Abstract
The contribution of penicillin-binding protein 5 (PBP5) and the PBP5 synthesis repressor (Psr) to the β-lactam resistance, growth, and cell autolysis of wild-type strain ATCC 9790 and resistant strain R40 of Enterococcus hirae was investigated by disruption or substitution of the corresponding pbp5 and psr genes by Campbell-type recombination. The resulting modifications were confirmed by hybridization and PCR. The low susceptibility of E. hirae to β-lactams was confirmed to be largely dependent on the presence of PBP5. However, against all expectations, inactivation of psr in ATCC 9790 or complementation of R40 cells with psr did not modify the susceptibility to benzylpenicillin or the growth and cell autolysis rates. These results indicated that the psr gene does not seem to be involved in the regulation of PBP5 synthesis and consequently in β-lactam resistance or in the regulation of cell autolysis in E. hirae.
The natural low susceptibility of enterococci to β-lactams is due to a low-affinity penicillin-binding protein (PBP) that is overproduced in moderately resistant laboratory mutants and some clinical strains (16, 17, 35, 45). Highly resistant clinical strains generally do not overproduce this low-affinity PBP but modify its primary structure to further reduce its binding capacity (21, 24, 35, 45). Conversely, when the low-affinity PBP is not synthesized, the cells become highly susceptible to benzylpenicillin (PenG) (18).
Sequencing of the enterococcal genes encoding low-affinity PBPs showed that these proteins are relatively similar (∼75 kDa) (13, 14, 33, 35, 45). Including PBP2′ of methicillin-resistant staphylococci (1) and PBP3 of Bacillus subtilis (28), these protein form subgroup B1 of the class B high-molecular-mass PBPs (19). When penicillin is present, they can take over the functions of most if not all the other PBPs. This has not been verified for PBP3 in B. subtilis. Thus, in enterococci and staphylococci these proteins appear to be multifunctional PBPs that are not essential for growth under laboratory conditions but synthesize peptidoglycan when the other PBPs are inhibited (5, 6, 7, 17, 18).
The study of Enterococcus hirae contributed a great deal to these observations. From wild-type strain ATCC 9790 (MIC of PenG, 0.6 μg/ml), resistant strain R40 (MIC of PenG, 60 μg/ml) was isolated by a four-step selection procedure on plates containing increasing PenG concentrations. The greater resistance of strain R40 was attributed to overproduction of the low-affinity PBP5 (17). However, it also appeared that R40 differed from parent strain ATCC 9790 by slower growth, faster autolysis, a lower cell wall rhamnose content, and greater susceptibility to lysozyme (27). PenG-hypersusceptible strain Rev14 (MIC of PenG, 0.015 μg/ml), which does not synthesize PBP5, was derived from R40 by chemical mutagenesis (18). Except for its high level of susceptibility to β-lactams, this strain has the properties described above for R40 (27).
Expression of the PBP5-encoding gene, pbp5, in E. hirae was proposed to be under control of psr, a 882-bp gene starting 1 kb upstream of pbp5, on the basis of the 87-bp deletion that overlapped the 5′end of psr (23, 26). It was also suggested that the psr gene has a larger regulatory function in control of the different cell wall-related properties described above (27).
It is also assumed that a similar situation exists in Enterococcus faecium strains; all of these strains except D63 have immediately upstream of pbp5 (pbp5fm) a 756-bp psr gene (psrfm), the product of which exhibits 79.9% identity with the Psr protein of E. hirae (P. Brouillard and J. Coyette, unpublished data). In D63, psrfm ends prematurely with a 597-bp sequence (45). However, in contrast to previous results, it was recently demonstrated that the amount of the pbp5fm transcript in a psrfm-deficient clone of E. faecium was similar to the amount in the wild-type strain (34). Finally, a psr-like gene was also detected in Enterococcus faecalis JH2-2, but in contrast to the psr genes of the two species described above, this gene appeared to be located several kilobases away from pbp4, the low-affinity PBP4-encoding gene. No indication that the psr-like gene is involved in the overproduction of PBP4 in a resistant clone derived from E. faecalis JH2-2 (13) was found.
As the mutants used in previous studies were not isogenic strains, we could not eliminate the possibility that they contained different unidentified genetic modifications that could have contributed to their different traits, including their higher levels of resistance. The purpose of this study was to reexamine the role of the psr gene in cell autolysis and β-lactam resistance of E. hirae by specifically disrupting or deleting the gene by homologous double-recombination techniques. Isogenic pbp5-deficient mutants were also prepared and used as controls.
(Parts of this work were performed during preparation of Ph.D. theses by F.S., C.F., and O.D.)
MATERIALS AND METHODS
Bacterial strains, media, growth conditions, and MIC determination.
The Escherichia coli and E. hirae strains used in this work are listed in Table 1. E. coli Top10F′ was used to generate and maintain recombinant plasmids in Luria broth (Difco, Erembodegem, Belgium) at 37°C (2). E. hirae strains were grown in brain heart infusion broth (Difco) or in Todd-Hewitt broth (Difco) supplemented with 2% glucose (THG) at 37°C except for isolation of recombinants as indicated below. Growth curves were established by measuring the optical density at 550 nm (OD550) at regular intervals. M17 medium (Difco) was also used to prepare electrocompetent E. hirae cells (42).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype and/or phenotypea | Source or reference |
|---|---|---|
| E. coli strains | ||
| Top10F′ | F′ [lac1q Tn10 (Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) φ80 lacZΔ′M15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 nupG | Invitrogen |
| BL21(DE3) | F′ [ompT hsd SB(rB− mB−) gal dcm(DE3)] | Novagen |
| E. hirae strains | ||
| AS21 | Pens mutant derived from ATCC 9790 (MIC of PenG, 0.075 μg/ml) | 11 |
| AS21R2 | Penr mutant derived from AS21 (MIC of PenG, 30 μg/ml) | 11 |
| ATCC 9790 | Wild-type strain (MIC of PenG, 0.6 μg/ml) | ATCCb |
| Rev14 | Pens mutant derived from R40 (MIC of PenG, 0.015 μg/ml) | 18 |
| FS1 | R40 pbp5::Spcr | This study |
| FS2 | ATCC 9790 pbp5::Kmr | This study |
| FS3 | ATCC 9790 psr::Kmr (disruption) | This study |
| FS4 | ATCC 9790 psr::Kmr (substitution) | This study |
| FS5 | ATCC 9790 psr::Kmr (substitution) | This study |
| R40 | Penr mutant derived from ATCC 9790 (MIC of PenG, 60 μg/ml) | 17 |
| Plasmids | ||
| pBR322 | Apr TetrE. coli cloning vector | Amersham Pharmacia Biotech |
| pCR2.1 | Apr KmrE. coli TA cloning vector | Invitrogen |
| pDG792 | E. coli vector carrying the 1.5-kb Kmr gene cassette | 20 |
| pER924 | Apr ErmrE. coli-gram-positive shuttle vector(thermosensitive gram-positive replication) | 3 |
| pET22b(+) | KmrE. coli expression vector | Novagen |
| pGEM-3Zf(−) | AprE. coli cloning vector | Promega |
| pHP45Ω | AprE. coli vector carrying the 2.0-kb Smr-Spcr gene cassette | 32 |
| pIL252 | Ermr gram-positive cloning vector (low copy) | 39 |
| pIL253 | Ermr gram-positive cloning vector (high copy) | 39 |
| pSL1190 | AprlacZ E. coli cloning vector | Amersham Pharmacia Biotech |
| pUC18 | AprlacZ E. coli cloning vector | Amersham Pharmacia Biotech |
| pDML529 | pGEM-3Zf(−) carrying the 1.43-kb PCR-amplified psr gene | This study |
| pDML530 | pBR322 carrying the 6.9-kb EcoRI fragment (psr-pbp5) of ATCC 9790 | This study |
| pDML531 | pDML529-pIL253 fusion | This study |
| pDML532 | pDML530-pIL252 fusion | This study |
| pDML533 | pDML530-pIL253 fusion | This study |
| pDML534 | pET22b(+) carrying truncated psrN | This study |
| pDML535 | pUC18 carrying the 4.0-kb PstI psr::Kmr fragment | This study |
| pDML540 | pBR322 carrying the 6.9-kb EcoRI fragment (deleted psr-pbp5) of R40 | 14 |
| pDML541 | pBR322 carrying the 2.6-kb EcoRI/PvuI fragment (pbp5) derived from the pDML540 insert | 29 |
| pDML542 | pDML541-pIL252 fusion | 29 |
| pDML543 | pDML541-pIL253 fusion | 29 |
| pDML1600 | pUC18 carrying pbp5 (1.6-kb PCR fragment) | This study |
| pDML1601 | pUC18 carrying pbp5::Spcr | This study |
| pDML1602 | pUC18 carrying pbp5::Kmr | This study |
| pDML1603 | pSL1190 carrying psr (1.55-kb PCR fragment) | This study |
| pDML1604 | pSL1190 carrying psr::Kmr | This study |
| pDML1605 | pUC18 carrying an ftsW fragment (0.88-kb PCR fragment) | This study |
| pDML1606 | pSL1190 carrying pDML1605 insert | This study |
| pDML1607 | pUC18 carrying a pbp5 fragment (0.85-kb PCR fragment) | This study |
| pDML1608 | pSL1190 carrying pDML1605 insert-pDML1607 insert fusion | This study |
| pDML1609 | pSL1190 carrying pDML1608 insert::Kmr (KpnI) | This study |
| pDML1610 | pER924 carrying pDML1601 insert | This study |
| pDML1611 | pER924 carrying pDML1602 insert | This study |
| pDML1612 | pER924 carrying pDML1604 insert | This study |
| pDML1613 | pER924 carrying pDML1609 insert | This study |
Apr, ampicillin resistant; Ermr, erythromycin resistant; Kmr, kanamycin resistant; Penr, PenG resistant; Pens, PenG sensitive; Smr, streptomycin resistant; Spcr, spectinomycin resistant; Tetr, tetracycline resistant.
ATCC, American Type Culture Collection.
The antibiotics and concentrations used were as follows. Ampicillin (100 μg/ml; Squibb Bristol Myers, Brussels, Belgium), kanamycin (100 μg/ml; Calbiochem, Bierges, Belgium), and spectinomycin (100 μg/ml; Pharmacia Upjohn, Puurs, Belgium) were used for selection of E. coli transformants. Erythromycin (5 or 10 μg/ml; Fluka, Bornem, Belgium) was used for initial selection of E. hirae transformants after electroporation. Kanamycin (100 μg/ml) and PenG (0.1 μg/ml; Aventis, Lyon, France) were used to select E. hirae recombinants as indicated below. Other selective additives, such as 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 μg/ml) and isopropyl-β-d-thiogalactopyranoside (IPTG) (7 μg/ml) (both obtained from Immunosource, Zoersel-Halle, Belgium), were used or added to the media as required.
MICs were determined in 1 ml of brain heart infusion broth in 24-well culture plates in at least three independent experiments, essentially as previously described (9).
Cell autolysis.
Cells grown in THG and collected in the exponential (OD550, ≅1.0) and stationary (OD550, ≅1.7) phases were washed three times in cold water, resuspended in 10 mM sodium phosphate buffer (pH 6.8), and incubated at 37°C as previously described (27). Rates of autolysis were calculated from the exponential portions of lysis curves.
Plasmid construction.
Common E. coli plasmids (Table 1) were used for cloning purposes. The E. coli-E. hirae shuttle vectors were cointegrates of pIL252 or pIL253 (39) on the one side and pBR322 on the other side, usually at the EcoRI sites. Disruption or deletion experiments were performed by using the pER924 thermosensitive shuttle vector (3), into which resistance gene cassettes and specific DNA fragments were inserted.
General DNA methods.
Standard DNA methods were used (2, 36). Restriction endonucleases from Promega (Leiden, The Netherlands), Life Technologies (Merelbeke, Belgium), and Fermentas (St. Leon-Rot, Germany), calf intestinal alkaline phosphatase from Roche Diagnostics (Brussels, Belgium), T4 DNA polymerase from Life Technologies, and T4 DNA ligase from Boehringer Ingelheim Bioproducts (Verviers, Belgium) were used as recommended by the suppliers.
E. coli plasmids were extracted with a GFX Microplasmid prep kit (Amersham Pharmacia Biotech, Roosendall, The Netherlands). DNA fragments or PCR products were purified with a GeneClean Spin kit from Bio 101 Systems (Polylab, Antwerp, Belgium) or a FREE DNA kit from Millipore (Brussels, Belgium). The Enterococcus genomic DNA was purified by using a Wizard genomic DNA extraction kit (Promega) or as previously described (25).
Southern hybridization.
Transfer of DNA (4 μg per lane) from 0.8% (wt/vol) agarose gels to Hybond N+ membranes (Amersham Pharmacia Biotech) was performed by using the VacuGeneXL vacuum blotting system (Amersham Pharmacia Biotech) and standard protocols (36). Probe labeling, hybridization, and washing were done by using the AlkPhos direct labeling and detection systems (Amersham Pharmacia Biotech). The sizes of the hybridizing fragments of the chromosomal digestion products were determined by using the Smart ladder marker from Eurogentec (Seraing, Belgium) labeled by the same technique.
PCR.
PCR amplification was performed with a TRIO-Thermoblock Biometra thermocycler (Eurogentec). The Taq, Pwo, Vent, and Biotools DNA polymerases from Amersham Pharmacia Biotech, Eurogentec, New England Biolabs (Westburg, Leusden, The Netherlands), and Labsystem (Brussels, Belgium), respectively, were used according to the manufacturers' recommendations. Amplification was usually carried out by using 25 to 30 cycles. Each cycle consisted of 1 min at 95°C, 30 s at the melting temperature minus 3°C, and 1 min/kb at 72°C. The last cycle was followed by a 10-min elongation step at 72°C.
Nucleotide sequencing.
DNA sequencing was performed by using Autoread Thermo Sequenase sequencing kits (Amersham Pharmacia Biotech) with Cy5 primers and an ALF Express DNA sequencer (Amersham Pharmacia Biotech). The primers were chosen so that we obtained sequences that overlapped on both strands.
Transformation of E. hirae cells and isolation of recombinants.
E. coli competent cells were transformed as previously described (36). E. hirae electrocompetent cells were transformed by electroporation by using 0.1-cm cuvettes and a Gene Pulser (Bio-Rad, Nazareth-Eke, Belgium). The parameters used were as follows: 1,000 Ω, 25 μF, and 18 to 23 kV/cm (41).
Gene disruptions or deletions in E. hirae cells were obtained by double recombination of electroporated integration vectors. The recombinants grown at 28 and 42°C (the permissive and nonpermissive temperatures, respectively) were selected by replica plating (∼200 colonies/plate) on the basis of phenotypic susceptibility or resistance to PenG, erythromycin, kanamycin, or spectinomycin (4).
PBP detection by radioactive labeling and immunoblotting.
Preparation of the membranes of the different E. hirae clones, estimation of the protein contents, [14C]PenG PBP labeling, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and fluorography were performed as described elsewhere (45). However, in the fluorography procedure, the 2,5-diphenyloxazole scintillant used previously was replaced by a 1 M sodium salicylate solution (8). PBPs were detected by using a phosphorimager screen (Kodak-K screen) after 24 h of exposure at room temperature (FX-Imager; Bio-Rad).
Anti-PBP5 rabbit polyclonal antibodies that were prepared and partially purified by immunoadsorption, as previously described (14, 30), were used for detection of PBP5 by immunoblotting. Anti-Psr rabbit polyclonal antibodies were kindly provided by L. Daneo-Moore for detection of Psr by immunoblotting (27). Alkaline phosphatase-coupled goat anti-rabbit polyclonal antibodies were used as secondary antibodies (Immun-Blot colorimetric assay kit; Bio-Rad).
Electrophoretic mobility shift assay (EMSA).
Protein samples containing various amounts of PsrN (10 to 250 ng) were incubated with 20 ng of the Cy5-labeled probe in the presence of 2 to 4 μg of double-stranded poly(dI-dC) (Amersham Pharmacia Biotech) in 20 μl of a reaction buffer containing 25 mM HEPES (pH 7.5), 0.1 mM EDTA, 5 mM dithiothreitol, and 50 mM KCl. Incubation was carried out at room temperature or at 30°C for 20 to 60 min, and the reactions were stopped by adding 4 μl of the loading dye fluorescent sample buffer (Amersham Pharmacia Biotech). The samples were resolved on 4% (wt/vol) nondenaturing polyacrylamide gels in 0.89 M Tris-borate-20 mM EDTA buffer (pH 8.2) by using an Alf Express sequencer (Amersham Pharmacia Biotech). After electrophoresis (180 min, 10 W, 800 V, 45 mA), the gels were analyzed by using the Fragment Manager program (Amersham Pharmacia Biotech) (15).
Nucleotide sequence accession number.
The sequence of the 6.9-kb EcoRI insert of E. hirae ATCC 9790 has been deposited in the EMBL database under accession no. AJ429231.
RESULTS
Sequence analysis of the psr-pbp5 locus of E. hirae and its flanking regions.
The complete sequence of the 6.9-kb EcoRI insert of pDML540 bearing the psr and pbp5 genes, which was isolated previously (14) from the PenG-resistant (Penr) E. hirae R40 strain, was established for both strands. This sequence was compared to that of the allelic 6.9-kb EcoRI fragment of wild-type strain E. hirae ATCC 9790 carried by pDML530. This vector was selected from an EcoRI gene library made in the pUC18 Ready To Go vector (Amersham Pharmacia Biotech). Positive colonies were identified by PCR in the presence of specific psr and pbp5 primers O1 and O2*, O1 and O4*, and O3 and O4* (Table 2).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotideb | Sequencea | Gene | Position from ATG |
|---|---|---|---|
| O1 | 5′ GGAATCGCATATGAAAACATTTCAAAAAGTG 3′ | psr | −10 |
| O2* | 5′ AAATAAAATGTAAATCTCGAGGTCTAAAAACTG 3′ | psr | 900 |
| O3 | 5′ CGGTCGACTCTTGGGAAGTATTAAA 3′ | psr | 669 |
| O4* | 5′ AGCATCGGCATCAAGAGTTAATTT 3′ | pbp5 | 1052 |
| O5 | 5′ GAGTTTGGGTTGATAGGCGCA 3′ | psr | 431 |
| O6* | 5′ TTTGTCCCTCCGCTCACATTTTC 3′ | psr | 1001 |
| O7 | 5′ AAATGAATTCAAGCTTAGGCGAATTAAA 3′ | pbp5 | 361 |
| O8* | 5′ ATCAGGATCCACAGCAAATAGGAAG 3′ | pbp5 | 1928 |
| O9 | 5′ TAAGAAAATGTGAGCGGAGGG 3′ | pbp5 | −26 |
| O10* | 5′ TCAGGATTCACAGCAAATAGGAAGC 3′ | pbp5 | 1927 |
| O11 | 5′ ACGCTTAAGACCGAATCGCAATG 3′ | ftsW | 652 |
| O12* | 5′ AAGGATATCGCCTCTTTTAGC 3′ | pbp5 | 531 |
| O13 | 5′ AAGATCGACTGGTTGATTTTAGGCC 3′ | ftsW | 15 |
| O14 | 5′ TTTGAATTCTTTTTTTAGAAATGACCGCAGTGTTC 3′ | ftsW | 377 |
| O15 | 5′ CAAGCGCCCTTGTTTTGGTTCTTGCGGC 3′ | pbp5 | 49 |
| O16* | 5′ ACCGGGCCCAGGTATATCTACATAAGAATC 3′ | psr | 303 |
| O17* | 5′ CCGAACCTCTTACTTGAAATATAGCCA 3′ | ftsW | 1253 |
| O21 | 5′ AAAGGATCCTTAGCAATCAATCAGTTTTTAGA 3′ | pbp5 | −160 |
| O22* | 5′ GGTTGTACCCAACTTTGAGAGATAGCT 3′ | pbp5 | 703 |
| O23 | 5′ AGGATGAAGAGGATGAGGAGGC 3′ | aph3′ | 122 |
| O24* | 5′ CTCGTAGGCGCTCGGGAC 3′ | aph3′ | 1475 |
| O25* | 5′ AAATAAAATGTAAATCTCGAGGTCTAAAACTG 3′ | psr | 897 |
| O26 | 5′ GGTTACAACTCAAGAATTCTAAACTGTTAAA 3′ | psr | −123 |
| O27* | 5′ AGAATAACCGTGTACTTTTTGGAATGTTTTCATCTTCGAT 3′ | psr | −7 |
| O28 | 5′ AAAACATTCCAAAAAGTACACGGTTATTCTTCAACGAGTC 3′ | psr | 4 |
| O29* | Cy5-GCAAGAACCAAAACAAGGGCGC 3′ | pbp5 | 74 |
| O30 | 5′ CGGTCAACTCTTGGGAAGTATTAAAA 3′ | psr | 669 |
The underlined sequences are NdeI, AvaI, EcoRI, BamHI, and SmaI restriction sites.
*, complementary.
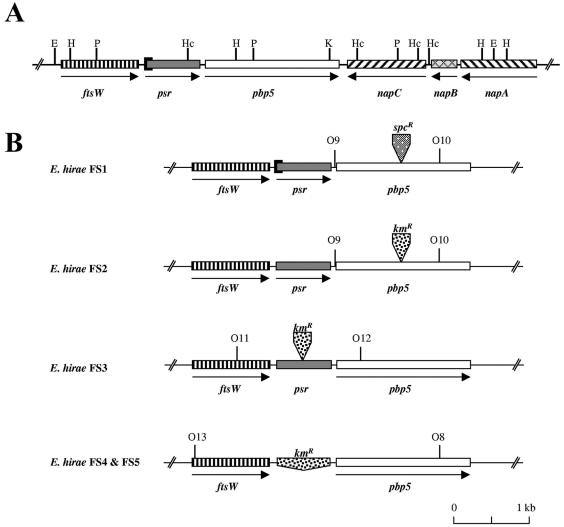
No difference except the 87-bp deletion identified previously (23, 26) encompassing the initial psr codon of E. hirae R40 was found when we compared the two inserts that contained four genes in addition to psr and pbp5 (Fig. 1A).
FIG. 1.
(A) Map of the ftsW-psr-pbp5 locus of E. hirae. (B) ftsW-psr-pbp5 loci of the different E. hirae recombinants. The arrows indicate the orientation of transcription. The solid rectangle at the origin of psr corresponds to the 87-bp deletion present in R40 and its FS1 derivative. The spcR and kmR inserts indicate the positions of the resistance cassettes. The hybridization sites of the O8 to O13 probes used in PCR are also shown. The restriction sites are indicated as follows: E, EcoRI; H, HindIII; P, PstI; Hc, HincII; K, KpnI.
Upstream of psr there was a 1,194-bp gene whose product exhibited 53, 35, 34, and 30.9% identity with the ftsW products of E. faecium, Streptococcus pneumoniae, Streptococcus pyogenes, and E. coli, respectively.
The three genes located downstream of pbp5 are the napA, napB, and napC genes identified previously on a 3.7-kb SauIIIA DNA fragment of E. hirae ATCC 9790 coding for an Na+/H+ antiporter activity (43). These genes appear to be oriented in the opposite direction compared to the orientation of pbp5. The product of the napC gene (1,203 bp) exhibits 41% similarity with the tetracycline resistance gene tet(H) from Pasteurella multocida (G. Guillaume, personal communication). In spite of this similarity, napC does not confer a resistant phenotype to the ATCC 9790 and R40 strains as both of these strains are normally susceptible to tetracycline (MIC, 1 μg/ml). The product of the napB gene (453 bp) exhibits 25 and 24% identity with two proteins of B. subtilis, one involved in secretion and the other related to the MraR regulator, respectively. A 517-bp fragment of napA forms the 3′ end of the pDML540 EcoRI insert shown in Fig. 1A. The 1,236-bp napA gene codes for an Na+/H+ antiporter (43).
Isolation of pbp5-deficient E. hirae clones.
To construct a pbp5 integration vector, a 1.6-kb pbp5 fragment extending from position 361 to position 1,929 was amplified by PCR by using the Taq polymerase, pDML540 as the template, and the O7 and O8* oligonucleotides (Table 2), which added EcoRI and BamHI restriction sites at the 5′ and 3′ ends of the DNA fragment, respectively (14). The purified PCR fragment was inserted into the EcoRI and BamHI sites of pUC18 to obtain pDML1600. The SstI site of the pDML1600 insert (position 1,144 from the ATG codon of pbp5) was blunt ended with the T4 DNA polymerase and used to introduce the 2.0-kb spectinomycin resistance marker (Spcr cassette), which was isolated by SmaI digestion of pHP45Ω. The pDML1600 derivative obtained in this way served as an intermediate for transfer of the disrupted pbp5 fragment into the EcoRI and BamHI sites of pER924. The final construct was designated pDML1610. This plasmid was used only once because the resistance gene did not confer a Spcr phenotype to E. hirae R40 recombinants. Nevertheless, one pbp5-deficient mutant designated FS1 (Fig. 1B) was identified by replica plating as a cefotaxime-sensitive clone among cefotaxime-resistant R40 revertants. In contrast to Penr R40 cells but like penicillin-susceptible (Pens) Rev14 cells, FS1 cells elongated when they were treated with 1 μM cefotaxime in liquid medium (as determined by microscopic examination) (10). This was an indication that the pbp5 gene had been interrupted.
To confirm these preliminary results, a second pbp5 integration vector, pDML1611, was constructed, in which a 1.5-kb kanamycin resistance marker (Kmr cassette) isolated by SmaI/NruI double digestion of pDG792 was inserted at the SstI site of pDML1600 instead of the Spcr cassette. Double recombinants derived from ATCC 9790 were selected as Kmr Erms (erythromycin-sensitive) clones by replica plating. One randomly chosen mutant, designated FS2, was selected for comparison with FS1 (Fig. 1B).
Isolation of psr-deficient E. hirae clones.
To obtain a psr integration vector, a 1.55-kb psr fragment, extending from position 652 in the ftsW gene and from position 882 in the psr gene, was amplified by PCR by using the Taq polymerase, pDML540 as the template, and the O2* and O11 primers (Table 2). The purified PCR fragment was first inserted into pCR2.1 and then transferred after purification into the EcoRI site of pSL1190 from which the NdeI site was first removed by EcoRV/NruI double digestion. The resulting construct, designated pDML1603, was used to insert the 1.5-kb Kmr cassette into the NdeI site (at position 864, almost in the middle of the PCR fragment) after the overhangs were blunt ended with the T4 DNA polymerase. The disrupted psr fragment obtained in this way (pDML1604) was then transferred into the EcoRI site of pER924 to generate pDML1612. ATCC 9790/pDML1612 transformants were isolated and used to obtain psr-inactivated mutants that were selected as Kmr Erms clones by replica plating. No Penr clones were identified on PenG-containing replica plates. One clone was randomly chosen for further analysis and was designated FS3 (Fig. 1B).
To confirm the results obtained with the FS3 mutant, a psr-deleting vector was constructed by inserting the Kmr cassette between a 0.88-kb ftsW fragment and a 0.86-kb pbp5 fragment into pER924. The ftsW fragment extending from position 377 to position 1,253 from the ATG site was amplified by PCR by using the Vent polymerase, pDML540 as the template, and the O14 and O17* primers (Table 2). The O14 primer added an EcoRI site at the 5′ end of the PCR fragment, which was cloned into the EcoRI and HincII sites of pUC18 to obtain pDML1605. Then the ftsW fragment was excised by EcoRI/SphI double digestion from pDML1605 and inserted into the corresponding sites of pSL1190 to generate pDML1606.
The 0.86-kb pbp5 fragment extending from position −160 to position 690 from the initial ATG site was amplified by PCR by using the Vent polymerase, pDML540 as the template, and oligonucleotides O21 and O22* (Table 2). The PCR fragment was cloned into the SmaI site of pUC18 (SURECLONE kit; Amersham Pharmacia Biotech) to obtain pDML1607, from which it was excised with the KpnI and HincII enzymes; it was then inserted into the KpnI and StuI sites of pDML1606 to generate pDML1608. The 1.5-kb SmaI/StuI-digested Kmr cassette (from pDG792) was then introduced into the KpnI site of pDML1608 after the KpnI overhangs were blunt ended with the T4 DNA polymerase. This construct, designated pDML1609, was digested with the EcoRI and SstI enzymes to isolate a 3.2-kb insert that was blunt ended with the T4 DNA polymerase and then ligated into the SmaI site of pER924 to form pDML1613. In this construct, the Kmr cassette started 60 bp downstream of the ftsW stop codon and ended 160 bp upstream of the pbp5 ATG codon.
With pDML1613, mutants of ATCC 9790 could be selected as Kmr Erms clones by psr, replica plating. Similar to the strains with an interrupted psr, none of the mutants with a deleted psr appeared to be resistant to PenG. Two of these mutants were randomly chosen for further analysis and designated FS4 and FS5.
PCR and hybridization analyses of the pbp5- and psr-deficient E. hirae mutants.
Insertion of the resistance cassettes into the different recombinants was verified by PCR by using primers (pbp5 primers O9 and O10*, “psr primers O11 and O12*,” and ftsW primer O13 and pbp5 primer O8*) that hybridized upstream and downstream of the insertion positions in both the pbp5 and psr genes (Fig. 1B). Comparison of the sizes of the PCR fragments indicated that for each recombinant an insertion or a deletion had occurred, as expected (results not shown).
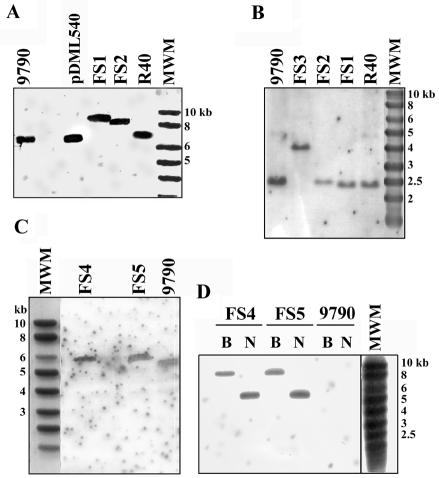
The presence of exogenous DNA inserts in the pbp5 and psr genes of the E. hirae deficient mutants was also confirmed by hybridization analysis. Southern blot analyses of restriction enzyme digestion products of the DNA of these mutants and their controls were performed with the specific pbp5 O15 and psr O16* probes or with a specific 1.2-kb probe derived by PCR (the O23 and O24* primers) from the Kmr cassette (from pDG792) and labeled with the AlkPhos Direct system.
Both the O15 and O16* probes revealed that the fragments originating from the FS1, FS2, and FS3 recombinants were systematically larger than those of the parental strains (Fig. 2). As shown in Fig. 2, the pbp5 O15 probe hybridized on one side with a 6.9-kb EcoRI fragment of R40 and on the other side with 8.9- and 8.4-kb EcoRI fragments of the FS1 and FS2 strains, respectively (Fig. 2A). These sizes were in agreement with insertion of the 2.0-kb Spcr and 1.5-kb Kmr cassettes into the 6.9-kb fragment, respectively. Similarly, the psr O16* probe hybridized with 2.5- and 4.0-kb PstI fragments of the ATCC 9790 and FS3 DNA digests, respectively (Fig. 2B). Again, this indicated that the 2.5-kb PstI fragment had integrated the 1.5-kb Kmr cassette. In addition, E. coli Top10F′ cells transformed with pDML535 (pUC18 bearing the 4.0-kb FS3 PstI fragment) were selected on plates supplemented with kanamycin. Sequencing of the insert confirmed that the psr gene was disrupted and that the Kmr gene was oriented in the direction opposite to that of psr.
FIG. 2.
Southern blots of restriction enzyme digestion products of the DNA of the E. hirae recombinants. The E. hirae ATCC 9790 (9790) and R40 strains, as well as pDML540 (6.9-kb EcoRI insert from R40) and the Smart ladder (MWM), were used as controls. (A) EcoRI digestion products probed with O15 (pbp5). (B) PstI digestion products probed with O16* (psr). (C) PvuII digestion products probed with O15 (pbp5). (D) BglII (lanes B) and NcoI (lanes N) digestion products hybridized with the Kmr probe.
Finally, the PvuII digestion products of FS4 and FS5 DNA that responded to the pbp5 O15 probe were 0.6 kb larger than the positive PvuII DNA fragments of ATCC 9790 (Fig. 2C). This size was expected if the 0.9-kb psr was replaced by the 1.5-kb Kmr cassette. Confirmation that the Kmr cassette had been inserted into the genome of the FS4 and FS5 recombinants was provided by the presence of one BglII fragment and one NcoI fragment in the digestion products of the FS4 and FS5 DNA that hybridized with the Kmr probe, while no hybridization was detected in ATCC 9790 (Fig. 2D).
PBP5 detection in the pbp5- and psr-deficient E. hirae mutants.
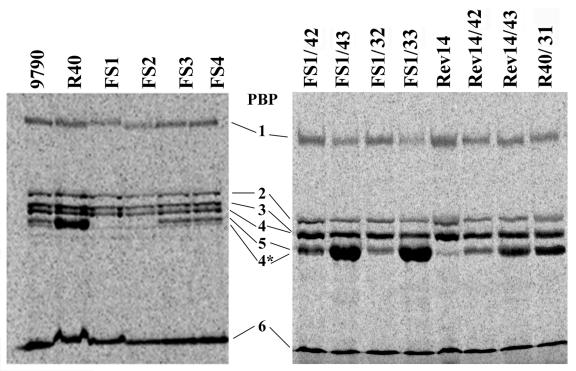
In order to determine whether inactivation of pbp5 or psr could modify the PBP profiles of E. hirae, membranes of the mutants were prepared, labeled with radioactive PenG, and compared with membranes of the ATCC 9790, R40, and Rev14 strains. As shown in Fig. 3 and in agreement with previous results (14, 17), the Penr R40 strain produced a larger amount of PBP5 than ATCC 9790 produced. In contrast, the Pens Rev14 strain did not synthesize PBP5, confirming the importance of this protein for β-lactam resistance (18).
FIG. 3.
PBP patterns of the pbp5- and psr-deficient E. hirae mutants. Membrane preparations (200 μg) were labeled for 60 min with 100 μM [14C]PenG and subjected to gel electrophoresis and fluorography. For simplicity, the last two digits of the plasmid designations are used to identify the different transformants (see Table 1). 9790, ATCC 9790.
As expected, the pbp5-deficient FS1 and FS2 mutants behaved like Rev14, since they were unable to synthesize PBP5. Synthesis of PBP5 was restored in these mutants by complementation with the pbp5-bearing pDML532, pDML533, pDML542, and pDML543 shuttle vectors. The protein was produced in amounts that were more or less related to the plasmid copy number, as pDML532 and pDML542 are low-copy-number vectors and pDML533 and pDML543 are high-copy-number vectors. Interestingly, membranes of the psr-deficient FS3, FS4, and FS5 mutants contained the same amount of PBP5 as membranes of the parental ATCC 9790 strain, indicating that neither the lack of Psr nor the presence of a resistance cassette inside or substituting for the psr gene modified PBP5 synthesis. In addition, when several copies of psr were reintroduced into R40 cells, PBP5 was still overproduced. The R40 transformants that were used in the latter experiment contained the pDML531 shuttle vector (Table 1) bearing a 1.4-kb psr fragment amplified from ATCC 9790 (primers O5 and O6* [Table 2]) and extending between position −431 and position 1,001 from the ATG site.
Western blot analysis with anti-PBP5 antibodies corroborated the results obtained by fluorography.
Penicillin susceptibility of the pbp5- and psr-deficient E. hirae mutants.
The results shown in Table 3 confirmed that the MICs of PenG for cells lacking PBP5, such as the cells of Rev14 (23) or the cells of FS1 and FS2, were markedly lower (20-fold lower for FS2, an ATCC 9790 derivative, and 4,000-fold lower for Rev14 and FS1, two R40 derivatives). When complemented with several copies of pbp5, these hypersusceptible strains regained a resistant phenotype, but at different levels. If the MIC for the Rev14 transformant (which should not have produced Psr) was the same as the MIC for R40, the FS1 and FS2 transformants were 6- to 12-fold less resistant. The FS3 clone with psr disrupted or the FS4 and FS5 clones with psr deleted were as susceptible as the parental ATCC 9790 strain. Moreover, in spite of a complementing psr gene (pDML531), the PenG resistance of R40 cells was not diminished. Thus, whether it was absent or present, the psr gene did not affect E. hirae susceptibility or resistance to PenG.
TABLE 3.
Susceptibility of E. hirae strains to PenG
| Strain | MIC (μg/ml) |
|---|---|
| ATCC 9790 | 0.6 |
| R40 | 60 |
| Rev14 | 0.015 |
| FS1 | 0.015 |
| FS2 | 0.025 |
| FS3 | 0.6 |
| FS4 | 0.6 |
| FS5 | 0.6 |
| R40/pDML531 | 60 |
| FS1/pDML532 | 10 |
| FS1/pDML533 | 20 |
| FS1/pDML542 | 5 |
| FS1/pDML543 | 10 |
| FS2/pDML542 | 5 |
| FS2/pDML543 | 10 |
| Rev14/pDML542 | 60 |
| Rev14/pDML543 | 60 |
Finally, Penr clones could be isolated from FS4 by four passages on plates containing increasing PenG concentrations. After the fourth step the mutant produced large amounts of PBP5 and was even more resistant than R40 (MIC, >70 μg/ml).
The resistant phenotypes could not be attributed to a β-lactamase activity encoded by the complementing vector as none of the extracts of the different clones could hydrolyze the chromogenic nitrocefin compound. In addition, Rev14 cells transformed with a pBR322/pIL252 fusion vector were as susceptible as the parental Rev14 cells.
Cell growth and autolysis of the psr-deficient E. hirae mutants.
The psr- and pbp5-deficient mutants isolated in this study were also tested to determine their growth rates in THG and their autolytic activities. Table 4 shows the mean generation times based on two or three independent cultures. On the basis of these results, we subdivided the clones into two groups, one containing ATCC 9790 and its psr- and pbp5-deficient FS2, FS3, and FS4 derivatives and one containing R40 and its FS1 and Rev14 derivatives. The strains of the first group grew faster than those of the second group.
TABLE 4.
Rates of cell growth and cell autolysis of the different E. hirae strains
| Strain | Generation time (min) | Autolysis rate k (h−1)
|
|
|---|---|---|---|
| Exponential-phase cells | Stationary-phase cells | ||
| R40 | 51 ± 1.12 | 1.25 ± 0.16 | 0.9 ± 0.08 |
| Rev14 | 45 ± 1.25 | 1.52 ± 0.17 | 0.28 ± 0.05 |
| FS1 | 48 ± 2.61 | 1.1 ± 0.17 | 0.27 ± 0.02 |
| ATCC 9790 | 27 ± 0.00 | 0.60 ± 0.28 | 0.10 ± 0.01 |
| FS2 | 29 ± 2.14 | 0.56 ± 0.21 | 0.14 ± 0.02 |
| FS3 | 23 ± 1.21 | 0.61 ± 0.27 | 0.17 ± 0.01 |
| FS4 | 28 ± 1.13 | 0.65 ± 0.39 | 0.16 ± 0.03 |
The autolysis rates determined in three separate experiments (Table 4) confirmed that exponential-phase cells of the different clones autolyzed more rapidly than stationary-phase cells. However, entrance into the stationary phase did not seem to reduce the autolysis rate of R40 cells (1.4-fold) as much as it reduced the autolysis rates of the other strains (3.6- to 6-fold). The results also indicated that the same two groups described above could be distinguished. ATCC 9790 and its derivatives autolyzed more slowly than FS1, Rev14, and R40; the rates were 2- to 2.5-fold and 3- to 9-fold lower than the rates for the exponential- and stationary-phase cells, respectively. Thus, in spite of an inactivated psr gene, the FS3 and FS4 mutants behaved more like FS2 and the parental ATCC 9790 strain than the Rev14 and R40 strains. We concluded that the inactivation of psr could not be responsible for the higher autolysis rates measured with R40.
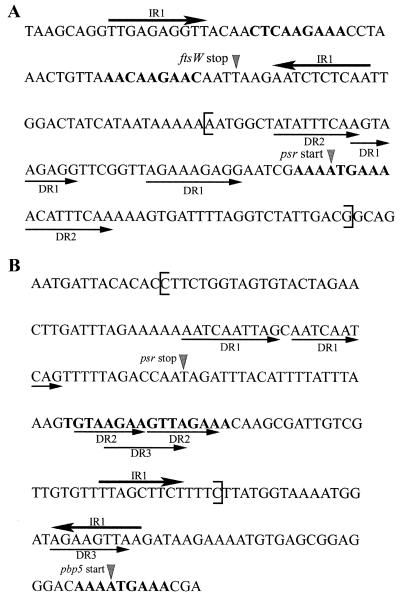
Analysis of the psr-pbp5 and ftsW-psr intergenic regions.
As shown in Fig. 4, direct repeats (DR) and inverted repeats (IR) are present upstream of pbp5 in E. hirae ATCC 9790. Three perfect DR, designated DR1, DR2, and DR3, and a perfect IR, designated IR1, are present in this region. The right arm of the perfect 9-bp IR1 is only 28 bp away from the pbp5 initial ATG codon. The 10-bp DR1 found at the 3′ end of psr terminates 12 bp upstream of its TAG stop codon. DR2 begins 20 bp downstream of the psr stop codon, and there is no separation between its 7-bp repeated sequences. In contrast, 53 nucleotides separate the left and right 8-bp sequences of DR3, which includes most of DR2 and the left IR1 sequence.
FIG. 4.
Sequences of the regions separating the 3′ end of ftsW and the 5′ end of psr (A) and the 3′ end of psr and the 5′ end of pbp5 (B). The initial codons and stop codons of the different genes are indicated by grey arrowheads. The thick and thin arrows indicate the IR and DR, respectively. The putative S1 and S2 sequences are indicated by boldface type. The brackets indicate the beginning and end of the deletion observed in R40 (A) and AS21R2 (B).
In addition, an imperfect 9-bp DR that exhibits some similarity with the DR proposed to be part of the regulating mechanism of the divergent lytR gene and lytABC operon of B. subtilis (22) appears to be present in the same region. The AAAATGAAA sequence overlapping the pbp5 initial ATG codon is identical to the sequence designated S1 in B. subtilis (Table 5). Two sequences (TGTAAGAAG and AGTTAGAAA) similar to the sequence designated S2 in B. subtilis are 90 and 83 bp, respectively, upstream of S1 and could form the other arm of the imperfect DR in E. hirae. Interestingly, each putative S2 sequence overlaps one of the DR2 arms.
TABLE 5.
Putative regulatory imperfect DR located upstream of the psr and pbp5 genes of three enterococcal species
| Species | Gene | S2 | S2-S1 distance (bp) | S1a |
|---|---|---|---|---|
| E. hirae | pbp5 | AGTTAGAAA | 83 | AAAATGAAA |
| TGTAAGAAG | 90 | AAAATGAAA | ||
| psr | AACAAGAAC | 83 | AAAATGAAA | |
| CTCAAGAAA | 103 | AAAATGAAA | ||
| E. faecium | pbp5fm | TTCCAGAAA | 101 | AAAATGAAA |
| psrfm | AAAGAGAAA | 96 | GCAATGAAA | |
| E. faecalis | pbp4 | ATTAAGAAA | 80 | TGAATGAAA |
| B. subtilis | lytR | ACAATGAAA | 78 | ATAATGAAA |
| lytABC | AGAATGAAA | 98 | AAAATGAAA | |
| Consensus | ----aGAAa | --AATGAAA |
The initial ATG codon of each gene is indicated by boldface type.
Figure 4 also shows that the region upstream of psr has several putative regulatory sequences. An almost perfect 10-bp IR (IR1) overlaps the 3′ end of ftsW, and its right sequence starts 1 bp downstream of the ftsW stop codon. IR1 could serve as an atypical ftsW rho-independent terminator. Between IR1 and the psr initial codon, there are also two 9-bp perfect repeats, which are designated DR1 and DR2. The right DR1 arm ends only 8 bp upstream of the psr initial ATG codon. In addition, DR1 is contained in the DNA segment separating the two DR2 arms. The left sequence of DR2 overlaps DR1 by one nucleotide, and its right sequence forms the third, fourth, and fifth psr codons.
Like pbp5, psr appears to have an imperfect 9-bp DR recalling the DR of the lytRABC divergon in B. subtilis (22). The same S1 sequence as in B. subtilis overlaps the psr initial ATG codon, and two sequences (CTCAAGAAA and AACAAGAAC) found 103 and 83 bp, respectively, upstream of S1, at the 3′ end of ftsW, could play the role of the S2 sequence (Table 5).
Identical or very similar sequences are found at the initial ends of pbp5fm and psrfm in E. faecium (24, 34, 45; and Brouillard and Coyette, unpublished data) and of pbp4 in E. faecalis (23) (Table 5). S2 sequences seem not to be as well conserved as S1 sequences, but in every case except pbp5fm in E. faecium there is a 9-bp sequence that recalls an S2 sequence about 80 bp upstream of S1. For pbp5fm, the only 9-bp sequence that exhibits similarity with S2 is 101 bp upstream of S1. E. faecium D63 differs from the other E. faecium strains not only by its shorter psrfm but also by the presence of a 202-bp insert 67 nucleotides upstream of pbp5fm (45; Brouillard and Coyette, unpublished data). Because of this insert, the putative S2 sequence, which is 101 bp away from S1 in the other strains, is shifted 202 bp farther away in D63. However, at the 3′ end of the 202-bp insert and 108 and 118 bp upstream of S1 are TAAAAGAAA and GTAAAGAAA sequences, respectively, which are very similar to S2 sequences.
EMSA with the truncated PsrN protein.
As the psr gene product was suspected to function as a regulator (23) and as the region separating the psr and pbp5 genes probably contains different signaling sequences (see above), attempts were made to verify that the Psr protein could interact with a 407-bp probe obtained from this region. The sequence extended from position −333 to position 74 from the initial ATG codon of pbp5 and was amplified by PCR by using E. hirae ATCC 9790 chromosomal DNA as the template and Cy5-labeled primers O29* and O30. Prediction analysis of the Psr protein sequence indicated that it has an N-terminal hydrophobic peptide that could serve as a membrane anchoring system. Thus, this protein was overproduced as a soluble derivative, designated PsrN, from which residues 7 to 26 were deleted. The 822-bp truncated psrN gene that was used for this purpose was derived from ATCC 9790 and had a 60-bp deletion (from position 19 to position 78) in the region coding for the hydrophobic peptide of Psr. It was amplified by PCR from the genomic DNA by using a mixture of the Taq and Pwo DNA polymerases and the O1 and O25* to O28 oligonucleotides combined to form three pairs of primers (Table 2). The first six psr codons were conserved, and two restriction sites (NdeI and AvaI) were introduced to facilitate insertion into the pET22b(+) expression vector. The recombinant vector obtained was designated pDML534.
Synthesis of PsrN with a polyhistidine peptide at the C-terminal end was induced by 1 mM IPTG in a 500-ml culture of E. coli BL21/pDML534 cells (OD550, ≅0.8) at 37°C. After 3 h of growth, the cells were harvested by centrifugation (6,000 × g for 10 min), washed in a 150 mM NaCl solution, and resuspended in 15 ml of conservation buffer containing 25 mM HEPES (pH 7.5), 5 mM MgCl2, 0.1 mM EDTA, 5 μM β-mercaptoethanol, 10% (wt/vol) glycerol, and 1 mM Pefabloc (Boehringer Ingelheim Bioproducts). They were then disintegrated at 1.35 × 108 Pa with cell disruption equipment (Constant Systems Ltd., Inceltech, Toulouse, France). Cellular debris was removed by centrifugation (17,000 × g for 25 min), and the clarified supernatant was used for an EMSA. The 32-kDa PsrN protein that was detected by immunoblotting and was produced only under inductive conditions represented about 20% of the cytoplasmic protein.
Partially purified PsrN preparations were eluted with an imidazole solution from an Ni2+-nitrilotriacetic acid agarose column (Ni-PDC kit; Affiland, Liège, Belgium) on which crude extracts of PsrN-overproducing E. coli cells were filtered. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed that the PsrN protein detected by Coomassie blue staining and Western blotting was the major component of the preparation (data not shown). However, no EMSA could be conducted with the partially purified PsrN as it had a strong propensity to precipitate. Thus, crude cytoplasmic extracts were used for the EMSA. Various experimental conditions were tested to detect a potential interaction between PsrN and the 407-bp Cy5 probe. The quantity of PsrN (as estimated by densitometry) varied from 10 to 250 ng, and two temperatures (room temperature and 30°C) and different incubation times (20 to 45 min), as well as the influence of β-mercaptoethanol, were tested. None of these conditions promoted any interaction between PsrN and the psr-pbp5 intervening sequence.
DISCUSSION
Enterococci have a low natural susceptibility to β-lactam antibiotics attributed to the constitutive synthesis of a low-affinity PBP usually designated PBP5, which is able to take over most functions of other PBPs when they are inhibited by β-lactams (17, 18). The genes of different strains of three species coding for these PBPs have been cloned and sequenced (13, 14, 31, 33, 38, 45). Moreover, with the DNA sequencing data collected recently from clones of E. hirae, E. faecalis, and E. faecium (13, 45; this study) and the determination of the genomic sequences of E. faecalis V583 (http://tigr.org) and E. faecium DO (http://genome.ornl.gov/microbial/efae/), it is possible to establish the genetic context of these genes.
In the genomes of E. hirae ATCC 9790 and E. faecium D63R, C68, and DO, genes similar to the ftsW gene in the dcw cluster of E. coli (23, 26, 34, 44, 45) are found upstream of psr genes, which in turn precede pbp5 genes. It can be presumed that in these organisms, these ftsW genes are required for cell division as, in contrast to E. coli, the organisms have no ftsW gene in their dcw clusters (12).
PBP5 functions in β-lactam resistance.
A point mutation introducing a stop codon in pbp5 was responsible for the lack of PBP5 in E. hirae Rev14 and for the 40- and 4,000-fold-higher susceptibility to PenG of this strain than of strains ATCC 9790 and R40, respectively (23). As expected, similar results were obtained with the FS1 and FS2 mutants (derived from R40 and ATCC 9790, respectively), in which pbp5 was inactivated by a Campbell-type recombination, as shown by PCR and hybridization analyses. Fluorography and Western blotting analyses confirmed that these mutants were unable to synthesize PBP5.
Resistance to PenG could be restored in Rev14, as well as in the FS1 and FS2 mutants, by complementation with a multicopy shuttle vector carrying pbp5. Whatever the copy number of psr (pDML532 or pDML533 and pDML542 or pDML543), the MICs of PenG for the FS1 transformants were not much different (there was a twofold difference). Similar results were obtained with complemented FS2 clones. Interestingly, after complementation, none of the FS1 and FS2 strains was as resistant as Rev14, although they seemed to produce as much, if not more, PBP5. Thus, we concluded that even if PBP5 is an essential element for substantially increasing the resistance to PenG, it is not sufficient to obtain optimal resistance. Similar conclusions were recently drawn from a comparison of different clones of E. faecium (37). Also, the pbp5 gene borne by pDML542 and pDML543 must have its own promoter as large amounts of PBP5 are synthesized in complemented FS2 strains. This is in agreement with the results previously obtained for E. coli (23).
Implication of Psr in cell growth and autolysis.
Psr, for which a repressor activity was proposed by Ligozzi et al. (23), exhibits 35.3% identity with the LytR protein of B. subtilis (26, 40). It has been suggested that LytR behaves like an attenuator of its own expression and expression of the N-acetylmuramoyl-l-alanine amidase (lytC) gene of B. subtilis. LytC is responsible for the bulk of the amidase activity during vegetative growth. However, a lytR-deficient mutant of B. subtilis showed the same turnover kinetics and cell wall autolysis as the parental strain (22).
Two types of psr-deficient mutants were obtained from the wild-type ATCC 9790 strain; FS3 was obtained by disruption, and FS4 and FS5 were obtained by deletion. The alterations were verified by PCR and hybridization analyses.
In spite of the psr deficiency, FS3, FS4, and FS5 grew and autolyzed at the same rates as ATCC 9790. This tells us that the psr gene product has no regulatory effect on these phenomena. Thus, we concluded that the slower growth and higher autolysis rates of R40, Rev14, and FS1 very likely do not result from the partial psr deletion but instead result from at least one other modification in the genome.
Implication of Psr in β-lactam resistance.
In contrast to R40, whose psr gene was inactivated by a short deletion, the three psr mutants constructed in this work synthesized the same amount of PBP5 as the wild-type ATCC 9790 strain. This demonstrated that neither of the two modifications in psr in FS3, FS4, and FS5 had a polar effect on expression of pbp5, which, as indicated above, appears to have its own promoter. When high-copy-number plasmids complementing psr were introduced into R40 cells, the level of synthesis of PBP5 was not reduced. In addition, the chromosomal psr gene of the pbp5-deficient FS2 mutant did not prevent overproduction of PBP5 when this strain was complemented with pDML542 or pDML543. It could be argued that in this case the gene could not act in trans. However, it cannot act in cis either. Indeed, the psr gene that is borne with pbp5 on the 6.9-kb fragment complementing FS1(pDML532 or pDML533) did not seem to prevent PBP5 overproduction. These different results are thus inconsistent with Psr regulation of PBP5 synthesis and consequently of penicillin resistance.
However, we point out that the impact of the 87-bp psr deletion in R40 on expression of pbp5 is different from the impact of the insertion or deletion in FS3, FS4, or FS5. This means that it is not the psr gene per se or its product that is important but that the region including the 87-bp sequence is involved in one way or another in the regulation of the psr-pbp5 locus. At this point, we have to consider that most probably pbp5 is regulated by one or several genes found elsewhere in the genome. This hypothesis explains why it was still possible to select PBP5-overproducing resistant clones from FS4 in spite of a deleted psr and why the moderately resistant E. faecium D344 strain overproduces PBP5fm even though no mutation could be detected in the psr-pbp5fm locus (35, 45; Brouillard and Coyette, unpublished).
In the search for a potential regulatory activity for Psr, EMSAs were performed to examine DNA-protein interactions between PsrN, a soluble Psr derivative, and a 407-bp probe corresponding to the DNA region located upstream of pbp5. These assays were unsuccessful in spite of the potential DR, IR, S1, and S2 regulatory sequences that were identified in that area.
Interestingly, in the E. hirae R40 and AS21R2 and E. faecium D63R Penr mutants (23, 45), overproduction of PBP5 or PBP5fm seems to be related to a deletion or an insertion that occurred in the upstream region of pbp5. In R40, the 87-bp deletion removed DR1 and DR2, as well as the S1 sequence overlapping the psr 5′ end. In AS21R2, the 137-bp deletion not only shortened the psr open reading frame at its 3′ end but also deleted DR2 and the left DR3, IR1, and S2 sequences that precede pbp5. Finally, in the resistant D63R strain derived from D63, an additional 1,330-bp DNA fragment was inserted 10 bp upstream of the 3′ end of the 202-bp insert that precedes pbp5fm in D63. As a result, the putative S2 sequences described above were moved farther away from S1 and no new potential S2 sequences were found in the −100 to −80 region.
The S1 and S2 sequences, as well as the IR and DR sequences, might thus be important for regulation of the psr and pbp5 genes in these organisms, but their significance remains to be determined.
Conclusion.
Because no change in PBP5 synthesis or cell autolysis occurred when the psr gene was inactivated, because the overproduction of PBP5 was not reduced when several psr copies were introduced into R40 cells, because the Psr protein did not interact with a DNA fragment amplified from the pbp5 promoter region, and because mutants as resistant as R40 could be selected from the psr-deficient FS4 strains, we concluded that the Psr protein has no regulatory effect either on the transcription of pbp5 or on the autolysis of E. hirae cells. Another regulatory mechanism of PBP5 and probably Psr synthesis should be considered and remains to be identified. The data could implicate one or several of the putative signaling sequences found upstream of pbp5 and psr. Also, functions complementary to synthesis of the low-affinity PBP5 protein are needed for optimal phenotypic resistance to PenG in E. hirae.
Acknowledgments
We are grateful to K. W. Bayles and A. M. Gherout-Fleury for their generous gifts of pER924 and pDG792, respectively, and to André Piette for his precious help in the final editorial step.
This work was supported in part by the Belgian Program of Interuniversity Poles of Attraction (PAI grant P4/03). C.F., F.S., and O.D. were fellows of the Fonds pour la Formation à la Recherche dans l'Industrie et l'Agriculture (FRIA), and A.A. was supported by a René Thalmann fellowship from Buenos Aires University and by an exchange program between the Argentinian SECyT and the Belgian FNRS.
REFERENCES
- 1.Archer, G. L., and D. M. Niemeyer. 1994. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 2:343-347. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2001. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Bayles, K. W., E. W. Brunskill, J. J. Iandolo, L. L. Hruska, S. Huang, P. A. Pattee, B. K. Smiley, and R. E. Yasbin. 1994. A genetic and molecular characterization of the recA gene from Staphylococcus aureus. Gene 147:13-20. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. F., and P. E. Reynolds. 1980. Intrinsic resistance to beta-lactam antibiotics in Staphylococcus aureus. FEBS Lett. 122:275-278. [DOI] [PubMed] [Google Scholar]
- 6.Canepari, P., M. Lleo, G. Cornaglia, R. Fontana, and G. Satta. 1986. In Streptococcus faecium penicillin-binding protein 5 alone is sufficient for growth at sub-maximal but not at maximal rate. J. Gen. Microbiol. 132:625-631. [DOI] [PubMed] [Google Scholar]
- 7.Canepari, P., M. Lleo, R. Fontana, and G. Satta. 1987. Streptococcus faecium mutants that are temperature sensitive for cell growth and show alterations in penicillin-binding proteins. J. Bacteriol. 169:2432-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamberlain, J. P. 1979. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal. Biochem. 98:132-135. [DOI] [PubMed] [Google Scholar]
- 9.Coyette, J., J. M. Ghuysen, and R. Fontana. 1980. The penicillin-binding proteins in Streptococcus faecalis ATCC9790. Eur. J. Biochem. 110:445-456. [DOI] [PubMed] [Google Scholar]
- 10.Coyette, J., A. Somzé, J. J. Briquet, J. M. Ghuysen, and R. Fontana. 1983. Function of penicillin-binding protein 3 in Streptococcus faecium, p. 523-529. In R. Hakenbeck, J. V. Höltje, and H. Labischinski (ed.), The target of penicillin. Walter de Gruyter and Co., Berlin, Germany.
- 11.Dardenne, O. 1996. Ph.D. thesis. University of Liège, Liège, Belgium.
- 12.Duez, C., I. Thamm, F. Sapunaric, J. Coyette, and J. M. Ghuysen. 1998. The division and cell wall gene cluster of Enterococcus hirae S185. DNA Seq. 9:149-161. [DOI] [PubMed] [Google Scholar]
- 13.Duez, C., W. Zorzi, F. Sapunaric, A. Amoroso, I. Thamm, and J. Coyette. 2001. The penicillin resistance of Enterococcus faecalis JH2-2r results from an overproduction of the low-affinity penicillin-binding protein PBP4 and does not involve a psr-like gene. Microbiology 147:2561-2569. [DOI] [PubMed] [Google Scholar]
- 14.El Kharroubi, A., P. Jacques, G. Piras, J. Van Beeumen, J. Coyette, and J. M. Ghuysen. 1991. The Enterococcus hirae R40 penicillin-binding protein 5 and the methicillin-resistant Staphylococcus aureus penicillin-binding protein 2′ are similar. Biochem. J. 280:463-469. [PMC free article] [PubMed] [Google Scholar]
- 15.Filée, P., M. Delmarcelle, I. Thamm, and B. Joris. 2001. Use of an ALFexpress DNA sequencer to analyze protein-nucleic acid interactions by band shift assay. BioTechniques 30:1044-1048, 1050-1051. [DOI] [PubMed] [Google Scholar]
- 16.Fontana, R., M. Aldegheri, M. Ligozzi, H. Lopez, A. Sucari, and G. Satta. 1994. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 38:1980-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fontana, R., R. Cerini, P. Longoni, A. Grossato, and P. Canepari. 1983. Identification of a streptococcal penicillin-binding protein that reacts very slowly with penicillin. J. Bacteriol. 155:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fontana, R., A. Grossato, L. Rossi, Y. R. Cheng, and G. Satta. 1985. Transition from resistance to hypersusceptibility to β-lactam antibiotics associated with loss of a low-affinity penicillin-binding protein in a Streptococcus faecium mutant highly resistant to penicillin. Antimicrob. Agents Chemother. 28:678-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 21.Klare, I., A. C. Rodloff, J. Wagner, W. Witte, and R. Hakenbeck. 1992. Overproduction of a penicillin-binding protein is not the only mechanism of penicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 36:783-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazarevic, V., P. Margot, B. Soldo, and D. Karamata. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J. Gen. Microbiol. 138:1949-1961. [DOI] [PubMed] [Google Scholar]
- 23.Ligozzi, M., F. Pittaluga, and R. Fontana. 1993. Identification of a genetic element (psr) which negatively controls expression of Enterococcus hirae penicillin-binding protein 5. J. Bacteriol. 175:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ligozzi, M., F. Pittaluga, and R. Fontana. 1996. Modification of penicillin-binding protein 5 associated with high-level ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 40:354-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loureiro Dos Santos, A. L., and A. Chopin. 1987. Shotgun cloning in Streptococcus lactis. FEMS Microbiol. Lett. 42:209-212. [Google Scholar]
- 26.Massidda, O., O. Dardenne, M. B. Whalen, W. Zorzi, J. Coyette, G. D. Shockman, and L. Daneo-Moore. 1998. The PBP 5 synthesis repressor (psr) gene of Enterococcus hirae ATCC9790 is substantially longer than previously reported. FEMS Microbiol. Lett. 166:355-360. [DOI] [PubMed] [Google Scholar]
- 27.Massidda, O., R. Kariyama, L. Daneo-Moore, and G. D. Shockman. 1996. Evidence that the PBP 5 synthesis repressor (psr) of Enterococcus hirae is also involved in the regulation of cell wall composition and other cell wall-related properties. J. Bacteriol. 178:5272-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray, T., D. L. Popham, and P. Setlow. 1996. Identification and characterization of pbpC, the gene encoding Bacillus subtilis penicillin-binding protein 3. J. Bacteriol. 178:6001-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Partoune, P. 1997. Ph.D. thesis. University of Liège, Liège, Belgium.
- 30.Piras, G., A. El Kharroubi, J. Van Beeumen, E. Coeme, J. Coyette, and J. M. Ghuysen. 1990. Characterization of an Enterococcus hirae penicillin-binding protein 3 with low penicillin affinity. J. Bacteriol. 172:6856-6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piras, G., D. Raze, A. El Kharroubi, D. Hastir, S. Englebert, J. Coyette, and J. M Ghuysen. 1993. Cloning and sequencing of the low-affinity penicillin-binding protein 3r-encoding gene of Enterococcus hirae S185: modular design and structural organization of the protein. J. Bacteriol. 175:2844-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 33.Raze, D., O. Dardenne, S. Hallut, M. Martinez-Bueno, J. Coyette, and J. M. Ghuysen. 1998. The gene encoding the low-affinity penicillin-binding protein 3r in Enterococcus hirae S185R is borne on a plasmid carrying other antibiotic resistance determinants. Antimicrob. Agents Chemother. 42:534-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice, L. B., L. L. Carias, R. Hutton-Thomas, F. Sifaoui, L. Gutmann, and S. D. Rudin. 2001. Penicillin-binding protein 5 and expression of ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 45:1480-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rybkine, T., J. L. Mainardi, W. Sougakoff, E. Collatz, and L. Gutmann. 1998. Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of β-lactam resistance. J. Infect. Dis. 178:159-163. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Sifaoui, F., M. Arthur, L. Rice, and L. Gutmann. 2001. Role of penicillin-binding protein 5 in expression of ampicillin resistance and peptidoglycan structure in Enterococcus faecium. Antimicrob. Agents Chemother. 45:2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Signoretto, C., M. Boaretti, and P. Canepari. 1994. Cloning, sequencing and expression in Escherichia coli of the low-affinity penicillin binding protein of Enterococcus faecalis. FEMS Microbiol. Lett. 123:99-106. [DOI] [PubMed] [Google Scholar]
- 39.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 40.Soldo, B., V. Lazarevic, C. Mauël, and D. Karamata. 1996. Sequence of the 305°-307° region of the Bacillus subtilis chromosome. Microbiology 142:3079-3088. [DOI] [PubMed] [Google Scholar]
- 41.Solioz, M., and M. Waser. 1990. Efficient electrotransformation of Enterococcus hirae with a new Enterococcus-Escherichia coli shuttle vector. Biochimie 72:279-283. [DOI] [PubMed] [Google Scholar]
- 42.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waser, M., D. Hess-Bienz, K. Davies, and M. Solioz. 1992. Cloning and disruption of a putative NaH-antiporter gene of Enterococcus hirae. J. Biol. Chem. 267:5396-5400. [PubMed] [Google Scholar]
- 44.Yura, T., H. Mori, H. Nagai, T. Nagata, A. Ishihama, N. Fujita, K. Isono, K. Mizobuchi, and A. Nakata. 1992. Systematic sequencing of the Escherichia coli genome: analysis of the 0-2.4 min region. Nucleic Acids Res. 20:3305-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zorzi, W., X. Y. Zhou, O. Dardenne, J. Lamotte, D. Raze, J. Pierre, L. Gutmann, and J. Coyette. 1996. Structure of the low-affinity penicillin-binding protein 5 PBP5fm in wild-type and highly penicillin-resistant strains of Enterococcus faecium. J. Bacteriol. 178:4948-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]