Abstract
The ligand-controlled retinoic acid (RA) receptors and retinoid X receptors are important for several physiological processes, including normal embryonic development, but little is known about how their ligands, all-trans and 9-cis RA, are generated. Here we report the identification of a stereo-specific 9-cis retinol dehydrogenase, which is abundantly expressed in embryonic tissues known to be targets in the retinoid signaling pathway. The membrane-bound enzyme is a member of the short-chain alcohol dehydrogenase/reductase superfamily, able to oxidize 9-cis retinol into 9-cis retinaldehyde, an intermediate in 9-cis RA biosynthesis. Analysis by nonradioactive in situ hybridization in mouse embryos shows that expression of the enzyme is temporally and spatially well controlled during embryogenesis with prominent expression in parts of the developing central nervous system, sensory organs, somites and myotomes, and several tissues of endodermal origin. The identification of this enzyme reveals a pathway in RA biosynthesis, where 9-cis retinol is generated for subsequent oxidation to 9-cis RA.
Retinoids (vitamin A derivatives) have important roles during embryonic development and normal physiological functions in adults. The physiological effects of retinoids are mediated by two classes of distantly related nuclear ligand-controlled transcription factors, the retinoic acid receptors (RAR α, β, and γ), able to bind all-trans and 9-cis stereo isomers of retinoic acid (RA), and the retinoid X receptors (RXR α, β, and γ) with ability to specifically bind 9-cis RA (reviewed in refs. 1 and 2). Heterodimers of RXR/RAR bind efficiently to RA response elements in vitro and form functional complexes in vivo (3). RXR also is known to heterodimerize with other nuclear hormone receptors, e.g., thyroid hormone receptors, the vitamin D3 receptor, peroxisome proliferator-activated receptors, and with several orphan nuclear receptors (reviewed in ref. 4). Thus, RXR may have a pleiotropic effect in several hormone signaling pathways by acting as a common heterodimeric partner. This possibility is supported by the broad expression patterns of the various RXR isoforms in developing embryos and in adult tissues (5, 6).
Gene targeting studies of RARs and RXRs have clearly confirmed the fundamental role of retinoids in embryonic development and that these two classes of nuclear receptors mediate retinoid signaling in vivo, because the induced phenotypes mimic the congenital developmental defects observed in fetuses from vitamin A-deficient dams (reviewed in ref. 7).
Direct identification of RA in tissues by HPLC techniques (8–13), or indirect measurements of the presence of RA by using RA-responsive reporter genes in cells or transgenic mice (14–17), has provided some clues to the potential roles of RA in regulation of transcription in various cell types and tissues. Given the limitations in our understanding of retinoid action, it is essential to get more information on the metabolic routes underlying biosynthesis of all-trans and 9-cis RA.
The generation of RA from retinol is a two-step reaction, the rate-limiting step being the oxidation of retinol into the intermediate retinaldehyde (reviewed in ref. 18). In developing limbs, the relative amounts of retinol and RA are correlated to different developmental stages, suggesting that this metabolic pathway is well controlled (11, 12). Two classes of unrelated enzymes have been implicated in the oxidation of retinol, the classical cytosolic medium chain alcohol dehydrogenases (reviewed in ref. 19) and recently identified microsomal members of the short chain alcohol dehydrogenase/reductase (SDR) superfamily (20–24). Enzymes from both groups are able to oxidize retinol in vitro but the relative role of the different enzymes in retinol oxidation in vivo is still enigmatic.
Further oxidation of formed retinaldehyde is believed to be catalyzed by several cytosolic aldehyde dehydrogenases (reviewed in refs. 19 and 25 and references therein). Several of these enzymes display a wide substrate specificity, except retinal aldehyde dehydrogenase type 2, which appears specific for retinal (26). The role(s) of the known aldehyde dehydrogenases in in vivo formation of RA remains to be established.
In this study we have isolated a retinol dehydrogenase (RDH) of the SDR family with ability to oxidize 9-cis retinol, but not all-trans retinol, into the corresponding aldehyde. This unique property and the expression of the enzyme in several adult and embryonic tissues imply a pathway for generating 9-cis RA.
MATERIALS AND METHODS
cDNA Synthesis, PCR Cloning, and Isolation of Full-Length cDNAs.
Total RNA was isolated from mouse gestation day (gd) 10 embryos by using standard procedures (27). cDNA was generated by priming 5–10 μg of RNA with dT18 using avian myelostosis virus reverse transcriptase (RT) (10 units) at 42°C for 30 min. Aliquots of the cDNA reactions were used as templates in a two-stage standard PCR. The conditions for the first reaction were; denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and elongation at 72°C for 2 min using 25 cycles. The primers in the first reaction were: 5′-ACGTGAATTCTG(TC)GA(TC)TC(N)GG(N)(TA)T(TC)GG (forward) and 5′-ACGTGAATTCTT(N)GC(AG)TCCCA(N)CC (reverse, an EcoRI site and a 5′-clamp are underlined). Aliquots of the first PCRs were used in a second PCR by using the reaction condition as described above with the exception that the annealing temperature was 55°C. The primers in the second reaction were: 5′- ACGTGAATTCGA(AG)GC(N)TT(CT)TC(N)GA (forward) and 5′-ACGTGAATTCCG(N)GT(N)C(TG)(N)GG(AG)TG (reverse, an EcoRI site and a 5′-clamp are underlined). The amplified approximately 300-bp band was isolated from an 1% low melting agarose gel, reamplified by using the same reaction conditions and primers as above, and cloned by using the TA cloning kit (Invitrogen). Plasmid DNA was prepared from transformants by standard protocols and analyzed by restriction analysis using EcoRI. Clones containing inserts of approximately 300 bp were sequenced by using Sequenase version 2.0 (Amersham) with vector specific primers. Clone 200 was identified as being closely related to 11-cis RDH (23).
To isolate a full-length cDNA, a mouse liver λ ZAP cDNA library (Stratagene) was screened by using the 300-bp insert of PCR clone 200 labeled with [32P]dCTP as the probes. The inserts from positive clones were subcloned into pBluescript SK(+) by in vivo excision as recommended by the supplier. Several of the cDNA clones were fully or partially sequenced (Sequenase 2.0, U.S. Biochemicals). The 1.2-kb insert of the full-length clone D2B1 was selected for further characterization. The insert of this clone encoded a protein designated RDH4.
Expression of RDH4 by Using Baculovirus Expression System and Analysis of the Enzymatic Properties of the Recombinant Protein.
The 1.2-kb EcoRI insert of clone D2B1 was subcloned into the baculovirus expression vector pFASTBAC1 as described by the supplier (GIBCO). Spodoptera frugiperda Sf9 insect cells, grown in TNM-FH medium (Nordcell) in 75 cm2 tissue culture flasks, were transfected with the recombinant vector by using Lipofectin (GIBCO). Sf9 cells, grown in 150 cm2 flasks, were infected with recombinant virus expressing RDH4 or with recombinant virus containing an unrelated gene (mock). The cells were harvested 72 hr postinfection, and total membrane fractions were prepared essentially as described (23).
Expression of RDH4 was analyzed by immunoblotting. Briefly, total membrane fractions from Sf9 cells expressing RDH4 or from mock-infected cells (5 μg of total protein) were subjected to SDS/PAGE and blotted on a Hybond ECL filter (Amersham). RDH4 was detected by ECL (Amersham) by using a rabbit antiserum raised against a fusion protein of glutathione S-transferase fused to residues 193–269 of RDH4.
To determine the enzymatic activity of RDH4, 25 μg of total membrane protein from the infected cells were incubated in PBS containing 100 μM of 9-cis retinol and all-trans retinol, respectively (a kind gift from Michael Klaus, Hoffman-La Roche AG, Basel). The final volumes of the samples were 200 μl. The cofactors, NAD+ or NADP, were separately included to a final concentration of 200 μM. After a 20-min incubation at 37°C, alkaline ethanol was added, and the mixture was extracted with 2.0 ml of n-hexane (28). The organic phase was carefully removed and dried under a stream of argon. The dried phases were dissolved in ethanol, and aliquots were analyzed by reversed-phase HPLC using a C18 column (Supelco 4,6 mm × 25 cm). The mobile phase was acetonitrile/water (85:15, vol/vol). Elution was at 1.0 ml/min, and the effluent was monitored at 350 nm (Applied Biosystems 783A Programmable Absorbance Monitor). Under the conditions used, 9-cis retinol and 9-cis retinaldehyde eluted at 12.2 and 16.6 min, respectively, whereas all-trans retinol and all-trans retinaldehyde eluted at 13.4 and 17.1 min, respectively. To assay for enzymatic activity against 9-cis, 11-cis and 13-cis isomers, membrane fractions from RDH4-expressing cells were incubated and analyzed as above by using 9-cis retinaldehyde (Sigma), 11-cis retinaldehyde (a kind gift of R. Crouch, Storm Eye Institute, Charleston, SC), or 13-cis retinaldehyde (Sigma) as the substrates with NADH as the cofactor. The amounts of generated 9-cis retinol, 11-cis retinol, or 13-cis retinol were determined. 11-cis Retinol and 11-cis retinaldehyde eluted at 12.4 and 16.5 min, respectively, whereas 13-cis retinol and 13-cis retinaldehyde eluted at 13.1 and 16.6 min, respectively.
Northern Blotting and RT-PCR Analysis.
A mouse multiple tissue Northern blot (CLONTECH) was probed by using PCR clone 200. The insert was labeled with [32P]dCTP to high specific activity by using a PCR amplification technique (29). The blots were hybridized overnight at 42°C by using 50% formamide, 6 × standard saline phosphate/EDTA buffer (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), 0.5% SDS, 2 × Denhardt’s solution, 100 μg/ml of salmon sperm DNA, and 1 × 106 cpm/ml of the labeled probe. The blots were washed at RT for 2 × 30 min in 2 × standard saline citrate (SSC) containing 0.1% SDS and then for 2 × 20 min at 50°C in 0.1 × SSC containing 0.1% SDS. Kodak XAR film was exposed overnight at −70°C by using intensifying screens.
For the RT-PCR amplifications, first-strand cDNA was generated from RNA isolated from mouse embryo gd 9, 11, 13, and 15 as described above. The conditions for the PCR amplifications were essentially as above. RDH4 transcripts were amplified by using a two-step procedure. The first reaction was carried out with primers 5′-TAGCTGGTATCATCGGGCCCA (forward) and 5′-AGAAACCAGGCAGCACTGG (reverse). Aliquots from the first amplification reaction were reamplified by using the same forward primer and a second reverse primer, 5′-GCTGGTCACCTTCGTTAGTTC (reverse). Glyceraldehyde-3-phosphate dehydrogenase transcripts were amplified by using 5′-TGGTATCGTGGAAGGACTCATGAC (forward) and 5′-ATGCCAGTGAGCTTCCCGTTCAGC (reverse). The amplified bands were analyzed on a 1.5% agarose gel and visualized by staining with ethidium bromide.
Nonradioactive in Situ Hybridization Analysis of RDH4 Expression in Mouse Embryos.
The breeding of the C57BL/6 mice, embedding, and sectioning of the embryos was performed as described elsewhere (30). In brief, the sections were dewaxed and rehydrated, and then pretreated with proteinase K (27 μg/ml in 10 mM Tris·HCL, pH 8.0 and 1 mM EDTA) for 7 min. After fixation in 4% paraformaldehyde in PBS, the probes, diluted in hybridization buffer (50% formamide/5 × SSC/50 μg/ml yeast RNA/1% wt/vol SDS/50 μg/ml heparin), were added. The digoxigenin (DIG)-labeled antisense probe was generated from a StuI-linearized cDNA template using T7 RNA polymerase according to the manufacturer’s recommendations. As a control a DIG-labeled sense probe was generated in the same manner, using a cDNA template of an unrelated clone. After overnight hybridization at 55°C, the sections were washed in 4 × SSC, followed by washes in 50% formamide, 5 × SSC, and 1% wt/vol SDS at 65°C and then washes in 50% formamide and 2 × SSC at 65°C followed by washes in 25 mM Tris·HCL, pH 7.5 containing 140 mM NaCl and 1% Tween-20. The pretreatment and application of the alkaline-phosphatase-conjugated Fab fragments of sheep anti-DIG antibodies, as well as the color development, was performed according to the manufacturer’s recommendations (Boehringer Mannheim). After color development, the sections were washed in PBS, dehydrated to 100% ethanol, cleared with xylene, and mounted in Pertex.
RESULTS
Identification of a RDH Closely Related to the Eye-Specific 11-cis RDH.
A RT-PCR-based approach was used to identify cDNAs encoding novel RDHs expressed in developing mouse embryos. One cDNA clone encoded a polypeptide highly related to 11-cis RDH. A full-length cDNA clone was isolated from a mouse liver cDNA library and found to encode a protein of 318 amino acid residues with a calculated mass of 34,830 Da (Fig. 1A), which was designated RDH4. An amino acid sequence alignment of RDH4, bovine 11-cis RDH, and three all-trans RDHs, revealed that RDH4 displayed 86% identity with bovine 11-cis RDH and 52–53% identity with all-trans RDHs (Fig. 1A). The catalytic domain of RDH4 displayed the amino acid sequence typical of SDR family members, including the cofactor binding motif G-X-X-X-G-X-G and the active site, Y-X-X-X-K (31).
Figure 1.
(A) Alignment of the amino acid sequence of RDH4 with bovine 11-cis RDH (b11-cis RDH) and three all-trans RDHs expressed in rat liver (ra-t RDHs). Residues identical to RDH4 are boxed and gaps have been introduced to optimize the alignment. The numbered bars indicate the positions of the oligonucleotide mixtures used in the first (#1), and in the second (#2) round of PCR amplifications of mRNA from mouse gd 10 embryos. (B) Phylogenetic analysis of the amino acid sequences of several RDHs. RDH4 is closely related to bovine and human 11-cis RDHs (t11-cis RDH and h11-cis RDH, respectively) suggesting that the cis-specific RDHs and the all-trans-specific RDHs form distinct subgroups within the RDHs of the SDR family. The analysis was done by using the PAM 250 distance table.
Phylogenetic analysis verified the close structural relationships between RDH4 and 11-cis RDHs whereas the group of all-trans RDHs were more distantly related (Fig. 1B).
RDH4 Is a 9-cis RDH.
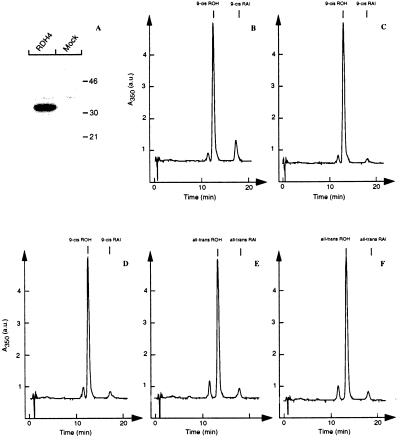
To explore the enzymatic properties of RDH4, the enzyme was expressed in baculovirus-infected Sf9 insect cells. Immunoblotting analyses of membrane fractions from infected cells showed that RDH4 was expressed as a 32-kDa membrane-associated protein whereas mock-infected cells lacked this protein (Fig. 2A).
Figure 2.
(A) Immunoblotting analysis of total membrane fractions from baculovirus-infected Sf9 insect cells expressing RDH4 or from mock-infected Sf9 insect cells. The results show that RDH4 is expressed as a 32-kDa membrane-associated polypeptide. (B-F) Reversed-phase HPLC profiles on the formation of 9-cis retinaldehyde (9-cis RAl) from 9-cis retinol (9-cis ROH) in membrane fractions from Sf9 insect cells overexpressing RDH4 in the presence of NAD+ (B), NADP (C), or from mock-infected cells in the presence of NAD+ (D). Identical analyses using all-trans retinol as the substrate when using membrane fractions from cells overexpressing RDH4 (E) or from mock-infected cells (F) show that all-trans retinol is not a preferred substrate for RDH4. The results demonstrate that RDH4 is a NAD+-dependent 9-cis RDH.
Membrane fractions from cells expressing RDH4 or mock-infected cells were incubated separately with 9-cis retinol in the presence of NAD+ or NADP as the cofactors. Analysis by reverse-phase HPLC showed that RDH4 was able to generate 9-cis retinaldehyde in the presence of NAD+ (Fig. 2B). When using NADP as the cofactor, RDH4 was unable to catalyze the generation of 9-cis retinaldehyde above the levels found in the mock-infected Sf9 cells (Fig. 2 C and D). Using all-trans retinol as the substrate for RDH4, no significant formation of all-trans retinaldehyde was observed when comparing membrane fractions from cells overexpressing RDH4 and from mock-infected cells (Fig. 2 E and F). Additional analyses showed that RDH4 was able to reduce 9-cis, 11-cis, and 13-cis retinaldehydes into the corresponding retinols in the presence of NADH as the cofactor (data not shown). The ability of RDH4 to oxidize 9-cis retinol and to reduce 9-cis retinaldehyde shows that the enzyme is an oxidoreductase. This finding suggests that RDH4 also can oxidize 11-cis and 13-cis retinols into the corresponding retinaldehydes.
Expression of RDH4 in Adult and Embryonic Mouse Tissues.
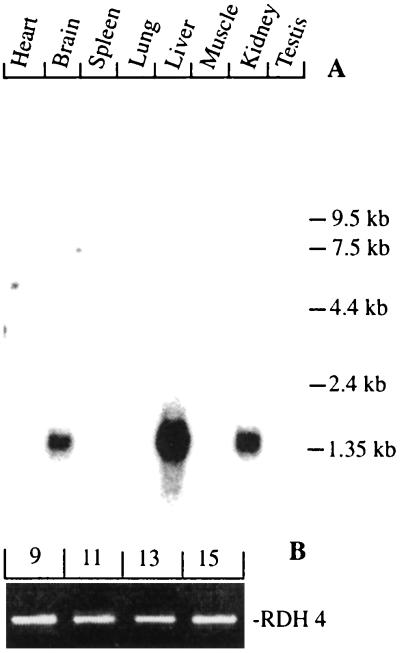
Northern blotting analysis revealed abundant expression of a 1.4-kb transcript in liver, kidney, and brain whereas several other tissues, including heart, lung, spleen, skeletal muscle, and testis appeared devoid of detectable expression of RDH4 (Fig. 3A). The tissue expression of RDH4 is thus different from the expression of the closely related 11-cis RDH, which is abundantly expressed only in retinal pigment epithelium (23). This finding suggests that RDH4 is not the mouse equivalent of 11-cis RDH.
Figure 3.
(A) Northern blotting analysis of transcripts for RDH4 in several adult mouse tissues. Expression of a single 1.4-kb transcript is seen in liver, kidney, and brain whereas expression levels in other tissues are much lower. (B) RT-PCR amplifications of a 420-bp fragment derived from RDH4 transcripts in mouse gd 9, 11, 13, and 15 embryos show that the enzyme is expressed during a large part of mouse embryonic development.
In early mouse embryos, the expression of RDH4 was examined by RT-PCR analysis using transcripts derived from gd 9, 11, 13, and 15 (Fig. 3B). The results show that RDH4 transcripts are present during a large part of murine embryonic development. Transcripts encoding glyceraldehyde-3-phosphate dehydrogenase were examined as positive controls and no amplified products were obtained when omitting the addition of templates to the PCRs (data not shown).
In Situ Localization of Transcripts for RDH4 in the Early Mouse Embryo.
Nonradioactive in situ hybridization of tissue sections of mouse embryos was carried out by using digoxigenin-labeled RDH4 antisense and sense cRNA probes. The general pattern of expression of RDH4 in mouse embryos is given in this paper, focusing on gd 10 and 11 (Fig. 4 and Table 1). A more detailed study, including other developmental stages, will be published separately (P.T., A.R., L.D., and U.E., unpublished observations).
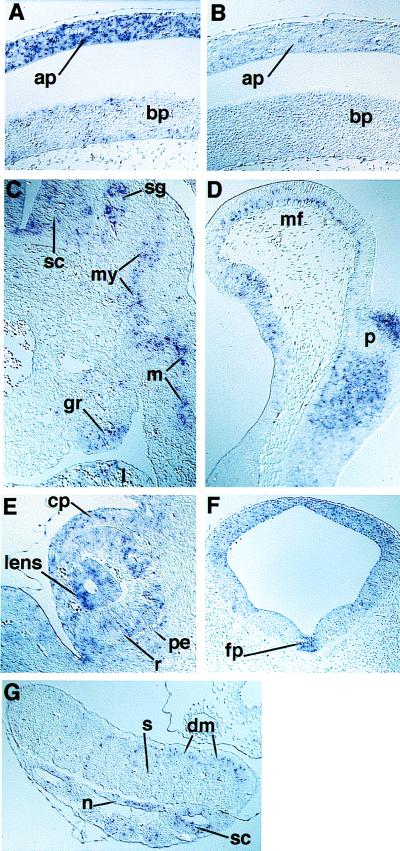
Figure 4.
Nonradioactive in situ hybridization analysis of RDH4 expression in mouse embryos during gd 10 and 11. (A) Sagittal section of a gd 10 embryo, at the level of the heart, showing expression of RDH4 in the alar plate of the spinal cord. (B) Section, parallel to that of A, hybridized with a sense probe showing absence of staining, thus verifying the specificity of the in situ hybridization technique. (C) Transverse section of a gd 11 embryo, at the level of the genital ridge, showing the expression of RDH4 in myotome and migrating muscle progenitor cells in the body wall. Expression also was seen in cells of the genital ridge, spinal ganglion, and the liver. (D) Sagittal section of a gd 11 embryo showing expression of RDH4 in the basal cells of the neuroepithelium of the mesencephalic flexure, and in collections of cells in the pons. (E) Frontal section of a gd 11 embryo, showing expression of RDH4 in the retina and lens, and also in the neural crest-derived cells of the anterior eye segment. (F) Frontal section of a gd 11 embryo, showing a dorso-ventral gradient of expression of RDH4 in the mesencephalon and a distinct expression in the floor plate. (G) Section through the hind part (just behind the hind limb) of a gd 11 embryo showing strongly stained cells in the spinal cord and notochord, and in the cells of the dermomyotome in a pattern consistent with an expression in a myogenetic cell lineage. Note the lack of expression in the sclerotome. ap, Alar plate; bp, basal plate; cp, neural crest-derived corneal precursor mesenchyme; dm, dermomyotome; fp, floor plate; l, liver; m, myocytes; mf, mesencephalic flexure; my, myotome; pe, pigment epithelium; p, pons; n, notochord; r, retina; s, sclerotome; sc, spinal cord; sg, spinal ganglia; gr, genital ridge. Original magnifications were ×80 (A–E and G) and ×40 (F).
Table 1.
Summary of the expression pattern of RDH4 in gd 10 and 11 mouse embryos
| Organ | Specific sites of RDH4 expression |
|---|---|
| Neural tube and ganglia | Dorsally, esp. mesencephalon and neural tube caudal to hindbrain; cells close to central canal |
| Specific cells in the floor of the mesencephalic flexure, pons and isthmus | |
| Optic stalk | |
| Floor plate | |
| Cranial and dorsal root ganglia | |
| Sensory organs | Optic and otic vesicles |
| Retina and lens | |
| Neural crest-derived corneal precursor cells | |
| Endodermally derived organs | Endoderm of the primitive gut |
| Liver | |
| Trachea and pulmonary epithelia | |
| Circulatory system | Trabecular layer of heart ventricles and outflow tract |
| Dorsal aorta and cardinal veins | |
| Others | Notochord |
| Myotome and muscle progenitor cells | |
| Rathke’s pouch | |
| Genital ridge, mesonephric ducts, tubuli |
Predominant sites of expression of RDH4 were in the developing central nervous system and sensory organs (optic and auditory systems), cranial and spinal ganglia, and endoderm of both foregut and hindgut. At gd 8.5, the earliest stage investigated, the neuroepithelium was stained, apparently in a relatively even pattern, except for a lower staining in the forebrain (data not shown). There was no evidence of a rhombomere-specific labeling at this or at later stages in the hindbrain.
At gd 10, RDH4-expressing cells could be found along the entire neural tube, generally with strongest expression dorsally (Fig. 4A, Table 1). Positive cells also were found in the mid- and hindbrain floor and around the central canal of the brain vesicles and spinal cord. Other sites of expression were endoderm and endodermally derived structures such as the tracheal epithelium and the liver. Lung mesenchyme and the trabecular layer of the heart ventricles also showed strong expression.
At gd 11, the dorso-ventral gradient of the expression of RDH4 in the brain vesicles and along the spinal cord was still apparent, although less marked. Expression was found both in the telencephalic and mesencephalic roof (Fig. 4F). Strong staining was seen in the isthmus area, i.e., the cerebellar primordium, in collections of cells in the pons and in a defined layer of cells basal in the neuroepithelium of the mesencephalic flexure (Fig. 4D). Rathke’s pouch, spinal and cranial ganglia (Fig. 4C), and the floor plate also showed staining (Fig. 4F). Interestingly, the expression of RDH4 in the floor plate appeared discontinuous along the rostro-caudal axis of the neural tube, as analysis of series of transverse sections yielded a variable labeling in the floor plate (unpublished observation). Cells in the dermomyotomes (Fig. 4G) and migrating muscle progenitor cells (Fig. 4C) expressed transcripts for RDH4, as did cells in the notochord (Fig. 4G). Gastric and intestinal epithelium, liver, mesonephric ducts and tubuli, and the genital ridge (Fig. 4C) expressed RDH4 transcripts. In the eyes (retina and lens), and the optic stalks there was a strong expression, as well as in the neural crest-derived mesenchyme in the anterior eye segment (Fig. 4E). It is important to note that neural crest-derived cells, previously recognized as major targets for retinoids (reviewed in ref. 7) generally do not seem to express RDH4 in the early stages of embryonic development. However, this finding does not exclude a later expression in their derivatives at local differentiation (e.g., anterior eye segment shown in Fig. 4E) or that stained cells in tissues in part derived from the neural crest, like spinal or cranial ganglia, may be of neural crest origin.
Flank mesoderm and the limb buds showed no specific staining for RDH4 at gd 10 and 11. At later developmental stages, migrating limb muscle progenitor cells expressed RDH4 (unpublished observation). The cochlear portion of the otic vesicle showed strong expression of RDH4, whereas fewer cells of the vestibular portion were stained. Strong staining also was found in the endolymphatic duct.
DISCUSSION
RDH4 Is a Member of the SDR family of RDHs Expressed in Adult and Embryonic Tissues.
The physiological substrate for this enzyme is likely to be 9-cis retinol despite the ability of the enzyme to metabolize 11-cis and 13-cis retinols, because 11-cis retinoid isomers are exclusively associated with the visual processes of the eyes, and there are no known nuclear receptors for 11-cis or 13-cis RA. The close structural similarities between RDH4 and the eye-specific 11-cis RDH furthermore suggest that the cis-retinol specific RDHs, has evolved as a separate subgroup among the RDHs of the SDR family.
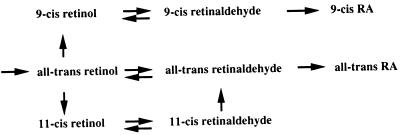
9-cis RA as a naturally occurring ligand for the RARs is widely accepted, but the mechanism underlying its biosynthesis has remained obscure. All-trans retinol is the ultimate precursor in the biosynthesis of both 9-cis and all-trans isomers of RA, but it has not been established whether the in vivo isomerization reaction from an all-trans to a cis isomer occurs on the level of retinol, retinaldehyde, or RA. In the visual system, with unique requirements for 11-cis stereo isomers, isomerization occurs on the level of all-trans retinol by a specific enzymatic process involving an isomerase-hydrolase activity (32). The identification of a 9-cis RDH expressed in embryonic and adult tissues suggests a metabolic pathway in vivo, where 9-cis retinol is generated from all-trans retinol by an isomerization reaction similar to that described in the visual system (see Fig. 5). Oxidation of 9-cis retinol into 9-cis RA then is likely to proceed similarly to the generation of all-trans RA from all-trans retinol. A consequence of this metabolic scheme would be that generation of the two isomers of RA is carried out by two independent pathways, each of which may be under separate metabolic control. This provides an alternative to the view where all-trans RA is believed to be the precursor in 9-cis RA biosynthesis (8, 33).
Figure 5.
Proposed metabolic pathways for synthesis of all-trans, 9-cis, and 11-cis retinoids. The existence of a 9-cis RDH suggests that 9-cis retinol is generated from all-trans retinol by using an evolutionary conserved mechanism similar to that involved in the generation of 11-cis retinol in the visual system. The proposed metabolic route generating 9-cis RA from 9-cis retinol is similar to that generating all-trans RA from all-trans retinol.
The expression pattern of RDH4 is only partially overlapping with other known retinoid-metabolizing enzymes expressed in developing embryos (34–36). At present, the functional implications of this observation is unclear but it may suggest that several different enzymes/enzyme classes are involved in regulation of RA homeostasis during development.
In adult mice RDH4 is coexpressed with RARα and β in liver, brain, and kidney (37), with RXRα and γ in liver, and with RXRβ and γ in brain. All three RXRs are expressed in kidney (6). It is notable that RDH4 is not expressed in adult heart, muscle, and several other tissues with abundant expression of RARs and RXRs. This finding suggests that enzymes, other that RDH4, may act as 9-cis RDHs or that 9-cis RA is not generated in these tissues. It should be noted that 9-cis RA has been directly demonstrated in adult mouse liver and kidney (8), sites of high expression of RDH4, and urodele wound epidermis (38).
The restricted expression pattern of RDH4 in adult mouse tissues contrasts its widespread expression during embryonic development. Abundant RDH4 expression was seen in parts of the central nervous system, the sensory organs, somites and somite derivatives, and organs of endodermal origin. Again, all RDH4-expressing tissues express RARs and RXRs (e.g., the widely expressed RARα and β and RXRα and β). There is a partial overlap in the expression with RXRγ whereas coexpression with RARγ appears minimal (39). Notable coexpression of RDH4 and RARβ is seen in the dorsal region of the early central nervous system (40, 41) and there is a marked colocalization of RDH4 and RXRγ in muscle progenitor cells in somites and in the developing myotome (6).
Null mutations for all six RARs and RXRs have been generated. Deletion of RAR genes, particularly different combinations of RAR double mutants, frequently results in ocular and heart abnormalities, among other defects, which also are commonly seen in fetuses from vitamin A-deficient animals (reviewed in ref. 7). RXRα−/− mice suffer from impaired heart development and ocular abnormalities (42, 43). Notable expression of RDH4 is seen in the trabecular layer of the developing ventricular wall of the heart and a “spongy myocardium” develops in RXRα−/− animals and in RARα/γ double mutants. Similarly, RDH4 is highly expressed in the retina and lens and in neural crest-derived cells contributing to the anterior segment of the developing eye. RXRα−/− and some RAR double-mutant mice develop malformations of the anterior segments of the eye, lens agenesis, and a shortening of the ventral retina (42, 44). Thus, RDH4 is expressed in tissues affected by the receptor deletions and with known requirements for retinoids.
The expression of RDH4 in the neural system and in muscle progenitor cells suggests that 9-cis RA affects gene expression in these tissues and thus it may play a role in neuronal patterning and myogenesis. The expression of RDH4 in the notochord and floor plate of the central nervous system is particularly intriguing. The floor plate is known to synthesize active retinoids (13), and both the notochord and the floor plate have an instructive role in genesis of motor neurons in the spinal cord (reviewed in ref. 45).
The spatially and temporally well-controlled expression of RDH4 suggests that the capacity to generate 9-cis RA is strictly regulated. This feature contrasts with the widespread expression of the nuclear RARs, and it remains to be shown whether expression of RDH4 is an indicator of local synthesis of 9-cis RA. By using molecular genetic approaches it will be possible to obtain valuable insights into the role(s) of RDH4, and other retinoid-metabolizing enzymes, in RA homeostasis during embryonic development and in adult tissues.
Acknowledgments
We thank Barbara Åkerblom and Raili Engdahl for expert technical assistance and Dr. Ivor Mason for introducing us to the in situ hybridization technique. This work was supported by grants from the Swedish Medical Research Council (K97–03P-12070–01A to U.E. and K97–03X-07899–11B to L.D.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: gd, gestation day; RA, retinoic acid; RAR, retinoic acid receptor; RXR, retinoid X receptor; RDH, retinol dehydrogenase; SDR, short chain alcohol dehydrogenase/reductase; RT, reverse transcriptase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF013288).
References
- 1.Chambon P. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 2.Mangelsdorf D J, Umesono K, Evans R M. In: The Retinoid Receptors. Sporn M B, Roberts A B, Goodman D S, editors. New York: Raven; 1994. pp. 319–349. [Google Scholar]
- 3.Kastner P, Mark M, Ghyselinck N, Krezel W, Dupe V, Grondona J M, Chambon P. Development (Cambridge, UK) 1997;124:313–326. doi: 10.1242/dev.124.2.313. [DOI] [PubMed] [Google Scholar]
- 4.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 5.Dollé P, Fraulob V, Kastner P, Chambon P. Mech Dev. 1994;45:91–104. doi: 10.1016/0925-4773(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 6.Mangelsdorf D J, Borgmeyer U, Heyman R A, Zhou J Y, Ong E S, Oro A E, Kakizuka A, Evans R M. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 7.Kastner P, Mark M, Chambon P. Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 8.Heyman R, Mangelsdorf D, Dyck J, Stein R, Eichele G, Evans R, Thaller C. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 9.Horton C, Maden M. Dev Dyn. 1995;202:312–323. doi: 10.1002/aja.1002020310. [DOI] [PubMed] [Google Scholar]
- 10.Hunter K, Maden M, Summerbell D, Eriksson U, Holder N. Proc Natl Acad Sci USA. 1991;88:3666–3670. doi: 10.1073/pnas.88.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thaller C, Eichele G. Nature (London) 1987;327:625–628. doi: 10.1038/327625a0. [DOI] [PubMed] [Google Scholar]
- 12.Thaller C, Eichele G. Nature (London) 1990;345:815–819. doi: 10.1038/345815a0. [DOI] [PubMed] [Google Scholar]
- 13.Wagner M, Thaller C, Jessell T M, Eichele G. Nature (London) 1990;345:819–822. doi: 10.1038/345819a0. [DOI] [PubMed] [Google Scholar]
- 14.Colbert M C, Linney E, LaMantia A S. Proc Natl Acad Sci USA. 1993;90:6572–6576. doi: 10.1073/pnas.90.14.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendelsohn C, Ruberte E, LeMeur M, Morriss-Kay G, Chambon P. Development (Cambridge, UK) 1991;113:723–734. doi: 10.1242/dev.113.3.723. [DOI] [PubMed] [Google Scholar]
- 16.Rossant J, Zirngibl R, Cado D, Shago M, Giguere V. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- 17.Wagner M, Han B, Jessell T M. Development (Cambridge, UK) 1992;116:55–66. doi: 10.1242/dev.116.1.55. [DOI] [PubMed] [Google Scholar]
- 18.Napoli J L. Clin Immunol Immunopathol. 1996;80:S52–S62. doi: 10.1006/clin.1996.0142. [DOI] [PubMed] [Google Scholar]
- 19.Duester G. Biochemistry. 1996;35:12221–12227. doi: 10.1021/bi961176+. [DOI] [PubMed] [Google Scholar]
- 20.Chai X, Boerman M H E M, Zhai Y, Napoli J L. J Biol Chem. 1995;270:3900–3904. doi: 10.1074/jbc.270.8.3900. [DOI] [PubMed] [Google Scholar]
- 21.Chai X, Zhai Y, Popescu G, Napoli J L. J Biol Chem. 1995;270:28408–28412. doi: 10.1074/jbc.270.47.28408. [DOI] [PubMed] [Google Scholar]
- 22.Chai X, Zhai Y, Napoli J L. Gene. 1996;169:219–222. doi: 10.1016/0378-1119(95)00833-0. [DOI] [PubMed] [Google Scholar]
- 23.Simon A, Hellman U, Wernstedt C, Eriksson U. J Biol Chem. 1995;270:1107–1112. [PubMed] [Google Scholar]
- 24.Mertz J R, Shang E Y, Piantedosi R, Wei S H, Wotgemuth D J, Blaner W S. J Biol Chem. 1997;272:11744–11749. doi: 10.1074/jbc.272.18.11744. [DOI] [PubMed] [Google Scholar]
- 25.McCaffery P, Dräger U C. Adv Exp Med Biol. 1995;372:173–183. doi: 10.1007/978-1-4615-1965-2_23. [DOI] [PubMed] [Google Scholar]
- 26.Zhao D, McCaffery P, Ivins K J, Neve R L, Hogan P, Chin W W, Dräger U C. Eur J Biochem. 1996;240:15–22. doi: 10.1111/j.1432-1033.1996.0015h.x. [DOI] [PubMed] [Google Scholar]
- 27.Chirgwin J, Przybyla A, MacDonald R, Rutter W. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 28.Posch K C, Boerman M H E M, Burns R, Napoli J L. Biochemistry. 1991;30:6224–6230. doi: 10.1021/bi00239a021. [DOI] [PubMed] [Google Scholar]
- 29.Konat G W, Laszkiewicz I, Grubinska B, Wiggins R C. In: Generation of Labeled DNA Probes by PCR. Griffin H G, Griffin A M, editors. Boca Raton, FL: CRC; 1994. pp. 37–42. [Google Scholar]
- 30.Gustafson A-L, Dencker L, Eriksson U. Development (Cambridge, UK) 1993;117:343–351. doi: 10.1242/dev.117.2.451. [DOI] [PubMed] [Google Scholar]
- 31.Jörnvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 32.Rando R R, Bernstein P S, Barry R J. In: New Insights into the Visual Cycle. Osborne N N, Chader G J, editors. Vol. 12. Oxford: Pergamon; 1991. pp. 161–178. [Google Scholar]
- 33.Levin A, Storzenbecker L, Kazmer Z, Bosakowski T, Huselton C, Allenby G, Speck J, Kratzeisen C, Rosenberger M, Lovey A, Grippo J. Nature (London) 1992;355:259–261. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]
- 34.Ang H, Deltour L, Zgombic-Knight M, Wagner M, Duester G. Alcohol Clin Exp Res. 1996;20:1050–1064. doi: 10.1111/j.1530-0277.1996.tb01946.x. [DOI] [PubMed] [Google Scholar]
- 35.Ang H L, Duester G. Dev Dyn. 1997;208:536–543. doi: 10.1002/(SICI)1097-0177(199704)208:4<536::AID-AJA9>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 36.Niederreither K, McCaffery P, Dräger U C, Chambon P, Dollé P. Mech Dev. 1997;62:67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- 37.Zelent A, Krust A, Petkovich M, Kastner P, Chambon P. Nature (London) 1989;339:714–717. doi: 10.1038/339714a0. [DOI] [PubMed] [Google Scholar]
- 38.Viviano C, Horton C, Maden M, Brockes J P. Development (Cambridge, UK) 1995;121:3753–3762. [Google Scholar]
- 39.Dollé P, Ruberte E, Leroy P, Morriss-Kay G, Chambon P. Development (Cambridge, UK) 1990;110:1133–1151. doi: 10.1242/dev.110.4.1133. [DOI] [PubMed] [Google Scholar]
- 40.Ruberte E, Dollé P, Chambon P, Morriss-Kay G. Development (Cambridge, UK) 1991;111:45–60. doi: 10.1242/dev.111.1.45. [DOI] [PubMed] [Google Scholar]
- 41.Ruberte E, Friederich V, Chambon P, Morriss-Kay G. Development (Cambridge, UK) 1993;118:267–282. doi: 10.1242/dev.118.1.267. [DOI] [PubMed] [Google Scholar]
- 42.Kastner P, Grondona J M, Mark M, Gansmuller A, LeMeur M, Decimo D, Vonesch J L, Dollé P, Chambon P. Cell. 1994;78:987–1003. doi: 10.1016/0092-8674(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 43.Sucov H M, Dyson E, Gumeringer C L, Price J, Chien K R, Evans R M. Genes Dev. 1994;8:1007–1018. doi: 10.1101/gad.8.9.1007. [DOI] [PubMed] [Google Scholar]
- 44.Mendelsohn C, Lohnes D, Decimo D, Lufkin T, LeMeur M, Chambon P, Mark M. Development (Cambridge, UK) 1994;120:2749–2771. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- 45.Tanabe Y, Jessell T M. Science. 1996;274:1115–1123. doi: 10.1126/science.274.5290.1115. [DOI] [PubMed] [Google Scholar]