Abstract
Salmonella enterica, in contrast to Escherichia coli K12, can use 2-deoxy-d-ribose as the sole carbon source. The genetic determinants for this capacity in S. enterica serovar Typhimurium include four genes, of which three, deoK, deoP, and deoX, constitute an operon. The fourth, deoQ, is transcribed in the opposite direction. The deoK gene encodes deoxyribokinase. In silico analyses indicated that deoP encodes a permease and deoQ encodes a regulatory protein of the deoR family. The deoX gene product showed no match to known proteins in the databases. Deletion analyses showed that both a functional deoP gene and a functional deoX gene were required for optimal utilization of deoxyribose. Using gene fusion technology, we observed that deoQ and the deoKPX operon were transcribed from divergent promoters located in the 324-bp intercistronic region between deoQ and deoK. The deoKPX promoter was 10-fold stronger than the deoQ promoter, and expression was negatively regulated by DeoQ as well as by DeoR, the repressor of the deoxynucleoside catabolism operon. Transcription of deoKPX but not of deoQ was regulated by catabolite repression. Primer extension analysis identified the transcriptional start points of both promoters and showed that induction by deoxyribose occurred at the level of transcription initiation. Gel retardation experiments with purified DeoQ illustrated that it binds independently to tandem operator sites within the deoQ and deoK promoter regions with Kd values of 54 and 2.4 nM, respectively.
Numerous bacteria can use the 2-deoxy-d-ribosyl moiety of 2′-deoxyribonucleosides as the sole carbon and energy source (15). Genetic and biochemical studies, using Escherichia coli and Salmonella enterica serovar Typhimurium as model organisms, have shown that the catabolic pathway involves four enzymes: thymidine phosphorylase (EC 2.4.2.4), purine nucleoside phosphorylase (EC 2.4.2.1), phosphopentomutase (EC 5.4.2.7), and deoxyriboaldolase (EC 4.1.2.4) (Fig. 1), encoded by the deoA, deoD, deoB, and deoC genes, respectively. In both organisms the four genes are organized as an operon, the deoCABD operon, located at approximately 99 centisomes on the chromosomes. The operon is transcribed from two promoters located 600 bp apart. Transcription initiation of the operon is highly regulated by a complex interplay of three regulatory proteins, of which two, DeoR and CytR, are repressors and the third is the cAMP-receptor protein (CAP). The genes encoding these proteins are unlinked to each other and to the deoCABD operon. The internal inducer for expression of the operon is 2-deoxy-d-ribose 5-phosphate (dRib5P) inactivating DeoR (8, 15) and cytidine (in S. enterica also uridine) inactivating CytR (27, 34)
FIG. 1.
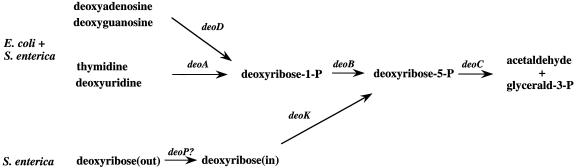
2-Deoxy-d-ribose and deoxynucleoside catabolism by S. enterica and E. coli. The individual enzymes are identified by the corresponding gene symbols: deoA, thymidine phosphorylase; deoB, phosphopentomutase; deoC, deoxyriboaldolase; deoD, purine nucleoside phosphorylase; deoK, deoxyribokinase; deoP?, inferred deoxyribose permease. Glycerald-3-P stands for glyceraldehyde 3-phosphate.
In contrast, only a few bacteria have been shown to be able to ferment 2-deoxy-d-ribose (dRib). They include Lactobacillus plantarum (11, 13), Selenomonas ruminantium (30), and serovar Typhimurium (17). Notably, E. coli K12 cannot use dRib as the sole carbon and energy source.
Deoxyribokinase, the first enzyme involved in dRib utilization, catalyzes the phosphorylation of deoxyribose to dRib5P. Its synthesis is induced in S. enterica following addition of dRib to the growth medium (17). Subsequently, dRib5P is cleaved by deoxyriboaldolase to glyceraldehyde 3-P and acetaldehyde (Fig. 1). The genes mediating fermentation of dRib were cloned from S. enterica serovar Typhi and shown to be located at 82.94 centisomes on the chromosome (38). The dRib system involved four genes, of which three, deoK, deoP, and deoX, were transcribed in one direction and the fourth, deoQ, was transcribed in the opposite direction. The deoK gene was shown to encode deoxyribokinase. In silico analyses determined that deoP encoded an inner-membrane protein, having 34.5% sequence identity with FucP of E. coli, and therefore most likely corresponded to the deoxyribose permease described by Hoffee (17). The deoQ gene was deduced to encode a putative regulatory protein with 37.4% amino acid sequence identity to the DeoR repressor of E. coli. No clear matches for the deoX gene product were found in the databases. The deoQKPX cluster is absent from the E. coli K12 genome.
In the present work we describe the transcriptional organization of the deoQKPX locus from serovar Typhimurium and show that deoK, deoP, and deoX constitute an operon and that transcription of both the deoQ gene and the deoKPX operon is negatively controlled by the deoQ gene product through binding of DeoQ to the intercistronic region between deoQ and deoK.
MATERIALS AND METHODS
Bacterial strains and plasmid vectors. E. coli K12 strain MC1061 (araD139 Δ(araABIOC leu)7697 ΔlacX74 galU galK rpsL hsdR) (31) was used in all clonings, and CSH26 [F− ara Δ(gtp-pro-cod-lac)] (26) and its deoR derivative, MEC011, were used as hosts for expression of the Salmonella deo genes. For overproduction of recombinant proteins, strain BL21(DE3) was used (37). In most clonings, the low-copy-number vectors pWSK29 and pWSK129 (6 to 8 copies per cell), encoding resistance to ampicillin (Apr) and kanamycin (Kmr), respectively, were employed (39). For lacZ transcriptional fusions, the very-low-copy-number vector pFZY1 (9.6 kb) was used (21). It contains a promoterless lacZ gene and part of lacY from E. coli. Upstream of the lacZ gene is a multiple cloning region followed by a leader with stop codons in all three reading frames. The replication origin of pFZY1 is oriF, and the plasmid is present in one to two copies per genome and harbors a gene for ampicillin resistance. From pFZY1, pFLC (5.5 kb) was constructed by deleting the lac DNA (see below). Translational fusions were introduced using a single-copy system with λgal8 as a vector (12).
Media and growth conditions.
Strains were grown at 37°C unless stated otherwise. Lysogenic strains harboring λgal8 derivatives were grown at 32°C. Media were NZY broth (modified LB medium [18]) or AB minimal medium (5) containing 0.2% of either glucose, glycerol, or 2-deoxy-d-ribose as the carbon source, thiamine (0.5 μg/ml), and proline (50 μg/ml). When used, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside was added at a concentration of 40 μg/ml; and isopropyl-β-d-thiogalactopyranoside was added at 0.5 mM. Antibiotics were added at the following final concentrations: ampicillin, 100 μg/ml, except with the very-low-copy-number plasmids, where the concentration was reduced to 30 μg/ml; tetracycline, 10 μg/ml; kanamycin, 30 μg/ml; and chloramphenicol, 30 μg/ml. Solid media were prepared by adding 1.5% agar to the respective liquid medium.
DNA techniques.
E. coli was made competent by treatment with CaCl2 (31). Plasmids were isolated by the alkaline-sodium dodecyl sulfate lysis procedure (3). Endonuclease digestion, ligation of DNA, and 5′-end phosphorylation of DNA by polynucleotide kinase was done according to the suppliers' recommendations. All PCR-generated fragments were sequenced by the chain termination method (32) using the BigDye terminator cycle sequencing kit (PE Applied Biosystems, Warrington, Great Britain) and an ABI PRISM 310 genetic analyzer (PE Applied Biosystems).
Cloning the deoK locus and plasmid constructions.
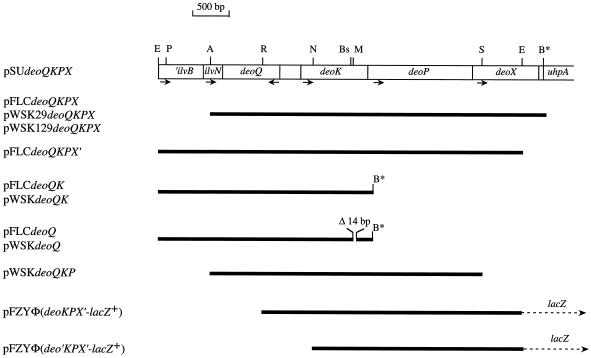
A plasmid library of serovar Typhimurium LT2 DNA containing from 8- to 15-kbp Sau3AI fragments in pBR328 was obtained from C. G. Miller (16) and amplified by one cycle of phage P22 propagation. The library was transformed into E. coli MC1061, and transformants capable of using dRib as the sole carbon source were selected. Restriction endonuclease mapping of plasmids isolated from dRib+ candidates indicated that they harbored the entire deoQKPX locus with the exception of the last 200 bp of the 3′ end of deoX. The missing DNA was obtained by PCR, making use of a unique EcoRI site located 230 bp from the 3′ end of deoX, and the entire region, including ilvN and part of ilvB, was assembled as a 5.4-kbp fragment in pSU19 (2), yielding pSUdeoQKPX (Fig. 2).
FIG. 2.
Structure of specific plasmids. The open box represents the 5.4-kb serovar Typhimurium DNA cloned in pSU19. Above the box, relevant restriction enzyme sites are shown: A, AclI; B*, BamHI introduced by PCR; Bs, BssHII; E, EcoRI; M, MluI; N, NotI; P, PshAI; R, RsrII; S, SacII. Arrows below the box indicate the direction of transcription. The thick solid lines pertain to cloned serovar Typhimurium DNA present in the individual plasmids. Dotted arrows indicate the plasmid-borne lacZ reporter gene.
pFLC derivatives.
The single-copy vector pFLC (5.5 kbp) was constructed by deleting all lac DNA (approximately 4.4 kbp) from pFZY1 by digesting pFZY1 with AflII and HindIII, filling the 5′ overhangs using the Klenow fragment, and ligating the blunt ends. A 4.7-kbp AclI/BamHI fragment, containing the entire deoQKPX region from pSUdeoQKPX, was blunt ended with the Klenow fragment and ligated into filled-in BamHI-digested pFLC, yielding pFLCdeoQKPX (Fig. 2). The plasmid pFLCdeoQKPX′, with 230 bp deleted from the 3′ end of deoX, was constructed by ligating the 5.1-kbp EcoRI fragment of pSUdeoQKPX into the unique EcoRI site of pFLC (Fig. 2). The plasmid pFLCdeoQK (Fig. 2) was obtained by a three-point ligation of BamHI/EcoRI-digested pFLC with the 2.2-kbp EcoRI/NotI fragment of pSUdeoQKPX, containing deoQ and the proximal part of deoK, and a PCR fragment of the entire deoK gene with a built-in BamHI site in the 3′ primer, and digested with NotI/BamHI. The plasmid pFLCdeoQ was constructed from pFLCdeoQK by deleting an internal 14-bp MluI/BssHII fragment from the deoK gene (Fig. 2).
pWSK derivatives.
Plasmids pWSK29deoQKPX and pWSK129deoQKPX were obtained by cloning the 4.7-kbp AclI/BamHI fragment from pSUdeoQKPX into AccI/BamHI-digested pWSK29 and pWSK129, respectively. Digestion of pWSK129deoQKPX with SacII (one SacII site is located in the plasmid immediately downstream of the 3′ end of deoX) resulted in deletion of almost the entire deoX gene, yielding pWSKdeoQKP. Plasmids pWSKdeoQK and pWSKdeoQ were obtained by transferring the deo fragments from pFLCdeoQK and pFLCdeoQ into pWSK129 as EcoRI/BamHI fragments (Fig. 2).
Translational fusions.
The intercistronic region between deoQ and deoK, including the first seven deoQ codons and the first two deoK codons, was amplified by PCR, and the 357-bp amplicon was separately cloned in each orientation into the SmaI site of the translational fusion vector pRAK80 (20). The resulting plasmids, pRAK80Φ(deoK-lacZ)hybrid and pRAK80Φ(deoQ-lacZ)hybrid, expressed the DeoK-LacZ and DeoQ-LacZ fusion proteins from the deoK and deoQ promoters located in the intercistronic region. The 3.7-kbp EcoRI/EcoNI fragments of pRAK80Φ(deoK-lacZ)hybrid and pRAK80Φ(deoQ-lacZ)hybrid, containing the intercistronic region and the entire deo-lacZ fusions, were blunt ended using the Klenow enzyme and ligated into the unique Ecl136II site of pGD519. pGD519 is a derivative of pGD56 (GenBank accession number X67018), with a unique Ecl136II site located between the B region and the galK gene of pGD519. This facilitated the transfer of the deo-lacZ gene fusions to λgal8 (λgalETK+ cI857) by in vivo recombination (25). The resulting recombinant λ phages were used to construct single lysogens of CSH26 as described previously (7).
Transcriptional fusions.
The 357-bp intercistronic region between deoQ and deoK, including the first few codons of the genes, was isolated from pRAK80Φ(deoK-lacZ)hybrid and from pRAK80Φ(deoQ-lacZ)hybrid as EcoRI-BamHI fragments and ligated into the transcriptional fusion vector pFZY1 upstream of the promoterless lacZ gene, yielding pFZYΦ(deoK-lacZ+) and pFZYΦ(deoQ-lacZ+), respectively. The insertions were such that translation initiating from the deoK or deoQ start codon would terminate at the same stop codon within the lacZ upstream region. Plasmid pFZYΦ(deoKPX'-lacZ+), containing the deoQ-deoK intercistronic region, deoK, deoP, and part of deoX (Fig. 2), was obtained from pSUdeoQKPX by digestion with PshAI and RsrII, filling of the 5′ overhang of the latter site, and religating. From the resulting plasmid, the desired region was excised using EcoRI and cloned into the EcoRI site of pFZY1 upstream of lacZ (Fig. 2). Plasmid pFZYΦ(deo′KPX′-lacZ+), lacking the deoQ-deoK intercistronic region and the first 46 codons of deoK, was constructed by deleting the PshAI-NotI fragment from pSUdeoQKPX, filling the 5′ overhangs, ligating, and moving the EcoRI fragment as described above into pFZY1 (Fig. 2).
Preparation of cell extracts and enzyme assays.
Cell extracts were prepared by sonic disruption of cells suspended in 0.1 M potassium phosphate buffer (pH 7.5)-0.5 mM EDTA, followed by centrifugation to remove cellular debris. β-Galactosidase activity was determined at 28°C as described by Miller (26). Thymidine phosphorylase activity was measured at 37°C as described by Schwartz (33). One unit is defined as 1 nmol of product formed per min. Specific activities are given as units per optical density at 436 nm (OD436) of the bacterial suspension prior to sonication.
Mapping of the transcriptional start sites.
Total cellular RNA was extracted from exponentially growing cells of MEC011/pWSK129deoQKPX using hot phenol, essentially as previously described (35). Primer extension analysis of the 5′ ends of the deoQ and deoKPX transcripts was carried out using avian myeloblastosis virus reverse transcriptase (36). The primers used were complementary to codons 21 to 29 of deoQ (5′CGCGCGGCGTCTTTCAGATGGATTCG) or to codons 16 to 25 of deoK (5′CCCCTTCTTTGGGCATCTGGTTGGTG) and were 5′ labeled using [γ-32P]ATP and T4 polynucleotide kinase. The extension products were resolved on a 6% polyacrylamide-7 M urea gel alongside DNA sequence ladders produced with the same labeled primers.
Expression and purification of the DeoQ protein.
The deoQ gene from serovar Typhi strain Ty2 was amplified by PCR using the following primers: forward, 5′ GGCACGCATATGGAGACCAAGCAAAAAGAACG (NdeI restriction site underlined), and reverse, 5′ AGGACTCGAGTTCACTTTCCGAATCAGGCGTG (XhoI restriction site underlined). (The amino acid sequence of the DeoQ protein from serovar Typhi is 100% identical with the DeoQ protein of serovar Typhimurium.) The amplicon was cleaved and inserted between the same two sites (NdeI-XhoI) in the expression vector pET22b (Novagen), creating an in-frame fusion between the 3′ end of the deoQ gene and six histidine codons. The resulting plasmid, pAOT9b, was transformed into E. coli BL21(DE3). The recombinant His-tagged DeoQ protein was overproduced from cells growing exponentially at room temperature in NZY medium containing Ap. At an OD600 of 1.0, isopropyl-β-d-thiogalactopyranoside (0.5 mM) was added, and incubation continued for 2 h, after which time the cells were harvested by centrifugation. Cells were suspended in 50 mM sodium phosphate (pH 8.0)-0.5 M NaCl and sonically disrupted, and the cellular debris was removed by centrifugation. The supernatant was applied to a Ni-CAM (Sigma, St. Louis, Mo.) affinity column equilibrated in sonication buffer containing 25 mM imidazole, and the column was washed with 7 volumes of the same buffer. The DeoQ protein was eluted with 200 mM imidazole in sonication buffer and dialyzed against sonication buffer to remove the imidazole. Following addition of glycerol to 10%, the preparation was stored at −20°C. The protein concentration was calculated from the absorbance at 280 nm.
Gel filtration.
The native molecular mass of purified DeoQ was estimated by gel filtration on a Superose 12HR 10/30 gel filtration column (Amersham Pharmacia Biotech) attached to a fast protein liquid chromatography apparatus (Bio-Rad). The column was equilibrated, and elution was carried out with 50 mM sodium phosphate (pH 6.5), 0.6 M NaCl, 2 mM EDTA, 1 mM dithiothreitol (DTT). Carbonic anhydrase (29 kDa), bovine serum albumin (66 kDa), and β-amylase (200 kDa) were used as molecular mass markers.
Gel retardation assays.
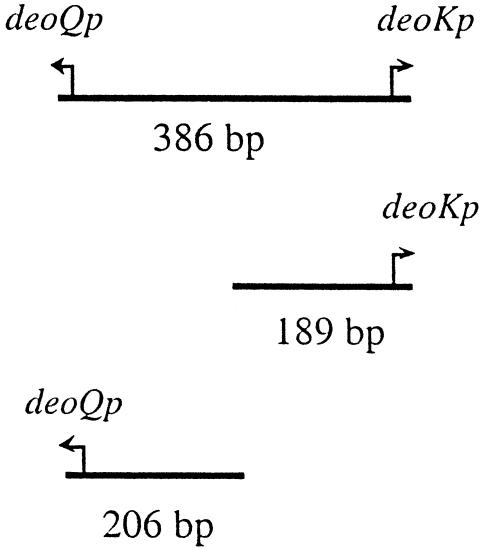
The DNA probes used (a 386-bp fragment including the entire intergenic region between deoQ and deoK, a 189-bp fragment including the deoK promoter and 70 bp of its upstream region, and a 206-bp fragment with the deoQ promoter and 90 bp of its upstream region) were generated by PCR and radiolabeled at the 5′ ends using [γ-32P]ATP and T4 polynucleotide kinase. The binding reaction contained, in a 10-μl volume, the following: 10 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, 1 mM DTT (pH 8.2), 250 μg of bovine serum albumin per ml, 5% glycerol, and 200 to 400 pM labeled probe. Different concentrations of DeoQ were used for each binding reaction. In the binding stoichiometry experiments, 0.2 μM nonradioactive DNA of the same fragment was added in addition to the radiolabeled fragment. After incubation at 37°C for 15 min, the samples were electrophoresed at room temperature on a 5% polyacrylamide gel (10 cm) for 3 h at 8 V/cm. Dried gels were quantitated with a Packard cyclone storage phosphor system. Kd values were determined as the concentration of DeoQ required for 50% of the labeled probe to be retarded on the gel.
DNA sequences.
The nucleotide sequence of the deoQKPX locus is located at coordinates 3991219 to 3995621 on the serovar Typhimurium LT2 genome (GenBank accession no. NC_003197) and at coordinates 3853107 to 3857498 on the serovar Typhi genome (accession no. NC_003198). The coordinates of the homologous regions of other bacteria are found under the following accession numbers: Salmonella enterica serovar Enteritidis, accession no. NC_002962; E. coli CFT073, accession no. NC_004431, coordinates 300643 to 305020; L. plantarum, accession no. NC_004567; coordinates 442825 to 448118; Pseudomonas syringa, accession no. NC_002949; Agrobacterium tumefaciens, accession no. NC_003304, coordinates 2020602 to 2029321.
RESULTS
Cloning and phenotypic characterization of the deoQKPX region of serovar Typhimurium.
The capacity of serovar Typhi to ferment dRib was previously shown to be encoded on a 4,392-bp DNA fragment located between ilvN and uhpA (38). No homologous DNA is present in the E. coli K12 genome, and accordingly, E. coli K12 cannot grow with dRib as the sole carbon source. The nucleotide sequence of the serovar Typhimurium genome (24) indicated that this organism contained a homologous 4,403-bp DNA fragment, likewise located between ilvN and uhpA. The nucleotide sequence of this region is 98.7% identical with that of serovar Typhi, and the deduced amino acid sequences of the four open reading frames within the fragment are 100% identical for DeoQ, DeoK, and DeoP and 98.5% for DeoX.
From a partial Sau3AI library of serovar Typhimurium LT2 DNA in pBR328, most of the deoQKPX region was obtained by complementation in E. coli. The missing DNA was obtained by PCR, and the entire region was assembled as a 5.4-kb EcoRI/BamHI fragment in pSU19, yielding pSUdeoQKPX. From this plasmid, all other plasmids used in the present work were derived as described in Materials and Methods and in Fig. 2.
It was previously shown that deoxyribokinase is required for growth of S. enterica or E. coli K12 with dRib as the sole carbon source (17, 38). To establish the importance of the deoP and deoX gene products in dRib utilization, various regions of the deoQKPX cluster were subcloned into the single-copy plasmid pFLC, and the ability of these plasmids to support growth of E. coli K12 with dRib as the carbon source was tested in liquid culture. As shown in Table 1, the presence of both deoP and deoX was required for the maximal growth rate with 0.08% dRib as the carbon source, whereas no significant difference was observed with 0.2% dRib.
TABLE 1.
Effect of the deoK, deoP, and deoX genes on the growth rate of E. coli K12 with dRib as the carbon source at 37°C
| Plasmida | Doublings/h with:
|
|
|---|---|---|
| 0.08% dRib | 0.2% dRib | |
| pFLC | <0.10 | <0.10 |
| pFLCdeoQKPX | 0.68 | 0.63 |
| pFLCdeoQKP | 0.32 | 0.57 |
| pFLCdeoQK | 0.22 | 0.55 |
The plasmids were contained in E. coli CSH26.
deoK, deoP, and deoX constitute an operon.
Sequence analysis of the deoQKPX DNA suggested that the deoK, deoP, and deoX genes were transcribed as a single transcript from a promoter located in the deoQ-deoK intercistronic region. Accordingly, we observed that expression of β-galactosidase activity from the deoX-lacZ transcriptional fusion carried on pFZYΦ(deoKPX′-lacZ+) (Fig. 2) was 46.8 U/OD436, whereas it was only 0.8 U/OD436 from pFZYΦ(deo′KPX′-lacZ+), which is lacking the deoQ-deoK intercistronic region and the first 46 codons of deoK (Fig. 2). A putative rho-independent transcriptional terminator, with a calculated stability of the stem-loop structure of −17.6 kcal per mol (23), was found 22 bp downstream of the deoX stop codon within the 74-bp intergenic region between deoX and uhpA.
Regulation of deoQ and deoKPX expression.
To identify the divergent promoters responsible for expression of deoQ and deoKPX, the intergeneric region between deoQ and deoK (including the first few codons of the genes) was fused in-frame to lacZ in each orientation, and the fusions were transferred to a lambda phage and integrated in single copy into the λ attachment sites of E. coli MEC011 and CSH26. The resulting four strains were grown in minimal medium with different carbon sources, and the levels of β-galactosidase activity, to monitor expression of deoK/deoQ, and thymidine phosphorylase (deoA) activity, to monitor expression of the DeoR controlled deoCABD operon, were determined. The results (Table 2) showed that expression of β-galactosidase from the two fusions was the same in the deoR genetic background (experiment 1) and that this expression was repressed by the presence of the deoQ gene in trans, the effect on the deoK fusion being the strongest (experiments 2 and 3). Addition of dRib to the growth medium resulted in about 15-fold induction of β-galactosidase synthesis from each fusion, whereas addition of thymidine had no effect (experiments 3, 4, and 5). The β-galactosidase activity levels observed when cells were grown with glucose as the carbon source (Table 2, experiment 6) indicated that deoK expression but not deoQ expression was regulated by catabolite repression.
TABLE 2.
Effect of DeoQ and DeoR on the synthesis of β-galactosidase from the translational deoK-lacZ and deoQ-lacZ fusions and of thymidine phosphorylase from deoAa
| Expt no. | Relevant host genotype | Relevant plasmid genotypeb | Carbon sourcec | Activity (units/OD436)
|
||
|---|---|---|---|---|---|---|
| β-Galactosidase
|
Thymidine phosphorylase (deoA)d | |||||
| deoK-lacZ | deoQ-lacZ | |||||
| 1 | deoR | Parental | Gly | 8.8 | 9.0 | 140 |
| 2 | deoR | deoQ+ | Gly | 0.3 | 1.3 | —e |
| 3 | deoR | deo(QKPX)+ | Gly | 0.08 | 0.4 | 116 |
| 4 | deoR | deo(QKPX)+ | Gly + dRib | 1.3 | 5.8 | — |
| 5 | deoR | deo(QKPX)+ | Gly + dT | 0.05 | 0.5 | — |
| 6 | deoR | Parental | Glu | 1.2 | 8.7 | — |
| 7 | deoR+ | Parental | Gly | 0.3 | 1.4 | 3.0 |
| 8 | deoR+ | Parental | Gly + dRib | — | — | 3.3 |
| 9 | deoR+ | Parental | Gly + dT | 5.8 | 6.1 | 145 |
| 10 | deoR+ | deo(QKPX)+ | Gly | 0.08 | 0.4 | 3.2 |
| 11 | deoR+ | deo(QKPX)+ | Gly + dRib | 1.4 | 4.8 | 178 |
| 12 | deoR+ | deo(QKPX)+ | Gly + dT | 0.05 | 0.3 | 150 |
MEC011 (deoR) or CSH26 (deoR+) lysogenized with phage λ carrying either the deoK-lacZ or the deoQ-lacZ translational fusion and the indicated plasmid was grown exponentially for at least three generations at 32°C in minimal medium with the indicated carbon sources. Cell extracts were assayed for β-galactosidase and thymidine phosphorylase activities.
All plasmids were derivatives of pWSK29. Parental indicates the vector with no insert. The serovar Typhimurium genes carried on the plasmids are indicated in italics.
All carbon sources were added at 0.2%.
Thymidine phosphorylase activity was measured only in strains carrying the deoQ-lacZ fusion.
—, not measured.
The significant amino acid sequence identity between DeoQ and DeoR (36.3% for serovar Typhimurium) led us to look for possible cross-activity between the two repressors and their targets. Thus, the regulation of expression of the two fusions was also investigated in a deoR+ genetic background (lower part of Table 2). The E. coli DeoR repressor was able to repress both deoK and deoQ expression to about the same extent as DeoQ (experiment 7 versus experiments 2 and 3). As expected, addition of thymidine, an effective inducer of the deoCABD operon, resulted in induction of DeoR-mediated repression (experiment 9). Since thymidine did not induce β-galactosidase synthesis in the presence of both DeoQ and DeoR (experiment 12), it appeared that DeoR was subordinate to DeoQ in repression of deoK and deoQ expression. In contrast, dRib, which is not an inducer of DeoR-mediated repression (experiment 8), was effective as an inducer of deoK and deoQ expression in a deoQ+ deoR+ genetic background (experiment 11 versus experiment 4). It is noteworthy that even though DeoR was capable of repressing expression of deoK, deoQ, and deoA (experiment 7), the reciprocal was not observed (experiment 3).
Strength of the deoK and deoQ promoters.
Results obtained for translational fusions of different promoters to a reporter gene cannot be directly compared, since they reflect not only promoter strength but also different ribosomal binding sites and different β-galactosidase hybrid proteins. To gain more reliable information about the relative strengths of the deoQ and deoK promoters, transcriptional fusions were constructed. The intergenic region between deoQ and deoK including the first few codons of the genes was cloned in either orientation into pFZY1. The plasmids were transformed into MEC011, and the β-galactosidase activity levels were determined. As shown in Table 3, expression from the deoK promoter was 10-fold higher than from the deoQ promoter when cells were grown with glycerol as the carbon source (experiments 1 versus 3). In glucose medium, however, a significant repression of deoK-lacZ expression but not of deoQ-lacZ expression was observed (experiments 5 versus 3).
TABLE 3.
Regulation of β-galactosidase synthesis from transcriptional deoK-lacZ and deoQ-lacZ fusions and specificity of inductiona
| Expt no. | Genotype of pFZY-plasmid | Relevant genotype of pWSK129-plasmid | Carbon source | β-Galactosidase (units/OD436) |
|---|---|---|---|---|
| 1 | deoQ-lacZ+ | Vector | Gly | 8.2 |
| 2 | deoQ-lacZ+ | Vector | Glu | 13 |
| 3 | deoK-lacZ+ | Vector | Gly | 81 |
| 4 | deoK-lacZ+ | Vector | Gly + dRib | 76 |
| 5 | deoK-lacZ+ | Vector | Glu | 24 |
| 6 | deoK-lacZ+ | deoQ+ | Gly | 0.9 |
| 7 | deoK-lacZ+ | deoQ+ | Gly + dRib | 0.7 |
| 8 | deoK-lacZ+ | deo(QK)+ | Gly | 0.9 |
| 9 | deoK-lacZ+ | deo(QK)+ | Gly + dRib | 23 |
| 10 | deoK-lacZ+ | deo(QKP)+ | Gly | 0.9 |
| 11 | deoK-lacZ+ | deo(QKP)+ | Gly + dRib | 17 |
| 12 | deoK-lacZ+ | deo(QKPX)+ | Gly | 1.0 |
| 13 | deoK-lacZ+ | deo(QKPX)+ | Gly + dRib | 15 |
| 14 | deoK-lacZ+ | deo(QKPX)+ | Gly + ribose | 13 |
Cells of MEC011 carrying the transcriptional fusions pFZYφ(deoK-lacZ+) or pFZYφ(deoQ-lacZ+) and the indicated pWSK129-based plasmids were grown exponentially for at least three generations at 37°C in minimal medium with various carbon sources. Cell extracts were assayed for β-galactosidase activity.
Nature and specificity of the inducer.
Introduction of a second plasmid, carrying deoQ and various amounts of the deoKPX locus into MEC011/pFZYΦ(deoK-lac+), showed (Table 3) that the presence of deoQ resulted in strong repression (80- to 90-fold) and that induction by dRib was entirely dependent on the simultaneous presence of deoK. Neither deoP nor deoX were required for induction to occur. This suggested that the endogenous inducer of deoKPX was a metabolite of deoxyribose, rather than the free sugar itself. Under no conditions did the induced level reach the fully constitutive levels.
The ability of l-fucose, d-xylose, l-arabinose, and d-ribose to induce the DeoQ-mediated repression of deoK-lacZ expression was investigated using cultures of MEC011/pFZYφ(deoK-lac+)/pWSKdeoQKPX grown in glycerol minimal medium in the presence of 0.2% of the putative inducer. Of the compounds tested, only d-ribose was capable of stimulating β-galactosidase synthesis (Table 3, experiment 14).
Mapping of the transcriptional start sites.
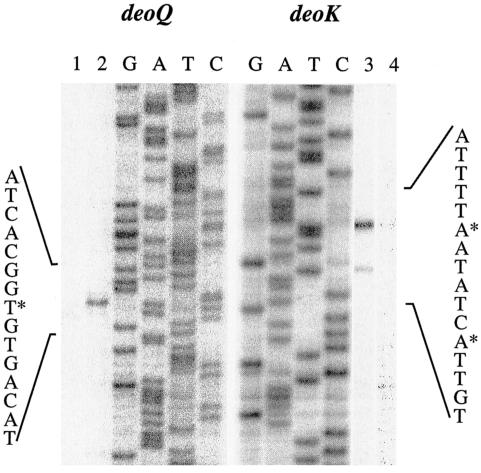
The transcriptional start site for each of the two promoters within the intergenic region was established by primer extension analysis. Total RNA was isolated from log-phase cultures of MEC011/pWSK129deoQKPX grown in minimal medium with either 0.2% glycerol or 0.2% dRib as the carbon source. As shown in Fig. 3, initiation of dRib-inducible deoK transcription occurred with ATP, 39 bp upstream of the DeoK start codon (Fig. 4). A weaker signal, which also appeared to be dRib regulated, was observed 6 bp downstream of the strong signal. Upstream of both of these sites, with the correct spacing, sequences resembling the consensus extended −10 element (TGNTATAAT) of putative promoters were identified (22). A dRib-regulated transcriptional start site signal was also observed for deoQ, although about 20-fold-higher levels of the same total RNA was required to obtain intensities similar to those observed for the deoK transcripts. The start site corresponded to an ATP start, 72 bp upstream of the deoQ start codon, and is preceded by a correctly spaced −10 element.
FIG. 3.
Primer extension analysis with total RNA from cells carrying pWSK129deoQKPX and grown in the absence (lanes 1 and 4) and presence (lanes 2 and 3) of 0.2% dRib. For the deoQ analyses, 20-fold more total RNA was used. Transcriptional start points are indicated with asterisks.
FIG. 4.
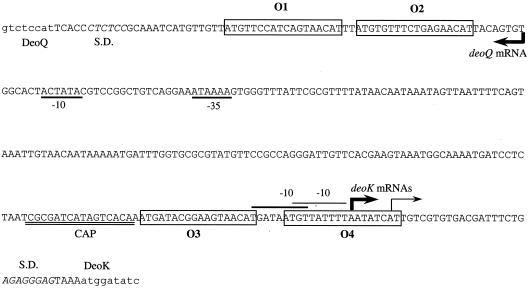
Nucleotide sequence of the deoQ-deoK intercistronic region. Major transcriptional starts are indicated with heavy arrows, and secondary starts with thin arrows. Coding regions are indicated with lowercase lettering. Deduced −10 elements for deoK are indicated by a line above them, and −10 and −35 elements for deoQ are underlined. Putative operator sites are boxed and numbered O1 through O4, and the inferred cAMP-CRP binding site is labeled CAP. Shine-Dalgarno elements are in italics and indicated by S.D.
DeoQ binding to the intergenic DNA.
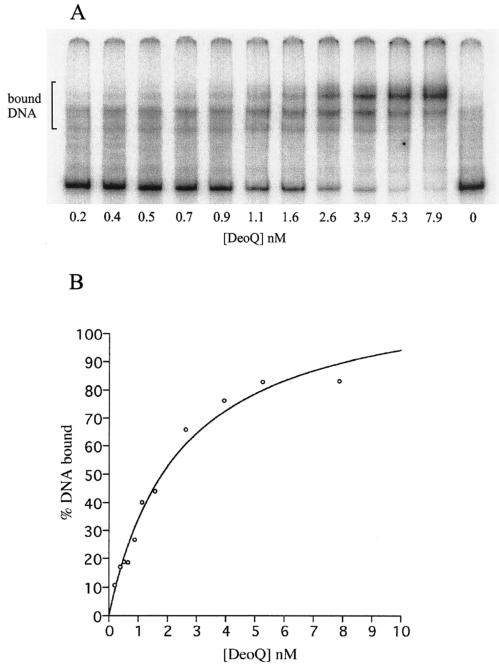
The binding of purified His-tagged DeoQ to the 324-bp deoQ-deoK intergenic region was determined using an electrophoretic gel mobility shift assay. The radiolabeled oligodeoxynucleotide was a 386-bp fragment containing the intergenic region and the first few codons of DeoQ and DeoK. As shown in Fig. 5A, multiple species of DeoQ-DNA complexes were observed, suggesting more than one binding site on the DNA. By determining the percentages of total DNA bound at different DeoQ concentrations, an apparent Kd of 2.4 nM was determined from the binding curve depicted in Fig. 5B. To estimate the contribution to this binding from each of the divergent promoter-operators, the binding of purified DeoQ to the individual promoter regions was determined, using radiolabeled PCR fragments of each half of the intergenic region, i.e., the 189-bp deoKp fragment and the 206-bp deoQp fragment (data not shown). The results (Table 4) indicated that the affinity of DeoQ to the deoK proximal operator was 18-fold higher than for the deoQ proximal operator. Inclusion of a 1 mM concentration of either dRib, dRib5P, or ribose in the mobility shift assays did not affect the percentage of DeoQ bound to the radiolabeled DNA probes (data not shown).
FIG. 5.
Binding of the DeoQ repressor protein to the 324-bp deoQ-deoK intergenic DNA. (A) Profile of gel retardation assays using 200 to 400 pM labeled probe and increasing concentrations of DeoQ. (B) Calculated binding curve for DeoQ with the intergenic DNA.
TABLE 4.
Binding of purified DeoQ to fragments of the deoQ-deoK intergenic regiona
Binding was determined using electrophoretic mobility shift assays as described in Materials and Methods.
DNA fragments used in the binding assays were prepared by PCR and labeled at the 5′ ends with 32P.
The apparent Kd values were determined as the concentration of DeoQ that gave 50% saturation of the operator sites.
[DeoQ], concentration of DeoQ based on the subunit size of 29 kDa.
ND, not determined.
Stoichiometry of DeoQ binding.
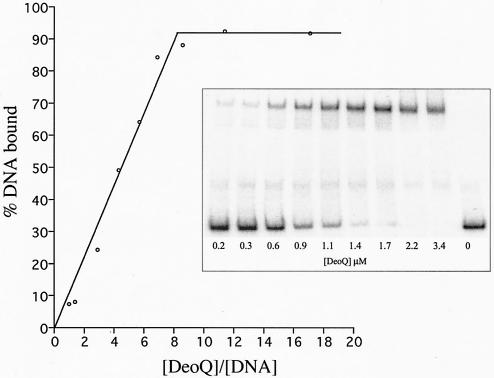
To determine the binding stoichiometry of DeoQ to the operator DNA, gel retardation assays were performed, using concentrations of operator DNA much higher than the Kd values. As shown for the deoKp fragment, containing only the deoK promoter/operator, approximately eight DeoQ subunits were bound per 189-bp fragment (Fig. 6). Similar titration experiments showed that approximately 16 DeoQ subunits were bound per 386-bp fragment when the full-length intergenic DNA was used as the probe (Table 4), suggesting that at saturation each promoter/operator is capable of binding one octameric DeoQ species.
FIG. 6.
DeoQ binding stoichiometry for the 189-bp deoK promoter/operator DNA. In each binding assay, a 200 nM nonradioactive 189-bp DNA fragment was present in addition to the radiolabeled DNA (200 pM).
Consistent with these findings, purified His-tagged DeoQ protein was observed to elute slightly ahead of the β-amylase (200 kDa) marker on a Superose 12HR gel filtration column, indicating that DeoQ in solution may exist as an octamer (8 × 29 kDa) (data not shown).
Two operator sites are required for efficient DeoQ binding.
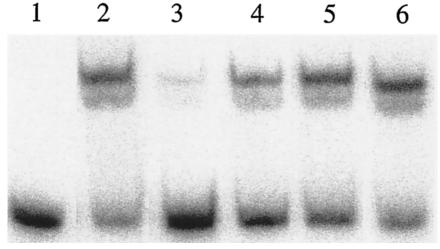
Inspection of the nucleotide sequence of the intergenic region between deoQ and deoK revealed a pair of closely spaced putative DeoQ binding sites within each of the two transcription initiation regions (Fig. 4). All four sites showed significant similarity with the consensus DeoR binding site (NTGTN10 ACAN [14]). One pair (O3-O4) included the entire deoK promoter and transcriptional start site, whereas the other (O1-O2) was located immediately downstream of the major transcriptional start site for deoQ mRNA. To determine whether O3 and O4 were involved in binding of DeoQ to the deoKp region, three 54-bp oligodeoxynucleotides were synthesized and used as competitor DNAs in gel retardation assays with radiolabeled deoKp and purified DeoQ. The unlabeled 54-mers contained either wild-type O3 and wild-type O4 (O3-O4), wild-type O3 and mutant O4 (O3-mutO4), or mutant O3 and wild-type O4 (mutO3-O4). The mutated operators harbored multiple base substitutions to destroy the palindromic nature of the sites (legend to Fig. 7). The concentration of DeoQ used in the experiment was chosen to give retardation of approximately 60% of the labeled DNA in the absence of competitor DNA. As shown in Fig. 7, the presence of a large excess of unlabeled O3-O4 resulted in a large decrease in the proportion of labeled DNA in the shifted position, whereas only minor competition was observed with the oligonucleotides harboring either one of the two operators in a mutated form. This suggested that both O3 and O4 are required for optimal binding of the DeoQ repressor to its DNA target.
FIG. 7.
Gel retardation assays of DeoQ binding to the 189-bp radiolabeled deoK promoter/operator DNA in the presence of synthetic, 54-bp, unlabeled operator DNA. All assays contained a 400 pM radioactive DNA fragment and an amount of DeoQ that gave an approximately 60% shift in the absence of competitor DNA. Lane 1, control with no DeoQ; lane 2, DeoQ with no competitor DNA; lane 3, DeoQ plus 1 μM unlabeled O3-O4 DNA; lane 4, DeoQ + 1 μM unlabeled O3-mutO4 DNA; lane 5, DeoQ + 1 μM unlabeled mutO3-O4 DNA; lane 6, DeoQ plus 0.02 A260 units of unlabeled poly(dI-dC). The 54-bp sequences of the competitor DNAs were as follows (the lowercase letters indicate mutations, and the putative operator sites are underlined): O3-O4, GTCACAAATGATACGGAAGTAACATGATAATGTTATTTTAATATCATTGTCGTG; O3-mutO4, GTCACAAATGATACGGAAGTAACATGATAgaaTcgTTTTAAcgTtgTTGTCGTG; mutO3-O4, GTCACAAgaaAcgCGGAAGgcAtgTGATAATGTTATTTTAATATCATTGTCGTG.
DISCUSSION
The deoQKPX gene system, responsible for the ability of serovar Typhimurium to utilize dRib as the sole carbon and energy source, was cloned for the purpose of a genetic characterization and a study of expression of the involved genes. In silico analysis led to the predictions that deoQ encoded a regulatory protein of the deoR-lacI family and that deoP encoded an inner-membrane protein involved in dRib transport (38). Recent studies of the purified DeoX protein have shown that it possesses mutarotase activity with high specificity towards deoxyribose (O. Barzu and A.-M. Gilles, unpublished results).
In the current study, complementation of the dRib− phenotype of E. coli with various regions of the deoQKPX locus showed that neither deoP nor deoX was required for growth on 0.2% dRib as carbon source. However, at suboptimal concentrations of dRib (0.08%), the presence of the two genes resulted in cumulative stimulation of growth (Table 1). Thus, both DeoP and DeoX are involved in facilitating the conversion of exogenous dRib to the endogenous substrate of deoxyribokinase, i.e., 2-d-deoxyribofuranose. This involves transport across the membrane and eventual interconversion of different steric forms of dRib.
The divergent deoQ and deoKPX promoters, deoQp and deoKp, are located at opposite ends of the deoQ-deoK intercistronic region, with deoKp being about 10-fold stronger than deoQp (Table 3). Initiation of transcription from each of the promoters is regulated negatively both by the DeoQ repressor and by DeoR, the repressor of the deoxynucleoside catabolic deoCABD operon. It should be noted, however, that DeoQ is not capable of repressing expression of the deoCABD operon, as shown by the high constitutive levels of thymidine phosphorylase in a deoR/pdeoQKPX strain (Table 2). In a deoR+/pdeoQKPX strain, which expresses both repressor proteins, addition of dRib resulted in induction of both deoQp and deoKp as well as the deoCABD operon. Thymidine, on the other hand, induced only the deoCABD operon. This indicates that dRib5P, which is the true inducer of the DeoR regulon (15), is not the endogenous inducer of DeoQ-mediated repression. Since we further observed (Table 3) that induction of deoKp by dRib required the presence of the deoK gene, we propose that deoxyribokinase, in addition to its metabolic function of catalyzing the conversion of dRib to dRib5P, may catalyze alternative transformations (phosphorylations) of dRib or dRib5P to produce the true inducer. This may resemble the induction pattern observed for the E. coli lac operon, where a functional β-galactosidase (lacZ) is required not only for cleavage of the substrate lactose but also for converting lactose to the true inducer of the operon, allolactose (19, 29).
Gel retardation assays with purified DeoQ protein showed that the repressor binds to each of the promoter-proximal halves of the deoQ-deoK intergenic DNA. In accordance with the in vivo results showing that DeoQ represses expression of deoK more effectively than expression of deoQ, the in vitro binding studies showed that the affinity of DeoQ for the deoKp region was much higher than for the deoQp region (Table 4). Although DeoQ is similar to DeoR from both E. coli and Bacillus subtilis in appearing to exist as an octamer in solution (28, 40) and containing a helix-turn-helix domain characteristic for DNA-binding proteins, it differs from the two DeoR proteins in that dRib5P is not the low-molecular-weight effector for the protein (see above).
Four putative operator sites resembling the E. coli DeoR-binding operators of the deoCABD operon were identified within the deoQ-deoK intergenic region (Fig. 4). They are organized in pairs, with one tandem operator, (O1-O2), located shortly after the transcriptional start point of deoQ, and the other (O3-O4) covering the entire deoK promoter. From competition experiments with synthetic mutant operator sequences (Fig. 7), we conclude that both the O3 and O4 wild-type operators are required for efficient binding of DeoQ to the deoK promoter. DeoR-mediated repression of the deoCABD operon, on the other hand, involves long-range cooperative binding to two or three operators located several hundred base pairs apart, resulting in looping of the interoperator sequences (6, 9). Thus, the observation that DeoR is an effective repressor of deoKPX expression suggests not only that the two repressor proteins may recognize the same operator sequences but also that DeoR binding may involve cooperation between operator sites from both deoQp and deoKp. The observation that DeoQ did not repress expression of the deoCABD operon may be explained simply by the lack of closely spaced tandem operators in the promoter region of the deoCABD operon.
In addition, transcription from deoKp, but not from deoQp, was observed to be subject to catabolite repression by glucose. Although direct evidence has not been obtained, the absence of a −35 element combined with the presence of a putative CAP binding site at the consensus position for a class II CAP-dependent promoter (centered at position −41 with respect to the transcription start) (Fig. 4) indicated that the glucose effect is likely to be mediated through the cAMP/CAP gene activation system (4, 10).
PSI-BLAST (1) searches were carried out to identify homologues of DeoX, DeoP, DeoK, and DeoQ. The entire deoQKPX locus, with the same organization as in S. enterica, was found in the genomes of serovar Enteritidis and the uropathogenic E. coli CFT073, whereas it is not present in the genome of the enterohemorrhagic E. coli O157:H7. P. syringae harbors a similar region except that the gene order is deoQ-deoX-deoP-deoK, whereas for L. plantarum, a bacterium known to ferment deoxyribose (11, 13), the four genes are found in a cluster together with the gene for deoxyriboaldolase (deoC). The gene order in L. plantarum is deoQ-deoC-deoP-deoX-deoK. The deoxyribose utilization genes are also organized together with deoC in the plant pathogen A. tumefaciens; however, in this bacterium the deoP gene is replaced by four genes encoding members of a putative ABC transporter system with specificity towards pentoses. The significance of the presence of the deoQKPX locus in only one of the three complete E. coli genomes is unknown, and the distribution of the operon in various E. coli strains is presently under investigation (C. Bernier-Febreau et al.; L. du Merle et al., unpublished data).
Acknowledgments
The expert technical assistance of Lisbeth Stauning is gratefully recognized.
This work was supported by a NATO International Collaborative research grant (to R.A.K. and J.N.) and grants from the Natural Sciences and Engineering Research Council of Canada (to R.A.K.), from the Centre National de la Recherche Scientifique (URA2185), and from Institut Pasteur (to O.B. and A.M.G.).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartolomé, B., J. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 5.Clark, D. J., and O. Maaløe. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 6.Dandanell, G. 1992. DeoR repression at-a-distance only weakly responds to changes in interoperator separation and DNA topology. Nucleic Acids Res. 20:5407-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dandanell, G., and K. Hammer. 1985. Two operator sites separated by 599 base pairs are required for deoR repression of the deo operon of Escherichia coli. EMBO J. 4:3333-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dandanell, G., and K. Hammer. 1989. The deoR repressor from E. coli and its action in regulation-at-a-distance, p. 79-91. In F. Eckstein and D. M. J. Lilley (ed.), Nucleic acids and molecular biology, vol. 3. Springer-Verlag, Berlin, Germany.
- 9.Dandanell, G., P. Valentin-Hansen, J. E. L. Larsen, and K. Hammer. 1987. Long-range cooperativity between gene regulatory sequences in a prokaryote. Nature (London) 325:823-836. [DOI] [PubMed] [Google Scholar]
- 10.De Crombrugghe, B., S. Busby, and H. Buc. 1984. Cyclic AMP receptor protein: role in transcription activation. Science 224:831-838. [DOI] [PubMed] [Google Scholar]
- 11.Domagk, G. F., and B. L. Horecker. 1958. Pentose fermentation by Lactobacillus plantarum. V. Fermentation of 2-deoxy-d-ribose. J. Biol. Chem. 233:283-286. [PubMed] [Google Scholar]
- 12.Feiss, M., S. Adyha, and D. L. Court. 1972. Isolation of plaque-forming, galactose-transducing strains of phage lambda. Genetics 71:189-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginsberg, A. 1959. A deoxyribokinase from Lactobacillus plantarum. J. Biol. Chem. 235:1292-1298. [PubMed] [Google Scholar]
- 14.Hammer, K., L. Bech, P. Hobolth, and G. Dandanell. 1993. DNA specificity of Escherichia coli deoP1 operator-DeoR repressor recognition. Mol. Gen. Genet. 237:129-133. [DOI] [PubMed] [Google Scholar]
- 15.Hammer-Jespersen, K. 1983. Nucleoside catabolism, p. 203-258. In A. Munch-Petersen (ed.), Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. Academic Press, Inc., Ltd., London, United Kingdom.
- 16.Hmiel, S. P., M. D. Snavely, C. G. Miller, and M. E. Maguire. 1986. Magnesium transport in Salmonella typhimurium: characterization of magnesium influx and cloning of a transport gene. J. Bacteriol. 168:1444-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffee, P. 1968. 2-Deoxyribose gene-enzyme complex in Salmonella typhimurium. I. Isolation and enzymatic characterization of 2-deoxyribose-negative mutants. J. Bacteriol. 95:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hove-Jensen, B., and M. Maigaard. 1993. Escherichia coli rpiA gene encoding ribose phosphate isomerase A. J. Bacteriol. 175:5628-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jobe, A., and S. Bourgeois. 1972. lac repressor-operator interaction. VI. The natural inducer of the lac operon. J. Mol. Biol. 69:397-408. [DOI] [PubMed] [Google Scholar]
- 20.Kelln, R. A., and J. Neuhard. 1988. Regulation of pyrC expression in Salmonella typhimurium: identification of a regulatory region. Mol. Gen. Genet. 212:287-294. [DOI] [PubMed] [Google Scholar]
- 21.Koop, A. H., M. E. Hartley, and S. Burgeois. 1987. A low-copy-number vector utilizing β-galactosidase for the analysis of gene control elements. Gene 521:245-256. [DOI] [PubMed] [Google Scholar]
- 22.Kumar, A., R. A. Malloch, N. Fujita, D. A. Smillie, A. Ishihama, and R. S. Hayward. 1993. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J. Mol. Biol. 232:406-418. [DOI] [PubMed] [Google Scholar]
- 23.Mathews, D. H., T. C. Andre, J. Kim, D. H. Turner, and M. Zuker. 1998. An updated recursive algorithm for RNA secondary structure prediction with improved free energy parameters. Am. Chem. Soc. Symp. Ser. 682:246-257. [Google Scholar]
- 24.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 25.McKenney, K., H. Shimatake, D. Court, U. Schmeissner, D. Brady, and M. Rosenberg. 1981. Analysis of nucleic acids by enzymatic methods, p. 383-415. In J. C. Chirikjian, and T. S. Papas (ed.), Gene amplification and analysis, vol. II. Elsevier-North Holland, Amsterdam, The Netherlands. [PubMed]
- 26.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Møllegaard, N. E., P. B. Rasmussen, P. Valentin-Hansen, and P. E. Nielsen. 1993. Characterization of promoter recognition complexes formed by CRP and CytR for repression and by CRP and RNA polymerase for activation of transcription on the Escherichia coli deoP2 promoter. J. Biol. Chem. 268:17471-17477. [PubMed] [Google Scholar]
- 28.Mortensen, L., G. Dandanell, and K. Hammer. 1989. Purification and characterization of the deoR repressor of Escherichia coli. EMBO J. 8:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller-Hill, B., H. V. Rickenberg, and K. Wallenfels. 1964. Specificity of the induction of the enzymes of the lac operon in Escherichia coli. J. Mol. Biol. 10:303-318. [DOI] [PubMed] [Google Scholar]
- 30.Rasmussen, M. A. 1993. Isolation and characterization of Selenomonas ruminantium strains capable of 2-deoxyribose utilization. Appl. Environ. Microbiol. 59:2077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz, M. 1978. Thymidine phosphorylase from Escherichia coli. Methods Enzymol. 51:442-445. [DOI] [PubMed] [Google Scholar]
- 34.Shin, M., S. Kang, S. J. Hyun, N. Fujita, A. Ishihama, P. Valentin-Hansen, and H. E. Choy. 2001. Repression of deoP2 in Escherichia coli by CytR: conversion of a transcription activator into a repressor. EMBO J. 20:5392-5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sørensen, K. I., and J. Neuhard. 1991. Dual transcriptional initiation sites from the pyrC promoter control expression of the gene in Salmonella typhimurium. Mol. Gen. Genet. 225:249-256. [DOI] [PubMed] [Google Scholar]
- 36.Stern, S., D. Moazed, and H. F. Noller. 1988. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164:481-489. [DOI] [PubMed] [Google Scholar]
- 37.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 38.Tourneux, L., N. Bucurenci, C. Saveanu, P. A. Kaminski, M. Bouzon, E. Pistotnik, A. Namane, P. Marlière, O. Barzu, I. L. de la Sierra, J. Neuhard, and A-M. Gilles. 2000. Genetic and biochemical characterization of Salmonella enterica serovar Typhi deoxyribokinase. J. Bacteriol. 182:869-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 40.Zeng, X., and H. H. Saxild. 1999. Identification and characterization of a DeoR-specific operator sequence essential for induction of dra-nupC-pdp operon expression in Bacillus subtilis. J. Bacteriol. 181:1719-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]