees are similar (Fig. 2A to E), and the lineage assignments are consistent with the VP1 and 2A relationships. Among the five trees with similar topologies, which are compared on the same scale, it is evident that genetic distances are generally substantially greater in the 5′-UTR hypervariable region (Fig. 2B) than in the other genomic intervals but that the tree topology is still conserved. The most likely explanation for this pattern is that the rate of nucleotide substitution in the hypervariable region is severalfold higher than those for the other intervals (see below). By contrast, genetic distances are shortest in the 5′-UTR conserved region, and the topology of the tree (Fig. 2A) is not fully congruent with those of the trees of the hypervariable and coding region sequences. The short branch lengths are a consequence of the relatively low evolution rate in this region, probably due to functional constraints, and the lack of comparable tree topologies is likely due to the small number of substitutions supporting the specific clustering, which is reinforced by low bootstrap values for many of the branches in this tree (data not shown). Trees comparing combined conserved and hypervariable sequences within the 5′-UTR (nt 186 to 742) have topologies very similar to those of coding region trees B to E (data not shown).
FIG. 2.
Maximum-likelihood trees of 34 wild-vaccine recombinant poliovirus isolates from China, constructed from nucleotide sequences of six genomic intervals. (A) 5′-UTR conserved region (nt 186 to 639); (B) 5′-UTR hypervariable region (nt 640 to 742); (C) complete VP4 and partial VP2 gene interval (nt 743 to 1176); (D) complete VP1 gene (nt 2480 to 3385); (E) complete 2A gene (nt 3386 to 3832); F, partial 3D gene sequence interval (nt 6011 to 6544). Each tree was rooted to the sequence of Hebei/91-2. Brackets, lineages (A to G) and sublineages (e.g., C1 and C2), labeled as previously described for the VP1 and 2A gene regions (23). Isolates representing each class of new recombinant 3D gene sequences are identified by symbols (R91-2, solid square; R91-3, solid circle; R91-4, open triangle; R91-5, solid triangle).
The lack of congruence of the 3D tree topology (Fig. 2F) with those of the other trees (Fig. 2A to E) has a different explanation. The topology and branch lengths of the 3D tree for most (25 of 34) of the isolates are similar to those for the other intervals. Nine 3D gene sequences, however, are much more divergent from the main group, as indicated by the long branches separating their sequences from the others. This discontinuity in 3D gene sequence relationships is most likely the result of recombination. A total of five distinct classes of 3D gene sequences were observed (defined as R91-1, R91-2, R91-3, R91-4, and R91-5; Fig. 2; Table 1).
Location of recombination sites in the five recombinant classes.
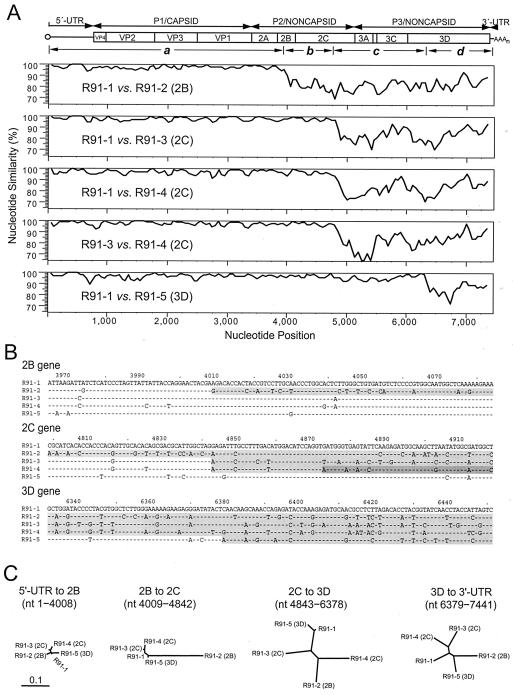
Complete genomic sequences were determined for a representative isolate of each inferred recombinant class (Hebei/91-2 [R91-1], Yunnan/92 [R91-2], Guangdong/92-2 [R91-3], Hainan/93-2 [R91-4], and Fujian/93-8 [R91-5]) (Fig. 3 and 4). Similarity plots of the complete genome sequences of these five isolates identified the approximate locations of the sites of recombination between the parental and recombinant viruses (Fig. 3A). Relative to the root genomic sequence (R91-1), recombination sites mapped to the 2B (R91-2), 2C (R91-3 and R91-4), and 3D (R91-5) genes (Fig. 3A). The upstream recombination sites for isolates Guangdong/92-2 (R91-3) and Hainan/93-2 (R91-4) mapped to similar locations within the 2C gene, but the sequences downstream from that site were quite dissimilar (84.8% nucleotide sequence identity; nt 4873 to 7441), indicating that they were derived from different donor viruses (Fig. 3A).
FIG. 3.
Mapping of recombination junction regions in representatives of each of the five R91 3D gene recombinant classes. (A) Nucleotide similarity plots comparing the complete genomic sequences of poliovirus type 1 wild-vaccine isolates representing each recombinant class: R91-1, Hebei/91-2; R91-2, Yunnan/92; R91-3, Guangdong/92-2; R91-4, Hainan/93-2; R91-5, Fujian/93-8. The similarity plots are aligned with a schematic of the poliovirus genome. The four intervals below the schematic (a, nt 1 to 4008; b, nt 4009 to 4842; c, nt 4843 to 6378; d, nt 6379 to 7441) correspond to genomic intervals bounded by recombination sites. The reference sequence in four plots is the early recombinant isolate Hebei/91-2, and the location of the crossover site in the sequence of the later recombinant isolate is in parentheses. Sequences of the closely related cluster C2 isolates Guangdong/92-2 and Hainan/93-2 were also directly compared. (B) Nucleotide sequence alignments of the sequences surrounding the predicted recombination sites in the nonstructural protein genes of wild-vaccine recombinant polioviruses. Newly incorporated nucleotide sequences derived from unknown sources are shaded and define the right (downstream) boundaries of crossover sites. Darker shade indicates nucleotide sequence derived from consecutive recombination in the 2C gene in group R91-4. (C) Unrooted maximum-likelihood trees summarizing sequence relationships across four genomic intervals bounded by recombination sites. The interval between the two recombination sites in the 2C gene was not included because it is too short for meaningful phylogenetic analysis. The input Ts/Tv ratios were estimated from program TREE-PUZZLE, version 5.0, to be 7.50, 3.40, 3.30, and 2.70 for the four intervals. The trees are plotted to the same scale, indicated by the scale bar at lower left.
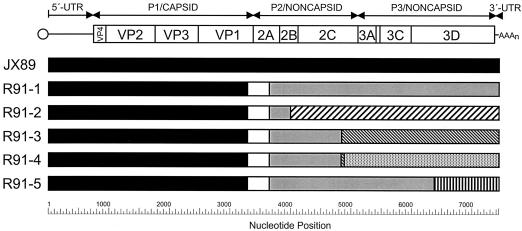
FIG. 4.
Approximate location of crossover sites of the five different classes of wild-vaccine recombinant polioviruses found in China from 1991 to 1993. Black bars, sequences derived from the indigenous type 1 wild poliovirus genotype JX89; white bars, Sabin 1-derived sequences; other patterns, sequences derived from enteroviruses other than the Sabin OPV strains. The bars symbolize the derivation of sequences of the wild parent, Henan/91-3 (genotype JX89), and representatives of each of the five 3D recombinant classes: R91-1, Hebei/91-2; R91-2, Yunnan/92; R91-3, Guangdong/92-2; R91-4, Hainan/93-2; R91-5, Fujian/93-8.
Alignment of sequences in the vicinity of the recombination sites facilitated localization of the likely sites of genetic exchange (Fig. 3B). The precise locations of the sites could not be determined unambiguously because the isolates available for comparison had also accumulated point nucleotide substitutions within those intervals. To better define the boundaries of the transitions, we aligned sequences of the most closely related pairs of isolates (on the basis of the trees in Fig. 2A to E) from AFP cases that occurred before and after the recombination event (data not shown). We identified the right (downstream) boundaries of the recombination sites at the positions of transition from high to low nucleotide sequence identity relative to the Hebei/91-2 or corresponding parental sequences (Table 2). The left (upstream) boundaries of the recombination sites were indeterminate because the parental sequences are not available for comparison. The predicted locations of the recombination sites relative to Hebei/91-2 were (i) before nt 4009 in Yunnan/92 (R91-2), (ii) before nt 4843 in Guangdong/92-2 (R91-3) and Hainan/93-2 (R91-4), and (iii) before nt 6379 in Fujian/93-8 (R91-5) (Fig. 3B and 4). Alignment of the Guangdong/92-2 and Hainan/93-2 sequences suggested that two successive recombination events had occurred in the 2C gene region. The earlier upstream crossover (before nt 4843) generated the R91-3 class of recombinants, represented by six isolates (Table 1; Fig. 2), and a subsequent nearby downstream crossover (before nt 4873) generated the R91-4 class of recombinant (represented by Hainan/93-2).
The recombination site in Fujian/93-8 (R91-5) occurred within the 534-nt interval used to compare 3D gene sequences. Only the downstream 166 nt of the Fujian/93-8 sequences within this interval are recombinant (relative to the R91-1 sequences), which explains the intermediate position of the Fujian/93-8 sequence in the tree of partial 3D gene sequences (Fig. 2F).
Noncapsid nucleotide sequences downstream of each recombination site were divergent across recombinant classes (Fig. 3C and 4; Table 2). Despite the diversity of the recombinant nucleotide sequences, the encoded amino acid sequences were very similar (97.6 to 99.4% amino acid identity). The nonrecombinant amino acid sequences (i.e., those derived from the parental root sequence, represented by Hebei/91-2) were generally even more closely related to each other (99.2 to 99.5% amino acid identity), reflecting their close evolutionary relationship.
The 3′-UTR sequences (nt 7370 to 7441) were nearly identical across all recombinant classes, as the maximum number of differences among the five representative isolates was only two base substitutions (97.2% nucleotide sequence identity).
Source of newly incorporated noncapsid region sequences.
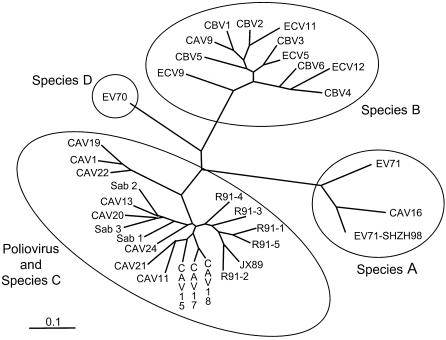
To identify the source of the recombinant noncapsid region sequences, we sequenced the partial 3D gene regions (nt 6011 to 6054) of 53 isolates representing type 1 wild polioviruses found in China from 1977 to 1994. No close matches to any of the recombinant 3D gene sequences were found (range: 78.1 to 90.6% nucleotide sequence identity) (data not shown). Because the number of distinct 3D gene sequences found to be associated with type 1 wild poliovirus isolates far exceeded the number of distinct type 1 wild poliovirus genotypes found in China (47) (H.-M. Liu, unpublished results), we reasoned that nonpolio enteroviruses (NPEVs) might be the source of the recombinant noncapsid sequences. We constructed a phylogenetic tree of the complete 3D gene sequences from a representative of each of the five wild-vaccine recombinant classes, a nonrecombinant type 1 wild poliovirus from China (Henan/91-3), the three reference OPV strains (Sabin 1 [28], Sabin 2 [42], and Sabin 3 [39, 42]), and 26 reference NPEVs (Fig. 5). The 3D gene sequences of the Chinese isolates clustered with those of the Sabin vaccine strains and members of human enterovirus (HEV) species C and were much more distantly related to the 3D gene sequences of viruses representing HEV species A, B, and D (Fig. 5). Phylogenetic analysis of the amino acid sequences encoded by these 3D gene sequences yielded a similar phylogenetic tree (data not shown). All five classes of 3D gene sequences of R91 viruses clustered most closely with each other and with those of Henan/91-3. It is likely that the donors of these new sequences are from other wild polioviruses or species C NPEVs circulating in China in 1991 to 1993.
FIG. 5.
Unrooted maximum-likelihood tree summarizing complete 3D gene sequence relationships among poliovirus and NPEV isolates and reference strains. In addition to the 3D gene sequences of the nonrecombinant JX89 isolate Henan/91-2 and of representative isolates of wild-vaccine recombinant classes R91-1 to R91-5 determined in this study, 3D gene sequences of the following reference strains were obtained from GenBank (accession numbers in parentheses): coxsackievirus A1 (CAV1; AF499635), CAV9 (D00627), CAV11 (AF499636), CAV13 (AF499637), CAV15 (AF499638), CAV16 (NC001612), CAV17 (AF499639), CAV18 (AF499640), CAV19 (AF499641), CAV20 (AF499642), CAV21 (D00538), CAV22 (AF499643), CAV24 (D90457), CBV1 (M16560), CBV2 (AF085363), CBV3 (M88483), CBV4 (X05690), CBV5 (X67706), CBV6 (AF105342), echovirus 5 (ECV5; AF083069), ECV9 (X92886), ECV11 (X80059), ECV12 (X79047), enterovirus 70 (EV70; D00820), EV71 (U22521), EV71 (China strain SHZH98; AF302996), Sabin 1 (Sab 1; J02285), Sab 2 (X00595), and Sab 3 (X00596).
Patterns of nucleotide variation within different genomic regions.
The patterns of nucleotide variation underlying the topologies of trees A to F (Fig. 2) were analyzed in more detail. All coding region and 3′-UTR sequences were colinear, suggesting that all genetic differences were base substitutions and that recombination did not alter the total number of noncapsid region codons. By contrast, variation in the 5′-UTR involved both substitution and insertion-deletion of bases.
(i) 5′-UTR.
Variation within the conserved 5′-UTR interval (nt 186 to 639) was primarily by fixation of point mutations. All 34 wild-vaccine recombinant isolates had >95.8% sequence identity in this interval, and the Ts/Tv ratio was estimated to be ∼5.8. Nucleotide substitutions occurred at only 64 different positions distributed across the 454-nt interval, and all were outside of the domains known to be invariant (data not shown). Three isolates (Shanxi/91 and Nei Mongol/91-2 [lineage F1] and Nei Mongol/91-1 [lineage E]) had a 1-nt deletion (U) in the U-rich loop (box A) of domain VI in the conserved part of the 5′-UTR (nt 569 to 572), relative to the other isolates (33, 40). Base substitutions occurred in both the predicted loop and stem regions, and compensating mutations to maintain base pairing were generally not found when substitutions occurred in stem structures.
The much greater variation within the hypervariable interval of the 5′-UTR (nt 640 to 742) also occurred primarily by fixation of point mutations. Nucleotide sequence identities within this interval were as low as 73.1% among the 34 isolates. Nucleotide substitutions occurred at 55 of 104 positions (52.9%), and the Ts/Tv ratio was estimated to be ∼4.3. Two closely related lineage G1 isolates (Fujian/91-1 and Fujian/91-2) had the same 16-nt deletion (nt 718 to 734), located 8 nt upstream of the initiation codon. All recombinant isolates described here (except Fujian/91-1 and Fujian/91-2) had the 1-nt insertion (U) between positions 728 and 729 in the hypervariable 5′-UTR interval found in the indigenous wild poliovirus isolate Henan/91-3 (and three other closely related JX89 isolates), which distinguished these viruses from most of the other type 1 polioviruses circulating in China from 1977 to 1994 (47; Liu et al., Abstr. 20th Annu. Meet. Am. Soc. Virol.).
(ii) Capsid region.
Capsid region sequences of all 34 isolates were very similar. Nucleotide sequence identities were ≥94.8% in the VP4-VP2 gene interval (nt 743 to 1176) and ≥93.5% in the VP1 gene (nt 2480 to 3385) (23). The large majority of the observed substitutions generated synonymous codons. The estimated Ts/Tv ratios for the capsid region (∼12.8 for the VP4-VP2 gene interval; ∼8.1 for the VP1 gene) were about twofold higher than those for the 5′-UTR. There is no evidence to infer recombination between the two sequenced portions of the capsid (nt 1177 to 2479), as indicated by the congruent trees for the VP4-VP2 and VP1 gene intervals in all isolates. Nucleotide sequence identities within the VP2-VP3 gene interval were high (≥95.8%) among the five representative isolates that were completely sequenced (Fig. 3A).
(iii) Noncapsid region.
As expected from previous findings (23, 42), noncapsid amino acid sequences were well conserved within and across recombinant classes (Tables 2 and 3). Within the R91-1 class, partial 3D gene (nt 6011 to 6544) nucleotide identities ranged from 94.4 to 100%, and the corresponding amino acid identities ranged from 94.9 to 100% (Table 3). The six class R91-3 isolates were also very similar to one another in the partial 3D gene interval (≥95.3% nucleotide sequence identity, ≥97.2% amino acid sequence identity). Across recombinant classes, partial 3D gene nucleotide identity ranged from 77.7 to 85.5%, but the corresponding amino acid identities remained high, varying from 93.8 to 99.4% (Table 3). The number of variable amino acid residues in the noncapsid proteins appears to have been strongly restricted, and saturation of variable sites had already begun within the R91-1 class during the ∼2.5 years of circulation (23).
TABLE 3.
Pairwise comparison of nucleotide and amino acid sequence identities of the partial 3D gene among the five 3D sequence classes of wild-vaccine recombinant poliovirus isolates
| Recombinant class | % Nucleotide sequence identity (amino acid sequence identity)
|
||||
|---|---|---|---|---|---|
| R91-1 | R91-2a | R91-3 | R91-4a | R91-5a,b | |
| R91-1 | 94.4-100 (94.9-100) | 81.3-83.5 (96.1-98.3) | 77.9-81.3 (93.8-97.8) | 79.4-81.5 (94.4-96.6) | 81.3-85.5 (96.4-98.2) |
| R91-2 | 79.6-81.1 (96.6-98.3) | 80.2 (97.2) | 81.9 (98.2) | ||
| R91-3 | 95.3-98.9 (97.2-99.4) | 81.1-83.0 (97.8-99.4) | 77.7-78.3 (94.6-96.4) | ||
| R91-4 | 80.1 (96.4) | ||||
| R91-5 | |||||
Recombinant class R91-2 is represented by the single isolate Yunnan/92, R91-4 is represented by the single isolate Hainan/93-2, and R91-5 is represented by the single isolate Fujian/93-8.
Because the recombination site in R91-5 is within the 3D interval sequencing window, only the 3′ 166 nt of this 3D interval were compared with those of other poliovirus R91 classes.
(iv) Rates of nucleotide substitution.
We had previously estimated the rate of fixation of synonymous substitutions in the VP1 and 2A genes of the wild-vaccine recombinants to be (3.45 ± 0.57) × 10−2 and (4.40 ± 0.70) × 10−2 synonymous substitutions per synonymous site per year, respectively (23). Different regions of the genome, however, evolved at different rates, as indicated by the variation in the mean branch lengths of nonrecombinant sequences in trees A to F (Fig. 2). Branch lengths in the coding region trees C (VP4-VP2 gene; average genetic distance, 0.028 nucleotide substitutions per site), D (VP1 gene; average genetic distance, 0.031), E (2A gene; average genetic distance, 0.028), and F (partial 3D gene; average genetic distance, 0.031; excluding branches representing the additional recombinant 3D gene sequences) were very similar, suggesting similar rates of base substitution for these different coding region intervals. Compared with the coding region trees, the mean branch lengths were shorter for tree A (5′-UTR conserved region; average genetic distance, 0.018) and much longer for tree B (5′-UTR hypervariable region; average genetic distance, 0.088) (Fig. 2). When evolution rates were estimated by linear regression, the rate of fixation of synonymous substitutions in the VP4-VP2 gene and partial 3D gene sequences were (4.23 ± 0.82) × 10−2 and (4.68 ± 1.32) × 10−2 synonymous substitutions per synonymous site per year, respectively (Table 4). When the evolution rates were expressed as variation in both synonymous and nonsynonymous sites, the four coding region intervals evolved at similar rates, ranging from (1.28 ± 0.21) × 10−2 to (1.84 ± 0.47) × 10−2 substitutions per site per year. By contrast, the evolution rate at all sites for the conserved 5′-UTR interval was (0.87 ± 0.27) × 10−2 substitutions per site per year, about 60% of the mean rate observed for the coding region intervals, whereas the evolution rate at all sites for the hypervariable 5′-UTR interval was a very high (3.86 ± 1.03) × 10−2 substitutions per site per year, over 2.5 times higher than the mean rate observed for the coding region intervals (Table 4). Although the evolution rate values are consistent with the genetic distances illustrated by trees A to F, the precision of the estimates is limited by the small number of isolates (n = 21) for which the dates of onset or sample collection were known and by the short time (∼2.5 years) for observing the evolution of the recombinant lineages.
TABLE 4.
Evolution rate of type 1 wild-vaccine recombinant poliovirus in different genomic intervals
| Genomic interval | Evolution rate (substitutions/nt/yr) at:
|
|
|---|---|---|
| All sites (R2)a | Synonymous sites (R2) | |
| 5′-UTR conserved region | (0.87 ± 0.26) × 10−2 (0.71) | |
| 5′-UTR hypervariable region | (3.86 ± 1.00) × 10−2 (0.79) | |
| VP4-VP2 | (1.33 ± 0.28) × 10−2 (0.83) | (4.23 ± 0.80) × 10−2 (0.86) |
| VP1 | (1.28 ± 0.21) × 10−2 (0.89) | (3.45 ± 0.57) × 10−2 (0.89) |
| 2A | (1.57 ± 0.27) × 10−2 (0.88) | (4.40 ± 0.70) × 10−2 (0.90) |
| 3D | (1.84 ± 0.46) × 10−2 (0.79) | (4.68 ± 1.29) × 10−2 (0.75) |
R2, linear correlation coefficient.
Approximate times of occurrence of subsequent recombination events.
We had previously estimated that the recombination event generating the first wild-vaccine recombinants (class R91-1) occurred in early 1991 (23). The approximate dates of the subsequent recombination events generating the four additional recombinant classes, R91-2 to R91-5, were estimated from the branch topology of trees A to E (Fig. 2) and the available epidemiologic record (Table 1). By this approach, we estimate the dates of the recombination events leading to the different recombinant classes to be between mid-1991 and the end of 1992 for R91-2 and R91-3, from the beginning of 1992 to the end of 1993 for R91-4, and in 1993 for R94-5. We are unable to estimate the dates more precisely because the number of isolates available for sequence analysis was small and the specific dates of onset or specimen collection for many isolates were unavailable (Table 1).
DISCUSSION
The appearance and spread of a type 1 wild-vaccine poliovirus recombinant in China provided a unique opportunity to investigate the nature of poliovirus recombination during widespread circulation in human populations (23). We had previously estimated that recombination between Sabin 1 and an indigenous wild-type 1 poliovirus from China occurred in early 1991, consistent with the findings that the earliest wild-vaccine recombinants (from April 1991) had no nucleotide substitutions in the rapidly evolving 367-nt block of sequences derived from Sabin 1 (23). From this single recombination event, at least four additional progeny lineages with differing noncapsid sequences emerged during the subsequent 2 to 3 years of transmission within China.
Our studies of natural recombination in poliovirus were facilitated by several factors. First, because type 1 poliovirus circulated widely in China in the early 1990s, the number of potential mixed poliovirus-NPEV infections was probably very large. A portion of these mixed poliovirus infections would have been with NPEVs of species C, some of which can apparently exchange noncapsid sequences with polioviruses (2). Moreover, enterovirus carriage rates were probably high in most of the provinces where the recombinants were found (total population, 450 million) because high population densities favor person-to-person contact and the prevailing tropical-to-subtropical conditions favor efficient enterovirus transmission during much of the year (23, 27). Second, because sensitive poliovirus surveillance had been implemented in China in support of polio eradication, numerous poliovirus clinical isolates were available for analysis (44). Third, because all recombinants described here were derived from a discrete 1991 recombination event (23), it was possible to estimate the timing of subsequent genetic exchanges leading to newly emergent recombinant lineages. Fourth, because the 367-nt block of sequence derived from Sabin 1 was conserved in subsequent recombinant genomes, all R91 isolates could be readily identified by the presence of this stable natural genetic marker (23).
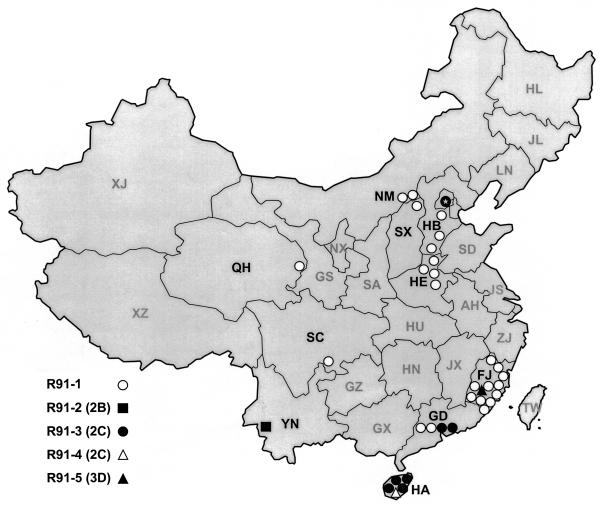
Our studies, however, were also limited by several factors, the most important of which was the unavailability of poliovirus isolates from several populous provinces where the disease was endemic (Fig. 6). AFP surveillance was not established in all provinces of China until 1993, and there were numerous gaps in the national polio surveillance system in the preceding years (46). Consequently, many circulating polioviruses, including type 1 wild-vaccine recombinants, were probably missed in China during 1991 and 1992. Moreover, only single recombinant isolates were available from the large provinces of Sichuan (population, ∼110 million) and Yunnan (population, ∼40 million) and from the smaller province of Qinghai (population, ∼4.5 million). The genetic distances on trees A to E between the sequences of these single isolates and those of the most closely related isolates from other provinces were relatively large, suggesting the existence of gaps in surveillance for polioviruses of recombinant lineages A2, F2, and F3. Accordingly, other recombinant lineages that are not represented in our study may have emerged. Although a more comprehensive collection of isolates may have yielded new classes of R91 recombinants, it may also have revealed the existence of additional R91 lineages within which no new recombinants emerged, and as a result the observed frequency of emergence of recombinants might be essentially unchanged. We were unable to monitor the evolution of the R91 recombinants after 1993 because their spread occurred during a time of sharply declining polio incidence in China in response to rapidly rising rates of polio immunization, leading to the eradication of all indigenous poliovirus lineages by early 1994 (44). The spread of the R91 lineages was probably also limited by competition with cocirculating nonrecombinant type 1 lineages during these last years of polio endemicity in China (20, 47). Recombinants could also have been missed due to upstream crossovers that would obscure all evidence of crossovers downstream, potentially leading to an underestimate of the total number of recombination events.
FIG. 6.
Geographic distribution of the 34 type 1 wild-vaccine poliovirus recombinant isolates that evolved to five different recombinant classes in China from 1991 to 1993. Province abbreviations: AN, Anhui; FJ, Fujian; GD, Guangdong; GS, Gansu; GX, Guangxi; GZ, Guizhou; HA, Hainan; HB, Hebei; HE, Henan; HL, Heilongjiang; HN, Hunan; HU, Hubei; JL, Jilin; JS, Jiangsu; JX, Jiangxi; LN, Liaoning; NM, Nei Mongol; NX, Ningxia; QH, Qinghai; SA, Shaanxi; SC, Sichuan; SD, Shandong; SX, Shanxi; XJ, Xinjiang; XZ, Xizang (Tibet); YN, Yunnan; ZJ, Zhejiang.
In view of the likely wide distribution of both wild-type 1 poliovirus and species C NPEVs in China from 1991 to 1993, it may seem surprising that only five R91 recombinant classes were observed and that most of the later R91 recombinants showed no evidence of additional genetic exchange. Although the conditions permitting emergence of poliovirus recombinants are unknown, random events appear to be significant contributors to the frequency of their emergence. The epidemiologic record revealed no evident differences among the recombinant lineages in their capacities to circulate and cause paralytic disease. Major differences in replicative capacity across recombinant lineages would predict that some lineages would rapidly displace others. What is observed instead is a pattern of cocirculation of different recombinant lineages within the same province. Only limited conclusions, however, can be drawn from the epidemiologic data, and systematic studies of the replicative fitness of different recombinants under natural conditions are not feasible. While recombination is unlikely to promote a net increase in poliovirus replicative fitness under natural conditions, it is possible that it could offset the effects of accumulation of deleterious mutations arising in some lineages (7, 26). Recombination appears to be a normal part of the evolution of circulating polioviruses (5, 15; Liu et al., Abstr. 20th Annu. Meet. Am. Soc. Virol.), and the likelihood of its occurrence along any chain of transmission increases with time (3, 5, 15, 23; Liu et al., Abstr. 20th Annu. Meet. Am. Soc. Virol.).
Unlike the initial R91 crossover event, which spanned the capsid-noncapsid junction (23), all of the subsequent crossovers occurred in the P2 and P3 noncapsid regions. Natural intertypic genetic exchange within the capsid region appears to be rare (1, 10, 25), probably because structural incompatibilities restrict the fitness of most intertypic recombinants. By contrast, recombination within the noncapsid region appears to be relatively common (3, 5, 6, 9, 12, 15, 18, 21, 22, 25, 36, 41). The high degree of similarity in noncapsid amino acid sequences among polioviruses and many species C NPEVs may favor intertypic recombination across homologous regions (2). Recombination within the conserved interval of the 5′-UTR has also been observed (45) but is less frequent, possibly because crossovers are less likely to occur over the shorter (∼640-nt) conserved 5′-UTR interval than over the approximately sixfold-longer (3,984-nt) noncapsid sequence interval, as would be predicted if the poliovirus recombination map (4) is proportional to the physical map (43).
We cannot identify which serotypes of HEV species C were the donors of the recombinant noncapsid sequences. Because of dynamic recombination in the noncapsid region, both for polioviruses and for species C NPEVs (2), the linkage between specific capsid and noncapsid lineages is transient. Thus, the patterns of poliovirus (23, 35, 38) and NPEV (29-31) circulation can be monitored unambiguously from comparisons of VP1 or other capsid sequences but not from comparisons of noncapsid sequences.
In addition to finding frequent recombination in the noncapsid region, we also found sharp differences in the rates of nucleotide substitution in different functional domains of the poliovirus genome. Substitution rates in the various coding region intervals sampled were similar. The conserved sequences in the 5′-UTR evolve more slowly than the coding region sequences. By contrast, the last 100 nt in the 5′-UTR are not only highly variable (34, 42) but also very rapidly evolving. The estimated rate of substitution for this interval is among the highest observed for any genetic sequence. Unlike the conserved 5′-UTR, where the primary sequence (34, 42) and secondary structure are both well conserved (33), or the coding region, whose encoded amino acid sequences are conserved, evolution of the hypervariable 5′-UTR is much less constrained, and both nucleotide substitutions and insertions-deletions appear to be common. Although this interval is dispensable for growth in cell culture (19), its retention in clinical isolates suggests that it contributes in some way to poliovirus replication in humans.
The pattern of natural poliovirus recombination described in this report is relatively simple and amenable to analysis. All recombinants were derived from a common 1991 recombination event, and all subsequent exchanges occurred independently along different evolutionary pathways or serially along a single evolutionary pathway. The characteristic block of Sabin 1-derived sequences was present in all isolates, as all observed subsequent crossovers occurred downstream from that block. Consequently, most, if not all, of the recent genetic record of the exchanges was preserved. A much more complex pattern of recombination in the noncapsid region is observed among other type 1 wild polioviruses previously indigenous to China (Liu et al., Abstr. 20th Annu. Meet. Am. Soc. Virol.). Taken together, our observations demonstrate that recombination between polioviruses and species C NPEVs is a normal feature of poliovirus evolution in nature.
Acknowledgments
We thank the virologists from the National Polio Laboratory Network of China for sharing their isolates with us, Jaume Jorba for helpful discussions on phylogenetic and statistical analyses, and Larry Anderson for review of the manuscript. The technical advice of Kaija Maher and Betty Brown (CDC) and the excellent technical assistance of Hong Zheng (Chinese CDC) and Naomi Dybdahl-Sissoko, Mary R. Flemister, Silvia Peñaranda, and A. J. Williams (CDC) are appreciated.
REFERENCES
- 1.Blomqvist, S., A. L. Bruu, M. Stenvik, and T. Hovi. 2003. Characterization of a recombinant type 3/type 2 poliovirus isolated from a healthy vaccinee and containing a chimeric capsid protein VP1. J. Gen. Virol. 84:573-580. [DOI] [PubMed] [Google Scholar]
- 2.Brown, B. A., M. S. Oberste, K. Maher, and M. Pallansch. 2003. Complete genomic sequencing shows that polioviruses and members of human enterovirus species C are closely related in the noncapsid coding region. J. Virol. 77:8973-8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cammack, N., A. Phillips, G. Dunn, V. Patel, and P. D. Minor. 1988. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology 167:507-514. [PubMed] [Google Scholar]
- 4.Cooper, P. D. 1968. A genetic map of poliovirus temperature-sensitive mutants. Virology 35:584-596. [DOI] [PubMed] [Google Scholar]
- 5.Dahourou, G., S. Guillot, O. Le Gall, and R. Crainic. 2002. Genetic recombination in wild-type poliovirus. J. Gen. Virol. 83:3103-3110. [DOI] [PubMed] [Google Scholar]
- 6.Driesel, G., S. Diedrich, U. Kunkel, and E. Schreier. 1995. Vaccine-associated cases of poliomyelitis over a 30 year period in East Germany. Eur. J. Epidemiol. 11:647-654. [DOI] [PubMed] [Google Scholar]
- 7.Escarmís, C., M. Dávila, N. Charpentier, A. Bracho, A. Moya, and E. Domingo. 1996. Genetic lesions associated with Muller's ratchet in an RNA virus. J. Mol. Biol. 264:255-267. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 9.Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated poliomyelitis. Virology 196:199-208. [DOI] [PubMed] [Google Scholar]
- 10.Georgescu, M.-M., F. Delpeyroux, and R. Crainic. 1995. Tripartite genome organization of a natural type 2 vaccine/nonvaccine recombinant poliovirus. J. Gen. Virol. 76:2343-2348. [DOI] [PubMed] [Google Scholar]
- 11.Georgescu, M. M., F. Delpeyroux, M. Tardy-Panit, J. Balanant, M. Combiescu, A. A. Combiescu, S. Guillot, and R. Crainic. 1994. High diversity of poliovirus strains isolated from the central nervous system from patients with vaccine-associated paralytic poliomyelitis. J. Virol. 68:8089-8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 14.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-123. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 15.Kew, O. M., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. André, E. Blackman, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Miyamura, H. van der Avoort, M. S. Oberste, D. Kilpatrick, S. Cochi, M. Pallansch, and C. de Quadros. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 16.Kew, O. M., and B. K. Nottay. 1984. Evolution of the oral polio vaccine strains in humans occurs by both mutation and intramolecular recombination, p. 357-362. In R. Lerner and R. Chanock (ed.), Modern approaches to vaccines. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 18.King, A. M. Q. 1988. Preferred sites of recombination in poliovirus RNA: an analysis of 40 intertypic cross-over sequences. Nucleic Acids Res. 16:11705-11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuge, S., and A. Nomoto. 1987. Construction of viable deletion and insertion mutants of the Sabin strain of type 1 poliovirus: function of the 5′ noncoding sequence in viral replication. J. Virol. 61:1478-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, J., T. Yoneyama, H. Yoshida, K. Yoshii, H. Shimizu, T. Miyamura, M. Hara, X.-H. Hou, H. Zheng, Y. Fang, L.-B. Zhang, and A. Hagiwara. 1995. Genetic analysis of wild type 1 poliovirus isolates in China, 1985-1993. Res. Virol. 146:415-422. [DOI] [PubMed] [Google Scholar]
- 21.Li, J., L.-B. Zhang, T. Yoneyama, H. Yoshida, H. Shimizu, K. Yoshii, M. Hara, T. Nomura, H. Yoshikura, T. Miyamura, and A. Hagiwara. 1996. Genetic basis of the neurovirulence of type 1 polioviruses isolated from vaccine-associated paralytic patients. Arch. Virol. 141:1047-1054. [DOI] [PubMed] [Google Scholar]
- 22.Lipskaya, G. Y., A. R. Muzychenko, O. K. Kutitova, S. V. Maslova, M. Equestre, S. G. Drozdov, R. Perez Bercoff, and V. I. Agol. 1991. Frequent isolation of intertypic poliovirus recombinants with serotype 2 specificity from vaccine-associated polio cases. J. Med. Virol. 35:290-296. [DOI] [PubMed] [Google Scholar]
- 23.Liu, H.-M., D.-P. Zheng, L.-B. Zhang, M. S. Oberste, M. A. Pallansch, and O. M. Kew. 2000. Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J. Virol. 74:11153-11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martín, J., E. Samoilovich, G. Dunn, A. Lackenby, E. Feldman, A. Heath, E. Svirchevskaya, G. Cooper, M. Yermalovich, and P. D. Minor. 2002. Isolation of an intertypic poliovirus capsid recombinant from a child with vaccine-associated paralytic poliomyelitis. J. Virol. 76:10921-10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller, H. J. 1964. The relation of recombination to mutational advance. Mutat. Res. 1:2-9. [DOI] [PubMed] [Google Scholar]
- 27.Nathanson, N., and J. R. Martin. 1979. The epidemiology of poliomyelitis: enigmas surrounding its appearance, epidemicity, and disappearance. Am. J. Epidemiol. 110:672-692. [DOI] [PubMed] [Google Scholar]
- 28.Nomoto, A., T. Omata, H. Toyoda, S. Kuge, H. Horie, Y. Kataoka, Y. Genba, Y. Nakano, and N. Imura. 1982. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc. Natl. Acad. Sci. USA 79:5793-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oberste, M. S., K. Maher, M. L. Kennett, J. J. Campbell, M. S. Carpenter, D. Schnurr, and M. A. Pallansch. 1999. Molecular epidemiology and genetic diversity of echovirus type 30 (E30): genotypes correlate with temporal dynamics of E30 isolation. J. Clin. Microbiol. 37:3928-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oberste, M. S., W. A. Nix, D. R. Kilpatrick, M. R. Flemister, and M. A. Pallansch. 2003. Molecular epidemiology and type-specific detection of echovirus 11 isolates from the Americas, Europe, Africa, Australia, southern Asia and the Middle East. Virus Res. 91:241-248. [DOI] [PubMed] [Google Scholar]
- 32.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 33.Palmenberg, A. C., and J.-Y. Sgro. 1997. Topological organization of picornaviral genomes: statistical prediction of RNA structural signals. Semin. Virol. 8:231-241. [Google Scholar]
- 34.Pöyry, T., L. Kinnunen, and T. Hovi. 1992. Genetic variation in vivo and proposed functional domains of the 5′ noncoding region of poliovirus RNA. J. Virol. 66:5313-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rico-Hesse, R., M. A. Pallansch, B. K. Nottay, and O. M. Kew. 1987. Geographic distribution of wild poliovirus type 1 genotypes. Virology 160:311-322. [DOI] [PubMed] [Google Scholar]
- 36.Rousset, D., M. Rakoto-Andrianarivelo, R. Razafindratsimandresy, B. Randriamanalina, S. Guillot, J. Balanant, P. Mauclère, and F. Delpeyroux. 2003. Recombinant vaccine-derived poliovirus in Madagascar. Emerg. Infect. Dis. 9:885-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 38.Shulman, L. M., R. Handsher, S.-J. Yang, C.-F. Yang, J. Manor, A. Vonsover, Z. Grossman, M. Pallansch, E. Mendelson, and O. M. Kew. 2000. Resolution of the pathways of poliovirus type 1 transmission during an outbreak. J. Clin. Microbiol. 38:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanway, G., P. J. Hughes, R. C. Mountford, P. Reeve, P. D. Minor, G. C. Schild, and J. W. Almond. 1984. Comparison of the complete nucleotide sequences of the genomes of the neurovirulent poliovirus P3/Leon/37 and its attenuated Sabin vaccine derivative P3/Leon 12a1b. Proc. Natl. Acad. Sci. USA 81:1539-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart, S. R., and B. L. Semler. 1997. RNA determinants of picornavirus cap-independent translation initiation. Semin. Virol. 8:242-255. [Google Scholar]
- 41.Tatem, J., C. Weeks-Levy, S. J. Mento, S. J. Di Michele, A. Georgiu, W. F. Waterfield, B. Sheip, C. Costalas, T. Davies, M. B. Ritchey, and F. R. Cano. 1991. Oral poliovirus vaccine in the United States: molecular characterization of Sabin type 3 after replication in the gut of vaccinees. J. Med. Virol. 35:101-109. [DOI] [PubMed] [Google Scholar]
- 42.Toyoda, H., M. M. Kohara, Y. Kataoka, T. Suganuma, T. Omata, N. Imura, and A. Nomoto. 1984. Complete nucleotide sequences of all three poliovirus serotype genomes: implication for genetic relationship, gene function and antigenic determinants. J. Mol. Biol. 174:561-585. [DOI] [PubMed] [Google Scholar]
- 43.Wimmer, E., C. U. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]
- 44.Yang, B., J. Zhang, M. W. Otten, Jr., K. Kusumoto, T. Jiang, R. Zhang, L. Zhang, and K. Wang. 1995. Eradication of poliomyelitis: progress in the People's Republic of China. Pediatr. Infect. Dis. J. 14:308-314. [PubMed] [Google Scholar]
- 45.Yang, C.-F., T. Naguib, S.-J. Yang, E. Nasr, J. Jorba, N. Ahmed, R. Campagnoli, H. van der Avoort, H. Shimizu, T. Yoneyama, T. Miyamura, M. A. Pallansch, and O. Kew. 2003. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J. Virol. 77:8366-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, J., L.-B. Zhang, M. W. Otten, Jr., T. Jiang, X.-L. Zhang, R.-Z. Zhang, and K.-A. Wang. 1997. Surveillance for polio eradication in the People's Republic of China. J. Infect. Dis. 175(Suppl. 1):S122-S134. [DOI] [PubMed] [Google Scholar]
- 47.Zheng, D.-P., L.-B. Zhang, Z.-Y. Fang, C.-F. Yang, M. Mulders, M. A. Pallansch, and O. M. Kew. 1993. Distribution of wild type 1 poliovirus genotypes in China. J. Infect. Dis. 168:1361-1367. [DOI] [PubMed] [Google Scholar]