Abstract
Topoisomerase IV, a C2E2 tetramer, is involved in the topological changes of DNA during replication. This enzyme is the target of antibacterial compounds, such as the coumarins, which target the ATP binding site in the ParE subunit, and the quinolones, which bind, outside the active site, to the quinolone resistance-determining region (QRDR). After site-directed and random mutagenesis, we found some mutations in the ATP binding site of ParE near the dimeric interface and outside the QRDR that conferred quinolone resistance to Streptococcus pneumoniae, a bacterial pathogen. Modeling of the N-terminal, 43-kDa ParE domain of S. pneumoniae revealed that the most frequent mutations affected conserved residues, among them His43 and His103, which are involved in the hydrogen bond network supporting ATP hydrolysis, and Met31, at the dimeric interface. All mutants showed a particular phenotype of resistance to fluoroquinolones and an increase in susceptibility to novobiocin. All mutations in ParE resulted in resistance only when associated with a mutation in the QRDR of the GyrA subunit. Our models of the closed and open conformations of the active site indicate that quinolones preferentially target topoisomerase IV of S. pneumoniae in its ATP-bound closed conformation.
Topoisomerase IV and DNA gyrase are bacterial type II topoisomerases, which modify DNA topology during the replication process by unlinking DNA and facilitating chromosome segregation (59). Topoisomerase IV forms a C2E2 tetramer involved in segregation of the chromosome at cell division (1, 30, 59). The ParC subunit contains the site of topoisomerization catalyzing the double-stranded DNA break (30, 53), while the ParE subunit catalyzes the hydrolysis of ATP, providing the free energy necessary for these reactions (2, 6, 11). The ParE and ParC subunits share extensive sequence homology with, respectively, the GyrB and GyrA subunits of the DNA gyrase A2B2 tetramer, which catalyzes negative DNA supercoiling during the initiation and elongation processes of DNA replication (16, 53).
Both topoisomerases are the targets of antibacterial molecules, such as the quinolones and the coumarins. The coumarins inhibit supercoiling and enzyme turnover by preventing the binding and hydrolysis of ATP (45). The quinolones form a ternary complex with the topoisomerases in the presence of DNA, resulting in lethal double-stranded DNA breaks (11). Enzymatic studies and binding assays have also shown that the quinolones can form a complex with the topoisomerases before binding DNA (15, 29); however, some binding data indicate the occurrence of a specific and higher level of binding to the enzyme-DNA complex rather than to the enzyme alone (43, 56). These families of molecules, in particular, the fluoroquinolones (FQs), are under continuous development as resistance to antibiotics in pathogenic bacteria has dramatically increased during the last decade (20, 22, 49).
In gram-negative as well as in gram-positive organisms, such as Streptococcus pneumoniae, a bacterial pathogen responsible for community-acquired pneumonia and meningitis, resistance to coumarins and FQs has been reported after selection in vitro as well as in vivo (9, 22, 26, 36, 49). Resistance is commonly associated with mutations in topoisomerase IV or DNA gyrase or mediated by efflux (4, 7, 19, 60) or is associated with both mechanisms (19, 22, 50). In gram-negative organisms, the GyrA subunit of gyrase and the ParC subunit of topoisomerase IV are, respectively, the primary and the secondary targets of FQs (19). In S. pneumoniae, the primary and the secondary quinolone targets are either the GyrA subunit or the ParC subunit, depending upon the compound used as for selection (38, 39, 50). Once a mutation is present in the primary target, higher levels of resistance may result from additional mutations in both the primary and the secondary targets (21, 38, 49, 50). These mutations generally cluster in the N-terminal part of GyrA (between Escherichia coli positions 67 and 106) (57), in the so-called quinolone resistance-determining region (QRDR), or in its homologous region in ParC (37). Different mutations involved in resistance have also been described for the EGDSA and the P(I/L)RGK) motifs identified as part of the QRDR in GyrB or in ParE (18, 39, 58), outside the ATP binding site.
In S. pneumoniae, in vitro-selected resistance to novobiocin and other coumarins has been reported to be associated with point mutations in the 43-kDa N-terminal domain of GyrB, which contains the ATPase domain as well as the binding site for coumarins (10, 36, 45).
Janoir et al. recently described an in vitro-selected, second-step ParE mutation in S. pneumoniae which, in association with a QRDR mutation in GyrA, resulted in increased susceptibility to novobiocin and decreased susceptibility to two FQs, moxifloxacin and sparfloxacin (27). The His103Tyr mutation responsible for the resistance was located outside the ParE QRDR at a position equivalent to His99 in the 43-kDa N-terminal domain of Escherichia coli GyrB. The crystal structures of the 43-kDa domain of GyrB have shown that this residue is involved in the intramolecular and intermolecular interactions of the two GyrB subunits, resulting in stabilization of the dimer in the presence of ATP or novobiocin (8, 32).
To explore further the interplay between the putative sites involved in the dimerization of the ParE subunit and the resistance to FQs, we performed site-directed and random mutagenesis of this region and found other mutations in the N-terminal domain of ParE which were associated with FQ resistance. We looked for the structural implications of these new mutations within a ParE model deduced from the structure of the homologous 43-kDa N-terminal domain of GyrB of E. coli (8).
MATERIALS AND METHODS
Bacterial strains, growth conditions, and antibiotic susceptibility tests.
S. pneumoniae R6 is a susceptible derivative of the nonencapsulated Rockefeller University strain R36A (46). TR6 is a GyrA (Ser81Phe) derivative of R6 (50). Strains were grown at 37°C either in Todd-Hewith broth supplemented with 0.5% yeast extract (Difco Laboratories, Detroit, Mich.) or in C medium (31) supplemented with 0.2% yeast extract at 37°C. MICs were determined on Mueller-Hinton agar plates supplemented with 5% horse blood by using a Steers replicator device with an inoculum of 104 CFU/spot (26). Plates were incubated for 24 h. Ciprofloxacin and moxifloxacin were obtained from Bayer Pharma, Puteaux, France; sparfloxacin and pefloxacin were obtained from Aventis, Vitry sur Seine, France; gatifloxacin was obtained from Grünenthal, Neuilly sur Seine, France; gemifloxacin was obtained from GlaxoSmithKline, Harlow, United Kingdom; and novobiocin was obtained from Sigma.
Transformation of S. pneumoniae.
Transformation of S. pneumoniae R6 or TR6 with either PCR-generated fragments or various plasmids was carried out as previously described (26, 31). Selection for FQ resistance was generally done with sparfloxacin at a concentration of 1.5 μg/ml.
DNA techniques.
Chromosomal DNA was isolated as previously described (26). Oligonucleotides were purchased from MWG-BIOTECH AG, Courtaboeuf, France. Amplification was done with Pwo DNA polymerase (Roche-Boehringer, Mannheim, Germany) and a PROGENE thermocycler (Techne, Cambridge, United Kingdom). DNA sequencing was done by the BigDye terminator cycle sequencing method with a Perkin-Elmer 3700 DNA analyzer.
Site-directed mutagenesis experiments and PCR-based hypermutagenesis.
Site-directed mutagenesis was performed by a two-step amplification procedure by the megaprimer method of PCR-based mutagenesis (44). The first PCR incorporates forward primer SpParE1 (5′-CTGCTGAAATTGTCACATCG-3′ [92 to 73 bp upstream of the GTG translation codon of parE; positions 1163 to 1182] [37]) and a mutagenic reverse primer (5′-TCCCTCCGGCATGAAGAATGGTAAA-3′ [295 to 319 bp downstream of parE; positions 1549 to 1573] [37]), in which CAT (underlined), encoding His, was replaced by TAT for Tyr and AAA for Phe to introduce the mutations. The 411-bp product was purified from agarose (Geneclean; Bio 101, Inc., La Jolla, Calif.) and used in a second PCR as a forward megaprimer with reverse primer SpParE4 (5′-CTGCAAGTGTTCTTCAGGA-3′ [996 to 1,014 bp downstream of parE; positions 2251 to 2268] [37]) to amplify a larger, 1,106-bp PCR fragment harboring the desired mutation. One microgram of DNA was then used for the transformation of strain TR6.
Random mutagenesis was done according to the previously described procedure of hypermutagenic-based PCR (51). A 529-bp PCR fragment (92 bp upstream of the GTG translation codon to 437 bp downstream of parE; positions 1163 to 1691 [37]) containing the N terminus of ParE (amino acids [aa] 1 to 129 [37]) was amplified by using primers SpParE1 and SpParE3 (5′-TTGAAACTGCGCCATCAC-3′ [420 to 437 bp downstream of parE; positions 1673 to 1691] [37]) and chromosomal DNA from wild-type R6. To generate mutations, a bias in the quantities of deoxynucleoside triphosphates was introduced (dTTP, 1,000 μM; dATP and dGTP, 50 μM; and dCTP, 1, 3, or 10 μM), resulting in three conditions of random mutagenesis. The reaction was performed with 10 mM Tris-HCl (pH 8.85)--2 mM MgCl2 in the presence of Taq polymerase (Amersham Biosciences, Orsay, France). Conditions for PCR were as follows: 50 cycles of denaturation at 95°C for 30 s, primer annealing at 54°C for 30s, and primer extension at 72°C for 10 min. The products were used to transform TR6.
Allelic replacements of parE in S. pneumoniae R6.
The ermB gene was amplified from pAT18 (47) and introduced into the pCR-blunt plasmid (Invitrogen). PCR fragments of 1,352 bp, including the promoter of parE (305 bp upstream of the GTG translation initiation codon to 1,047 bp downstream of parE; positions 940 to 2991 of parE [37]), were amplified from TR6 II4 (His103Tyr) and TR6 I1 (M31L) by using SpParE5 (5′-GTTGTTCTCGAGGTCAATCACAAAGGTTG-3′) and SpParE6 (5′-GTTGTTTTCTAGATTATCCTTGGTCTGTC-3′) (underlining indicates XbaI and XhoI sites). After digestion with XbaI and XhoI, the fragments were cloned into the pCR-blunt plasmid (ermB), producing plasmids pBJE103 and pBJE31. These plasmids were used for the disruption of wild-type parE by insertion duplication with erythromycin at 1 μg/ml for selection.
Modeling of ParE.
The E. coli, Thermus thermophilus, and S. pneumoniae N-terminal 43-kDa domains of ParE were manually aligned by using the program Seqlab of Wisconsin Package Version 10.2 (Genetics Computer Group, Madison, Wis.). The crystal structure of the E. coli 43-kDa GyrB domain in complex with ADPPNP (8) (Protein Data Base [PDB] accession number 1EI1) was used to generate the ParE model in the closed conformation, and the crystal structure of the T. thermophilus 43-kDa GyrB domain in complex with novobiocin (32) (PDB accession number 1KIJ) was used to generate the ParE model in the open conformation. For the open conformation, the first 14 residues of ParE were not included in the model because this region is disordered in the T. thermophilus complex with novobiocin. The resulting models of the ParE 43-kDa domain were generated by using Modeller Package Version 2.0 (42). The side chains of the residues implicated in the dimeric interface in the N-terminal part of the enzyme were manually built by using the program O6 (28).
RESULTS
Role of the amino acid at position 103 of ParE in susceptibility to FQs and novobiocin.
In a previous study (27) with a GyrA Ser81Phe QRDR mutant of S. pneumoniae, Janoir et al. selected on moxifloxacin a second-step mutant with a spontaneous His103Tyr mutation in the N-terminal ATPase domain of ParE outside of the QRDR. The mutant had a particular susceptibility phenotype: slight hypersusceptibility to novobiocin and decreased susceptibility to moxifloxacin, sparfloxacin, and grepafloxacin but not to other FQs, such as pefloxacin and ciprofloxacin. Since the ParE His103Tyr substitution was apparently associated with this phenotype only when associated with a mutation in the QRDR of GyrA, the role of all substitutions studied in this work was assayed with S. pneumoniae TR6, which harbors a Ser81Phe substitution in GyrA (Table 1).
TABLE 1.
MICs of FQs for various N-terminal ParE mutants of S. pneumoniae
| Straina | ParE amino acid mutation at the following positionb:
|
MIC (μg/ml) of the following FQc:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 24 | 28 | 31 | 43 | 103 | SPX | GRE | MFX | CIP | GAT | NOV | |
| R6 | A | A | R | M | H | H | 0.25 | 0.5 | 0.125 | 1 | 0.5 | 0.5 |
| TR6 | 1 | 2 | 0.5 | 1.5 | 1 | 0.5 | ||||||
| TR6 17d | H | 4 | 8 | 1.5 | 1.5 | 1 | 0.25 | |||||
| TR6 II1d | C | 3 | 8 | 1.5 | 1.5 | 1 | 0.25 | |||||
| TR6 I1d | L | 4 | 8 | 1.5 | 1.5 | 1 | 0.25 | |||||
| TR6 I2d | V | V | 3 | 6 | 1.5 | 1.5 | 1 | 0.25 | ||||
| TR6 I4d | V | L | 3 | 6 | 1.5 | 1.5 | 1 | 0.25 | ||||
| TR6 II2d | V | R | 3 | 6 | 1.5 | 1.5 | 1 | 0.25 | ||||
| TR6 II4d | Y | 3 | 8 | 1.5 | 1.5 | 1 | 0.25 | |||||
| TR6 M1e | Y | 3 | 8 | 1.5 | 1.5 | 1 | 0.25 | |||||
| TR6 M2e | F | 3 | 6 | 1.5 | 1.5 | 1 | 0.25 | |||||
R6 is a wild-type S. pneumoniae strain; all of the mutants harbor the GyrA (Ser81Phe) mutation present in TR6; only one mutant representative of each class is presented.
ParE positions are those from reference 37. Only amino acids different from those in wild-type pneumococcal strain R6 are shown.
SPX, sparfloxacin; GRE, grepafloxacin; MFX, moxifloxacin; CIP, ciprofloxacin; GAT, gatifloxacin; NOV, novobiocin.
Mutant obtained by PCR-based hypermutagenesis.
Mutant obtained by site-directed mutagenesis.
Using site-directed mutagenesis, we first tested whether the His103Tyr mutation was sufficient to produce the particular antibiotic resistance phenotype, and we examined the potential effect of a different substitution at this position. After site-directed mutagenesis, the ParE DNA fragments containing either Tyr103 or Phe103 were introduced into TR6 by transformation. After selection on 1.5 μg of sparfloxacin/ml, transformants were obtained at frequencies of about 10−3 to 10−4, while spontaneous sparfloxacin-resistant mutants of TR6, used as a control, were obtained at frequencies of less than 10−8. The MICs of the different FQs and novobiocin, an inhibitor of the ATPase activity of bacterial topoisomerases, for the transformants are shown in Table 1 (TR6 M1 and TR6 M2). Compared to the data for TR6, three- to fourfold increases in MICs were obtained for moxifloxacin, sparfloxacin, and grepafloxacin, while no significant increase was observed for ciprofloxacin and gatifloxacin. A reproducible twofold decrease in the MIC of novobiocin was observed for both transformants.
Other positions in the N terminus of ParE play a role in susceptibility to FQs and novobiocin.
It was previously shown that the N-terminal domain of E. coli GyrB (residues 2 to 220), in particular, the N-terminal arm (residues 2 to 16), the loop from positions 80 to 90, and the loop from positions 99 to 117, including key position 99 (homologous to position 103 in ParE of S. pneumoniae), played an essential role in the intermonomeric and intramonomeric interactions which stabilized the GyrB dimers of the gyrase in the presence of ATP (8). Since the GyrB subunit is homologous to the ParE subunit in topoisomerase IV, we used random mutagenesis to determine whether other positions in the equivalent region of ParE could impair susceptibility to FQs and novobiocin. Random mutagenesis of a region encompassing aa 1 to 129 of the N-terminal fragment of ParE was carried out by the PCR-based hypermutagenesis method. After transformation with the PCR products, no resistant transformants could be selected on sparfloxacin or other FQs with wild-type R6 as a recipient. In contrast, with TR6 as a recipient, transformants with a resistance phenotype similar to that found for the His103Tyr and His103Phe mutants (Table 1) were selected at frequencies of about 10−4 to 10−5.
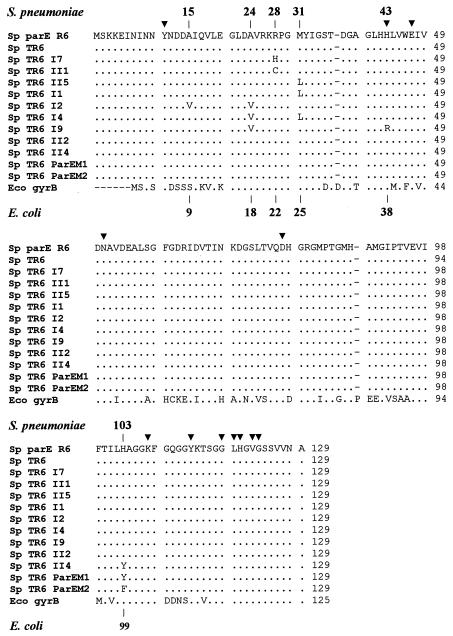
Sequencing of the parE region corresponding to aa 1 to 129 showed two clusters of mutations and diverse associations of mutations (Table 1 and Fig. 1). One cluster corresponded to mutations at previously identified position 103 (His103Tyr), while the other showed mutations at positions 9 to 43, most of which were at positions 24, 28, and 31. In the 20 clones sequenced, substitutions were not equally represented: Met31Leu was present nine times alone, His103Tyr was present four times alone, Arg28Cys and Arg28His were present two times each, and Ala24Val associated with either Ala15Val, Met31Leu, or His43Arg was present only once. To ensure that no other mutation had occurred during the transformation procedure, the sequence encoding the N-terminal fragment of ParE of one representative clone from each different class (Table 1) was PCR amplified and used to retransform Tr6. Resistant transformants with the same resistance phenotype were selected at frequencies of about 10−3 to 10−4, suggesting again that only one event of recombination had occurred in the absence of other mutations. While it is clear that the presence of a single substitution at position 28, 31, or 103 was responsible for the resistance phenotype observed, it is more difficult to attribute a specific weight to each substitution for the resistance phenotype when two substitutions were present, in particular, to substitutions at positions other than 28, 31, or 103.
FIG. 1.
Comparison of the ParE region from S. pneumoniae (Sp) for the different mutants with the GyrB region from E. coli (Eco). Triangles indicate residues from the E. coli GyrB region involved in ATP binding or hydrolysis (10, 45). Numbering above the sequences corresponds to that of ParE of S. pneumoniae; numbering under the sequences corresponds to the equivalent positions in GyrB of E. coli.
Roles of different mutations in wild-type strain R6.
Using mutated fragments of parE, it was not possible to transform wild-type strain R6 to FQ resistance (27) (data not shown). To ensure that the presence of the above ParE mutations alone did not alter the susceptibility phenotype of R6, we used an inactivation-insertion duplication technique to introduce a mutation positioned in each of the major clusters: His103Tyr and Met31Leu. This was done by selection on erythromycin after transformation of R6 with two plasmids, pBJE103 and pBJE31, which do not replicate in gram-positive organisms. After insertion duplication in the chromosome, one interrupted copy and one intact copy of the parE gene were obtained. Since no particular phenotype for FQs was expected, different clones were picked and sequenced by using primers amplifying the complete copy of the parE gene to identify those with the introduced mutation. Clones harboring either His103Tyr or Met31Leu mutations in the intact copy of the parE gene were found and tested for their susceptibility to antibiotics. No increase in the MICs of the different FQs tested (Table 1) was found, except for gatifloxacin, which showed a twofold increase in the MIC and that only for R6 containing the Met31Leu mutation (data not shown). Interestingly, and in contrast to the findings obtained when the GyrA mutation was present, no decrease in the MIC of novobiocin was found for these mutants. Again, this result stressed that a primary mutation in GyrA was necessary to express the full antibiotic resistance phenotype associated with the specific substitutions in ParE.
Modeling of ParE and effect of different substitutions.
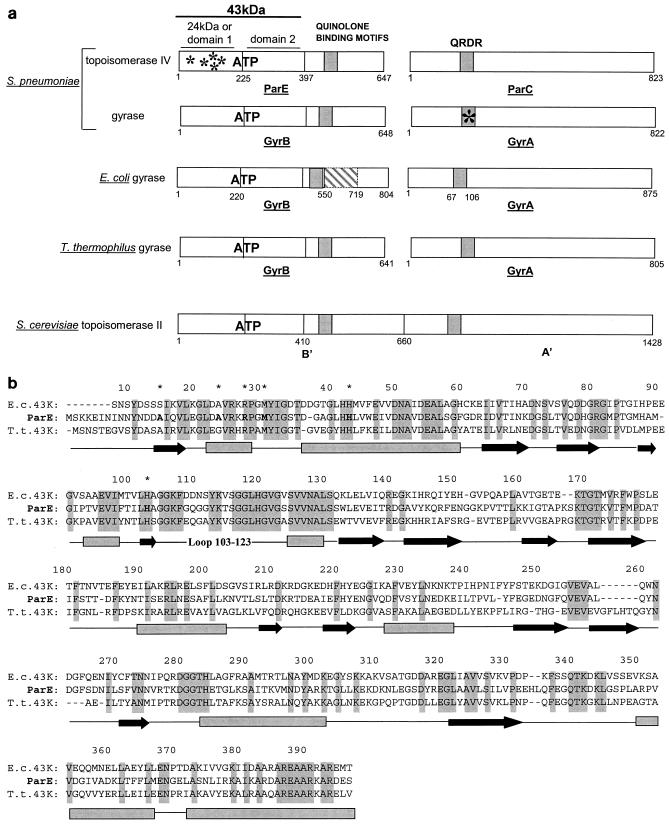
All type II prokaryotic topoisomerases are composed of two subunits forming a tetramer, whereas the eukaryotic enzyme contains regions homologous to the prokaryotic subunits within the same dimeric polypeptide (Fig. 2a). The crystal structures of the bacterial 24-kDa N-terminal domain of GyrB (domain 1) in complex with numerous coumarins have been solved. However, this domain alone is monomeric and cannot bind ATP (17) The crystal structures of the full dimeric ATP binding domain (domain 1 and domain 2), also called the 43-kDa domain, in complex with ADPPNP and more recently with novobiocin (8, 13, 32, 34, 48, 54) have been determined. Both structures show a dimeric organization of this enzyme. Furthermore, different conformations of a fragment of Saccharomyces cerevisiae topoisomerase II without the ATP binding site have been described (6). However, no structural information is available regarding either the entire prokaryotic topoisomerase II or the eukaryotic topoisomerase II containing both the 43-kDa domain and the QRDR.
FIG. 2.
Comparison of different topoisomerases and of the 43-kDa domains of GyrB and ParE. (a) Schematic representation of gyrase and topoisomerase IV subunits. The bacterial topoisomerases are composed of two subunits associated in an A2B2 tetramer (DNA gyrase) and a C2E2 tetramer (topoisomerase IV). The eukaryotic topoisomerase II contains in the same polypeptide sequences homologous to the ParE/GyrB (B′) and ParC/GyrA (A′) bacterial subunits. All topoisomerases contain an ATP binding site lying in the first 43-kDa domain of the ParE/GyrB subunit.E. coli gyrase B contains a large insertion (cross-hatched box) between positions 550 and 719. Quinolone binding motifs lie mainly in the N-terminal region of ParC/GyrA (QRDR) as well as in the C-terminal part of ParE/GyrB. In this study, S. pneumoniae mutations in the 24-kDa N terminus of ParE were found in quinolone-resistant strains associated with the Ser81Phe mutation in the QRDR of GyrA (asterisks). (b) Alignment of the 43-kDa domain sequences of E. coli (E. c.) and T. thermophilus (T. t) GyrB with the 43-kDa domain sequence of S. pneumoniae ParE. The residue numbering corresponds to that of the ParE sequence. The secondary structure elements deduced from the available crystal structures are shown below the sequences (boxes for helices and arrows for β sheets). Identical residues in all three sequences are highlighted in grey. ParE residues in which mutations resulted in quinolone resistance (this study) are indicated by asterisks.
The ATP binding site of S. pneumoniae ParE shares a high level of sequence homology with the E. coli and T. thermophilus GyrB 43-kDa ATP binding domains (49.4 and 47% identities, respectively), suggesting that ParE has the same fold as GyrB (Fig. 2b). Most of the residues implicated in quinolone resistance and residing in the ATP binding region of S. pneumoniae are conserved among the three protein sequences. The crystal structures of the E. coli GyrB N-terminal 43-kDa domain in complex with ADPPNP and of the T. thermophilus 43-kDa domain in complex with novobiocin (8, 32) show that the ATP binding site can adopt different conformations of the active site, closed and open, respectively. These conformations are defined by the position adopted by the loop closing the active site (from residues 99 to 119 in the E. coli sequence) and stabilized by the dimeric contacts between the subunits.
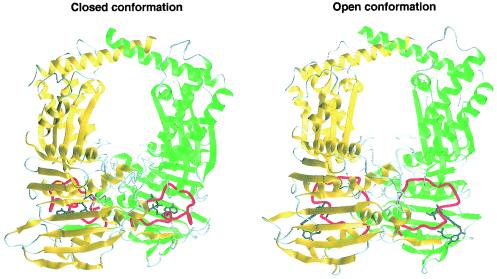
We built a model for the ParE N-terminal 43-kDa domain of S. pneumoniae in both the open and the closed conformations to examine the impact of the amino acid changes which led to FQ resistance (Fig. 3). Most of the residues implicated in quinolone resistance were located either at the end of the N-terminal arm of the protein or in the following helix (Fig. 4a), comprising most of the dimeric contacts in domain 1 (8). In both conformations, most of these residues are implicated in the hydrogen bond network or in Van der Waals contacts contributing to dimer formation.
FIG. 3.
Models of the ParE 43-kDa domain in closed and open conformations. In both conformations, monomer A is shown in green and monomer B is shown in yellow, whereas the loop closing the active site is shown in red. In the closed conformation, the loop from positions 103 to 123 (corresponding to the T. thermophilus loop from positions 98 to 118) tightly embraces the ADPPNP molecule (dark grey), whereas it embraces a larger volume in the open conformation, stabilized by the novobiocin molecule (dark grey).
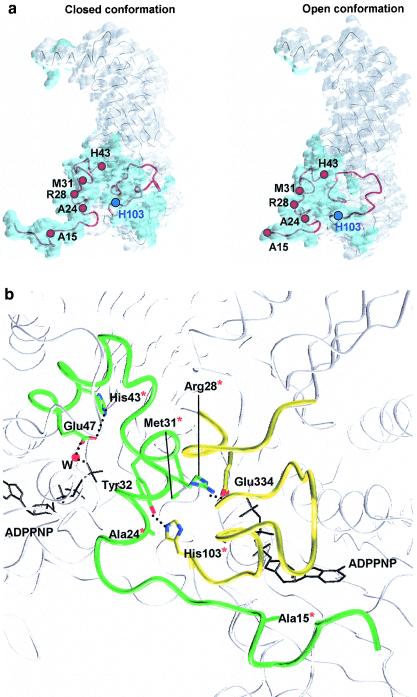
FIG. 4.
Localization of the mutated residues in the different conformations of the ParE 43-kDa domain and role in the network. (a) Localization of associated quinolone resistance mutations. The surface of monomer A in each conformation is shown in light grey, whereas the dimeric interface is shown in blue. The mutations are localized on the N-terminal arm, the following helix, and the loop from positions 103 to 123 (secondary structure elements are shown in red). In both conformations, the residues which are mutated are implicated in the contacts participating in the formation of the dimer. His103 is localized at the N-terminal end of the loop closing the active site. (b) Interaction network implicating the mutated residues. The E. coli 43-kDa complex with the ADPPNP (black) dimeric interface has elements of one monomer shown in yellow and the other shown in green. Red asterisks indicate the mutated residues in quinolone-resistant strains. For better clarity, the interactions of His43 and Glu47 with ADPPNP through a water molecule are shown on the green monomer side, although they are present in both monomers, both active sites being totally symmetrical.
The substitution of Ala15 with a bulkier hydrophobic residue, such as valine, destabilizes the anchoring of the N-terminal arm of one monomer on the other by steric hindrance. This effect is strengthened by the associated Ala24Val mutation placing a hydrophobic residue in a charged environment (Fig. 4b). These mutations predominantly affect the closed conformation, where the N-terminal residues play a key role in the formation of the active site (8), whereas in the open conformation, the dimer is less stable, with the first 10 residues being disordered in the crystal structure (32). In the same way, the substitution of Arg28 with either cysteine or histidine breaks the hydrogen bond linking this residue in one monomer to Glu334 in domain 2 of the other monomer (Fig. 4b). This interaction stabilizes the dimer in the closed conformation of the enzyme in the presence of ATP and is not maintained in the open conformation in the absence of ATP.
The substitution of Met31 with leucine alone or associated with the Ala24Val mutation also contributes to the destabilization of the dimer by introducing a bulky aromatic side chain in the hydrogen bond network, implicating Tyr32 and His103 and linking the dimeric interface to the ATP binding region (Fig. 4b). His103, which is among the most frequently substituted residues, is located in the N-terminal part of the loop from positions 103 to 123, equivalent to the loop from positions 98 to 118 of T. thermophilus (32), closing the active site and contacting the N-terminal arm and helix of the other monomer. In particular, His103 of one monomer interacts with Tyr32 of the other monomer either directly in the closed conformation or via a water molecule in the open conformation (Fig. 5). In both the open and the closed conformations, this conserved residue plays a key role in maintaining the interaction network leading to the formation of the dimer. Furthermore, the interaction via Tyr32 (E. coli Tyr26) of His103 with Gln342 (E. coli Gln337), a residue in domain 2 which interacts with the ATP γ-phosphate, links the conformational changes of the active site mediated by ATP hydrolysis to enzyme domain 2 and the dimeric interface (8, 32). Mutation of His103 to tyrosine or phenylalanine provokes the breaking of the hydrogen bond with Tyr32 and thus destabilizs the dimeric interface as well as the domain 1-domain 2 orientation.
FIG. 5.
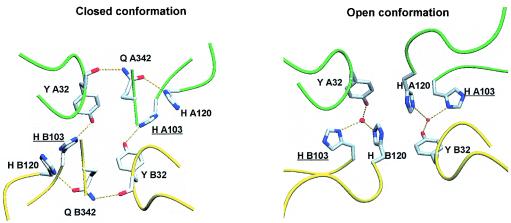
Hydrogen bond network implicating His103. The backbones of monomers A and B are, respectively, shown in green and yellow. In the closed conformation, Tyr32 interacts directly with the side chain of His103, whereas His120 contacts the side chain of Gln342 in the interaction with Tyr32. In the open conformation, the two histidines are implicated in a water-mediated H-bond network with Tyr32.
His43 is located at the interface between domain 1 and domain 2 (Fig. 4). Besides the destabilization of the dimeric interface, the substitution of His43 with arginine affects the domain 1-domain 2 orientation by introducing a very bulky charged side chain. Moreover, this residue corresponds to His38 in E. coli, a catalytic residue conserved among all prokaryotes and replaced by a lysine in eukaryotic topoisomerases. This residue participates in the hydrogen bond network contacting the γ-phosphate of the ATP molecule through Glu47 (Glu42 in E. coli) and a water molecule mediating the hydrolysis of the coenzyme (25). This mutation is associated with a substitution of Ala24, which is implicated in dimeric contacts (see above).
Finally, the intramonomeric contact occurring in this region participates in the stabilization of the overall dimeric conformation of the enzyme in both the closed and the open conformations.
DISCUSSION
This study shows that mutations outside the QRDR of ParE can interfere with the susceptibility of S. pneumoniae to FQs. Confirming the previous observation of Janoir et al. (27), we showed, by using an insertion duplication method, that in the absence of an associated mutation in the GyrA QRDR, the ParE mutations studied in this work did not lead to the expression of FQ resistance. Interestingly, by using as recipients derivatives of S. pneumoniae R6 harboring ParC mutated in the QRDR at position 79 (Ser79Tyr or Ser79Phe, corresponding to position 81 in the GyrA QRDR), we were unable to select resistant transformants containing the ParE substitution His103Tyr or Met31Leu (data not shown). These results suggested again that, in S. pneumoniae, the association of specific mutations in the N terminus of ParE required an additional QRDR mutation in GyrA for the full expression of antibiotic resistance. These results should be interpreted in the context of previous work that showed that first-step mutants resistant to sparfloxacin or moxifloxacin would first alter the QRDR of the GyrA subunit of gyrase and that only second-step resistant mutants would then alter the QRDR of topoisomerase IV (23, 38). Thus, in order to obtain the expression of the resistance phenotype, which could only be selected on sparfloxacin or moxifloxacin, the PCR products which harbored the different mutations in topoisomerase IV were introduced into TR6, a derivative of S. pneumoniae R6 harboring a GyrA Ser81Phe mutation (50). Interestingly in GrlB of Staphylococcus aureus, an Asp33Tyr mutation at a position equivalent to position 37 in ParE of S. pneumoniae was recently found to increase by twofold the MICs of FQs in the absence of other mutations in GyrA (24). This result would indicate that for S. aureus and in contrast to S. pneumoniae, a mutation in the N terminus of the ATPase domain of topoisomerase IV can be expressed in the absence of a GyrA mutation.
Site-directed mutagenesis showed that the His103Phe substitution resulted in a resistance pattern similar to that obtained with the His103Tyr substitution. This finding indicates an important role of His103 at this position, corresponding to position 99 in GyrB of E. coli. By random PCR mutagenesis and after transformation and selection on sparfloxacin, we were able to select four amino acid changes, most of which were located in the 43-kDa N-terminal amino acids of ParE. Apart from the His103Tyr substitution, the most representative selected substitutions were Ala24Val, Arg28His or Arg28Cys, and Met31Leu, which are located at positions equivalent to those of Ala18, Arg22, and Met25 in E. coli GyrB. Each of these positions once mutated, either alone or in association with other positions located in the vicinity (Table 1), resulted in the particular phenotype of resistance to FQs and hypersusceptibility to novobiocin observed with the His103Tyr or His103Phe substitution. None of these positions is located in the QRDR, mutations of which are thought to decrease the affinity of the topoisomerases for quinolones, a partial explanation of the mechanism of resistance to FQs (5, 40). Interestingly, in S. aureus, the Asn470Asp substitution near the conserved motif of the QRDR of ParE led to a similar phenotype, with decreased susceptibility to FQs and increased susceptibility to novobiocin (14). However, and in contrast to the phenotype of resistance observed in this work, this mutation, not located in the N terminus of ParE, was associated with decreased susceptibility to ciprofloxacin and no change in susceptibility to sparfloxacin.
The amino acids described in this work, in particular, Arg28, Met31, and His103, play a role in both the intermonomeric and the intramonomeric interactions between the two ParE subunits, as deduced from the crystal structures, of the E. coli GyrB 43-kDa dimer with ADPPNP (closed conformation) (8) and the T. thermophilus GyrB 43-kDa dimer with novobiocin (open conformation) (32). Thus, all of the above mutations lie in critical positions of the N-terminal domain, which plays a major role in the dimerization of the GyrB subunit (8, 32) and, by analogy, are very likely to play a role in the dimerization of ParE.
Previous work with E. coli GyrB has shown that an insertion in domain 2 outside the ATP binding region (mutant GyrB-225) increases quinolone sensitivity (18). It has been suggested that this mutation would strengthen GyrA-GyrB interactions and therefore further stabilize the conformation, a scenario which may enhance the inhibitory effect of the quinolones. Amino acid changes at the dimeric interface of the ParE ATP binding domain may, on the other hand, decrease the stability of the dimer and, as a consequence, destabilize the conformation targeted by quinolones.
Modeling of the ATPase domain of ParE indicates that substitution of His103 with tyrosine or phenylalanine would predominantly affect the ATP-bound, closed conformation of the active site because of the direct interaction between Tyr32 and His103 (Fig. 5). The similar effect of the mutation at Met31 further emphasizes the importance of this particular configuration of the local hydrogen bond network in the targeted conformation. Mutation of Arg28 would also disrupt intramonomeric or intermonomeric hydrogen bond interactions, specifically stabilizing the closed conformation. Moreover, mutation of His38 participating in the hydrogen bond network contacting the ATP molecule for catalysis would mainly affect the closed conformation, which is stabilized in the presence of ATP. Our mutational studies combined with structural observations indicate that quinolones would preferentially bind to the ATP-bound conformation of topoisomerase IV. Although it has been suggested that quinolones might impair ATP binding to E. coli topoisomerase IV (3), limited proteolysis experiments and ATP binding and hydrolysis assays with the homologous E. coli gyrase clearly indicate that quinolones are able to bind the ATP-bound topoisomerase (35), which can still hydrolyze ATP, although at a lower rate (29, 55). At least the destabilization of the dimer, the formation of which is necessary to bind ATP (8, 54), would weaken the subunit interaction, resulting in a smaller number of productive topoisomerase IV-DNA complexes and finally in decreased susceptibility to quinolones. Such a reduction in the amount of functional target enzyme-DNA complexes has been shown to cause resistance to topoisomerase II-targeting antitumor agents that, like quinolones, act as cellular poisons (33).
Previous enzymatic studies also showed that the homologous human topoisomerase II, also called TOP2, is targeted by antitumor drugs in its ATP-bound form (52). Human TOP2 contains in the same polypeptide sequences homologous to the prokaryotic ParE and ParC subunits and also a QRDR. Like quinolones, which are antitumor drugs, the antimicrobial agent ciprofloxacin acts in vitro as an ATP-sensitive TOP2 poison and also targets the QRDR of ATP-bound TOP2 (52).
This study and other studies cited above led us to propose a model in which quinolones would target topoisomerase IV in its ATP-bound conformation. However, since quinolones bind to a tetrameric holoenzyme-DNA complex, the conformation of the ParE dimer when complexed with ParC subunits may well show differences from the proposed structural model.
Besides, the quinolone-resistant S. pneumoniae strains show a slight concomitant increase in susceptibility to novobiocin. The crystal structures of the T. thermophilus 43-kDa domain of GyrB in complex with novobiocin have shown that the open conformation is the one targeted by novobiocin (31). Substitution of His103 with tyrosine or phenylalanine may affect this conformation to a lesser extent. In this case, the water-mediated hydrogen bond network may accommodate bulkier side chains and may allow the dimeric interface to be slightly reorganized. Taken together, these data suggest that there may be a displacement of the equilibrium in favor of the open conformation when His103 is mutated.
The crystal structures of yeast topoisomerase II have shown that the different domains of the enzyme are subject to large conformational changes. More recently, the crystal structures of the ATPase domain of T. thermophilus GyrB in complex with novobiocin have shown that conformational changes in the ATP binding site (also called domain 1) induce movements of domain 2. Our results suggest that the QRDR and the ATPase binding domain should, in some way, interact and that the conformation of the active site in the presence of ATP may induce a particular conformation of the ParE QRDR, which then binds the quinolones.
Concluding remarks.
In gram-negative organisms, as well as in S. pneumoniae, DNA gyrase was identified as the primary target at which first-step mutations occur for the expression of quinolones resistance, although this finding is dependent upon the FQ (19, 39). Most secondary mutations present in topoisomerase IV subunits were localized either in the equivalent QRDR of ParC or ParE (12, 41, 50). The present work shows that amino acid changes conferring quinolone resistance can also be localized at the dimeric interface in the N-terminal region of ParE near the ATP binding site. Modeling of the 43-kDa domain of ParE based on crystal structures of 43-kDa complexes with either ATP or novobiocin shows that substitution of the conserved His103 and His43 with other amino acids at the dimeric interface would preferentially affect the ATP-bound closed conformation of the enzyme. In agreement with data showing increased susceptibility to novobiocin, the models also suggest that the open conformation of the active site that is targeted by novobiocin may be less destabilized by the substitution of His103 at the dimeric interface. These results provide circumstantial evidence that the topoisomerase IV and, by analogy, the gyrase conformations targeted by the quinolones are those existing when ATP is bound. Additional structural data are necessary to study the conformations that the whole topoisomerase II would adopt in the presence of ATP, i.e., those which would bind the quinolones. Therefore, mutations in resistant strains may provide information not only about the quinolone binding site but also about distant sites that interact with this region.
Acknowledgments
This work was supported by institute funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, and the Université Louis Pasteur de Strasbourg.
We thank Pierre Oudet for careful reading of the manuscript.
REFERENCES
- 1.Adams, D. E., E. M. Shekhtman, E. L. Zechiedrich, M. B. Schmid, and N. R. Cozzarelli. 1992. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell 71:277-288. [DOI] [PubMed] [Google Scholar]
- 2.Ali, J. A., A. P. Jackson, A. J. Howells, and A. Maxwell. 1993. The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry 32:2717-2724. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, V. E., T. D. Gootz, and N. Osheroff. 1998. Topoisomerase IV catalysis and the mechanism of quinolone action. J. Biol. Chem. 273:17879-17885. [DOI] [PubMed] [Google Scholar]
- 4.Baranova, N. N., and A. A. Neyfakh. 1997. Apparent involvement of a multidrug transporter in the fluoroquinolone resistance of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1396-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnard, F. M., and A. Maxwell. 2001. Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser(83) and Asp(87). Antimicrob. Agents Chemother. 45:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, J. M., S. J. Gamblin, S. C. Harrison, and J. C. Wang. 1996. Structure and mechanism of DNA topoisomerase II. Nature 379:225-232. [DOI] [PubMed] [Google Scholar]
- 7.Brenwald, N. P., M. J. Gill, and R. Wise. 1998. Prevalence of a putative efflux mechanism among fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2032-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brino, L., A. Urzhumtsev, M. Mousli, C. Bronner, A. Mitschler, P. Oudet, and D. Moras. 2000. Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center. J. Biol. Chem. 275:9468-9475. [DOI] [PubMed] [Google Scholar]
- 9.Chen, D. K., A. McGeer, de Azavedo, J. C., and D. E. Low. 1999. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. Canadian Bacterial Surveillance Network. N. Engl. J. Med. 22:233-239. [DOI] [PubMed] [Google Scholar]
- 10.Contreras, A., and A. Maxwell. 1992. gyrB mutations which confer coumarin resistance also affect DNA supercoiling and ATP hydrolysis by Escherichia coli DNA gyrase. Mol. Microbiol. 6:1617-1624. [DOI] [PubMed] [Google Scholar]
- 11.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, M. J., Y. F. Jin, V. Ricci, and L. J. Piddock. 1996. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother. 40:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fass, D., C. Bogden, and J. Berger. 1999. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat. Struct. Biol. 6:322-326. [DOI] [PubMed] [Google Scholar]
- 14.Fournier, B., and D. C. Hooper. 1998. Mutations in topoisomerase IV and DNA gyrase of Staphylococcus aureus: novel pleiotropic effects on quinolones and coumarin activity. Antimicrob. Agents Chemother. 42:121-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Froelich-Ammon, S. J., and N. Osheroff. 1995. Topoisomerase poisons: harnessing the dark side of enzyme mechanism. J. Biol. Chem. 270:21429-21432. [DOI] [PubMed] [Google Scholar]
- 16.Gellert, M., K. Mizuuchi, M. H. O'Dea, and H. A. Nash. 1976. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. USA 73:3872-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert, E., and A. Maxwell. 1994. The 24 kDa N-terminal sub-domain of the DNA gyrase B protein binds coumarin drugs. Mol. Microbiol. 12:365-373. [DOI] [PubMed] [Google Scholar]
- 18.Heddle, J. G., T. Lu, X. Zhao, K. Drlica, and A. Maxwell. 2001. gyrB-225, a mutation of DNA gyrase that compensates for topoisomerase I deficiency: investigation of its low activity and quinolone hypersensitivity. J. Mol. Biol. 309:1219-1231. [DOI] [PubMed] [Google Scholar]
- 19.Hooper, D. C. 1999. Mechanisms of fluoroquinolone resistance. Drug Resist. Updates 2:38-55. [DOI] [PubMed] [Google Scholar]
- 20.Hooper, D. C. 2001. Emerging mechanisms of fluoroquinolone resistance. Emerg. Infect. Dis. 7:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooper, D. C. 2001. Mechanisms of action of antimicrobials: focus on fluoroquinolones. Clin. Infect. Dis. 15(Suppl. 1):S9-S15. [DOI] [PubMed] [Google Scholar]
- 22.Hooper, D. C. 2002. Fluoroquinolone resistance among Gram-positive cocci. Lancet Infect. Dis. 9:530-538. [DOI] [PubMed] [Google Scholar]
- 23.Houssaye, S., L. Gutmann, and E. Varon. 2002. Topoisomerase mutations associated with in vitro selection of resistance to moxifloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2712-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ince, D., X. Zhang, C. Siver, and D. C. Hooper. 2002. Dual targeting of the DNA gyrase and topoisomerase IV: target interaction of garenoxacin (BMS-284756, T-3811ME) a new desfluoroquinolone. J. Bacteriol. 46:3370-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson, A. P., and A. Maxwell. 1993. Identifying the catalytic residue of the ATPase reaction of DNA gyrase. Proc. Natl. Acad. Sci. USA 90:11223-11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janoir, C., V. Zeller, M. D. Kitzis, N. J. Moreau, and L. Gutmann. 1996. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob. Agents Chemother. 40:2760-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janoir, C., E. Varon, M. D. Kitzis, and L. Gutmann. 2001. New mutation in ParE in a pneumococcal in vitro mutant resistant to fluoroquinolones. Antimicrob. Agents Chemother. 45:952-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 29.Kampanis, S. C., and A. Maxwell. 1998. Hydrolysis of ATP at only one GyrB subunit is sufficient to promote supercoiling by DNA gyrase. J. Biol. Chem. 273:26305-26309. [DOI] [PubMed] [Google Scholar]
- 30.Kato, J., Y. Nishimura, R. Imamura, H. Niki, S. Higara, and H. Suzuki. 1990. New topoisomerase essential for chromosome segregation in E. coli. Cell 63:393-404. [DOI] [PubMed] [Google Scholar]
- 31.Lacks, S., and R. D. Hotchkiss. 1960. A study of the genetics material determining an enzyme activity in Pneumococcus. Biochim. Biophys. Acta 39:508-518. [DOI] [PubMed] [Google Scholar]
- 32.Lamour, V., L. Hoermann, J. M. Jeltsch, P. Oudet, and D. Moras. 2002. An open conformation of the Thermus thermophilus gyrase B ATP-binding domain. J. Biol. Chem. 277:18947-18953. [DOI] [PubMed] [Google Scholar]
- 33.Larsen, A. K., and A. Skladanowski. 1998. Cellular resistance to topoisomerase-targeted drugs: from drug uptake to cell death. Biochim. Biophys. Acta 1400:257-274. [DOI] [PubMed] [Google Scholar]
- 34.Lewis, R. J., O. M. Singh, C. V. Smith, T. Skarzynski, A. Maxwell, A. J. Wonacott, and D. B. Wigley. 1996. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 15:1412-1420. [PMC free article] [PubMed] [Google Scholar]
- 35.Li, T. K., and L. F. Liu. 1998. Modulation of gyrase-mediated DNA cleavage and cell killing by ATP. Antimicrob. Agents Chemother. 42:1022-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munoz, R., M. Bustamante, and A. G. de la Campa. 1995. Ser-127-to-Leu substitution in the DNA gyrase B subunit of Streptococcus pneumoniae is implicated in novobiocin resistance. J. Bacteriol. 177:4166-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan, X. S., and L. M. Fisher. 1996. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J. Bacteriol. 178:4060-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan, X. S., and L. M. Fisher. 1997. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob. Agents Chemother. 41:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan, X. S., and L. M. Fisher. 1998. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan, X. S., G. Yague, and L. M Fisher. 2001. Quinolone resistance mutations in Streptococcus pneumoniae GyrA and ParC proteins: mechanistic insights into quinolone action from enzymatic analysis, intracellular levels, and phenotypes of wild-type and mutant proteins. Antimicrob. Agents Chemother. 45:3140-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perichon, B., J. Tankovic, and P. Courvalin. 1997. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1166-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 43.Shen, L. L., W. E. Kohlbrenner, D. Weigl, and J. Baranowski. 1989. Mechanism of quinolone inhibition of DNA gyrase. Appearance of unique norfloxacin binding site in enzyme-DNA complexes. J. Biol. Chem. 264:2973-2978. [PubMed] [Google Scholar]
- 44.Smith, A. M., and K. P. Klugman. 1997. “Megaprimer” method of PCR-based mutagenesis: the concentration of megaprimer is a critical factor. BioTechniques 22:438-442. [DOI] [PubMed] [Google Scholar]
- 45.Smith, C. V., and A. Maxwell. 1998. Identification of a residue involved in transition-state stabilization in the ATPase reaction of DNA gyrase. Biochemistry 37:9658-9667. [DOI] [PubMed] [Google Scholar]
- 46.Tiraby, J. G., and M. S. Fox. 1973. Marker discrimination in transformation and mutation of Pneumococcus. Proc. Natl. Acad. Sci. USA 70:3541-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102:99-104. [DOI] [PubMed] [Google Scholar]
- 48.Tsai, F. T., O. M. Singh, T. Skarzynski, A. J. Wonacott, S. Weston, A. Tucker, R. A. Pauptit, A. L. Breeze, J. P. Poyser, R. O'Brien, J. E. Ladbury, and D. B. Wigley. 1997. The high-resolution crystal structure of a 24-kDa gyrase B fragment from E. coli complexed with one of the most potent coumarin inhibitors, clorobiocin. Proteins 28:41-52. [PubMed] [Google Scholar]
- 49.Varon, E., and L. Gutmann. 2000. Mechanisms and spread of fluoroquinolone resistance in Streptococcus pneumoniae. Res. Microbiol. 151:471-473. [DOI] [PubMed] [Google Scholar]
- 50.Varon, E., C. Janoir, M. D. Kitzis, and L. Gutmann. 1999. ParC and GyrA may be interchangeable initial targets of some fluoroquinolones in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vartanian, J. P., M. Henry, and S. Wain-Hobson. 1996. Hypermutagenic PCR involving all four transitions and a sizeable proportion of transversions. Nucleic Acids Res. 24:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, H., Y. Mao, N. Zhou, T. Hu, T. S. Hsieh, and L. F. Liu. 2001. ATP-bound topoisomerase II as a target for antitumor drugs. J. Biol. Chem. 276:15990-15995. [DOI] [PubMed] [Google Scholar]
- 53.Wang, J. C. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65:635-692. [DOI] [PubMed] [Google Scholar]
- 54.Wigley, D. B., G. J. Davies, E. J. Dodson, A. Maxwell, and G. Dodson. 1991. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351:624-629. [DOI] [PubMed] [Google Scholar]
- 55.Williams, N. L., A. J. Howells, and A. Maxwell. 2001. Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Biol. Mol. 306:969-984. [DOI] [PubMed] [Google Scholar]
- 56.Willmott, C. J., and A. Maxwell. 1993. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 37:126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshida, H., M. Bogaki, M. Nakamura, L. M. Yamanaka, and S. Nakamura. 1991. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35:1647-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zechiedrich, E. L., and N. R. Cozzarelli. 1995. Replications of topoisomerases IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 9:2859-2869. [DOI] [PubMed] [Google Scholar]
- 60.Zeller, V., C. Janoir, M. D. Kitzis, L. Gutmann, and N. J. Moreau. 1997. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1973-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]