Abstract
The virulence of Listeria monocytogenes is directly related to its ability to spread from cell to cell without leaving the intracellular milieu. During cell-to-cell spread, bacteria become temporarily confined to secondary vacuoles. Among the bacterial factors involved in escape from these vacuoles is a secreted broad-range phospholipase C (PC-PLC), the activation of which requires processing of an N-terminal prodomain. Mpl, a secreted metalloprotease of Listeria, is involved in the proteolytic activation of PC-PLC. We previously showed that, during intracellular growth, bacteria maintain a pool of PC-PLC that is not accessible to antibodies and that is rapidly released in its active form in response to a decrease in pH. pH-regulated release of active PC-PLC is Mpl dependent. To further characterize the mechanism regulating secretion of PC-PLC, the bacterial localization of PC-PLC and Mpl was investigated. Both proteins were detected in the bacterial supernatant and lysate with no apparent changes in molecular weight. Extraction of bacteria-associated PC-PLC and Mpl required cell wall hydrolysis, but there was no indication that either protein was covalently bound to the bacterial cell wall. Results from pulse-chase experiments performed with infected macrophages indicated that the rate of synthesis of PC-PLC exceeded the rate of translocation across the bacterial cell wall and confirmed that the pool of PC-PLC associated with bacteria was efficiently activated and secreted upon acidification of the host cell cytosol. These data suggest that bacterially associated PC-PLC and Mpl localize at the cell wall-membrane interface and that translocation of PC-PLC across the bacterial cell wall is rate limiting, resulting in the formation of a bacterially associated pool of PC-PLC that would readily be accessible for activation and release into nascent secondary vacuoles.
The virulence of Listeria monocytogenes is directly related to its ability to live and multiply in the cytosol of host cells and its efficacy to spread from cell to cell without leaving the intracellular milieu (14, 44). The steps involved in bacterial cell-to-cell spread include a host-derived actin-based mechanism of bacterial motility leading to the development of membrane protrusions and uptake of these protrusions by neighboring cells to produce double-membrane vacuoles, also called secondary vacuoles. A number of bacterial factors involved in escape from these vacuoles have been identified (7, 15, 27, 38, 47), and the kinetics of vacuolar lysis is likely to be related to the temporal and sequential activities of these factors (9). Among these factors are a broad-range phospholipase C (PC-PLC) and a zinc-dependent metalloprotease (Mpl). Both proteins are predicted to be secreted and are made as proenzymes, whose activation requires proteolytic cleavage of an N-terminal prodomain (31, 33). Mpl is involved (directly or indirectly) in the proteolytic activation of PC-PLC (27, 35). The mechanism of Mpl activation is unknown, although similar proteases are activated by an autocatalytic process (46). We are interested in the mechanisms regulating the activities of PC-PLC and Mpl during intracellular infection.
During intracellular growth, bacteria maintain a pool of PC-PLC that is nonaccessible to antibodies and that is rapidly secreted in its active form upon acidification of the intracellular milieu (28). Secretion of active PC-PLC occurs within minutes of a decrease in pH and is dependent on Mpl but independent of de novo protein synthesis. These results reveal the existence of a pH-dependent mechanism regulating the activities of PC-PLC and Mpl. For L. monocytogenes, intracellular survival relies on the precision of this mechanism, because the combination of premature activation and secretion of PC-PLC in the cytosol of infected cells is cytotoxic (28).
To further characterize the mechanism controlling PC-PLC secretion, the localization of PC-PLC and Mpl was investigated. Both proteins were detected in the bacterial supernatant and lysate, with no apparent changes in molecular weight. Bacterially associated PC-PLC and Mpl were not extractable from intact bacteria under denaturing and reducing conditions, but there was no indication that either protein was covalently bound to the bacterial cell wall. Results from pulse-chase labeling of de novo-made protein suggested that translocation across the bacterial cell wall was a rate-limiting step for the secretion of PC-PLC.
MATERIALS AND METHODS
Bacterial strains and reagents.
Wild-type L. monocytogenes strain 10403S (34) and isogenic mutant strains DP-L1075 (Tn917 insertion in PrfA) (12), DP-L1552 (ΔplcA) (7), DP-L1935 (ΔplcB) (38), DP-L1942 (ΔactA) (4), DP-L2296 (Δmpl) (26), and EJ-L12 (ΔinlA; kindly provided by J. F. Miller, University of California—Los Angeles) were used for this study. Wild-type L. monocytogenes EGD and Bug 1768 (ΔsrtA in EGD; kindly provided by Olaf Schneewind) (2) were used in one experiment.
Rabbit polyclonal antibodies to ActA, Mpl, and PrfA were provided by Daniel Portnoy. The mouse polyclonal antibody to InlA was provided by Terry Potter. The rabbit polyclonal antibodies to PC-PLC and PI-PLC were provided by Howard Goldfine. PC-PLC antibodies were affinity purified as described previously (27). Mpl antibodies were affinity purified with purified Mpl prodomain (amino acids 25 to 204 of Mpl plus a C-terminal His tag) and Mpl catalytic subdomain B (amino acids 353 to 510 plus a C-terminal His tag). The Mpl His tag constructs were expressed in Escherichia coli and affinity purified with Ni-nitrilotriacetic acid agarose (Qiagen) under denaturing conditions according to the manufacturer's instructions.
Modified Listeria-specific phage endolysin A118 was affinity purified as described previously (25). The endolysin is an endopeptidase that cleaves the peptidoglycan branching peptides.
Bacterial culture conditions.
For the in vitro experiments, expression of the PrfA-regulated genes was induced by growing bacteria in Luria-Bertani (LB) broth-50 mM MOPS (morpholinepropanesulfonic acid) adjusted to pH 7.3, 25 mM glucose-1-phosphate (G1P), and 0.2% activated charcoal (LB-MOPS-G1P) (36). Bacteria were grown at 37°C without shaking to an optical density at 600 nm (OD600) of 1.0 to 1.2. For the PrfA mutant, 10 mM glucose was substituted for G1P, because this mutant cannot use G1P as a source of carbon. For J774 infections, bacteria were grown overnight at 30°C in brain heart infusion (BHI) broth.
Preparation of bacterial fractions.
Bacteria were grown in LB-MOPS-G1P to an OD600 of 1 to 1.2 and washed in 50 mM Tris-HCl (pH 8.0)-10 mM MgCl2 (TM buffer). Bacterial cells were suspended in 0.5 M sucrose-50 mM Tris-HCl (pH 8.0)-10 mM MgCl2 (protoplast buffer) to a volume equivalent to a calculated OD600 of 10. AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride], bestatin, pepstatin A, E-64, and phosphoramidon were added as a premixed cocktail of protease inhibitors (Sigma) to a final dilution of 1/500, and sodium azide (3 mM) was added to block protein secretion. The bacterial cell wall was digested with purified Listeria-specific phage endolysin A118 (20 to 40 μg ml−1) at 37°C for 15 min. Protoplast formation was monitored by microscopy. Cell wall fraction and protoplasts were separated by centrifugation at 15,000 × g for 3 min at 4°C. The cell wall fraction was mixed with an equal volume of 2× sample buffer (4% sodium dodecyl sulfate [SDS], 20% glycerol, 125 mM Tris-HCl [pH 6.8], 10% β-mercaptoethanol [βME], bromophenol blue). The protoplasts were suspended in protoplast buffer to the original reaction volume, and an equal volume of 2× sample buffer was added. Bacterial lysates were generated by adding an equal volume of 2× sample buffer to bacterial cells treated with phage endolysin, and the samples were boiled for 5 min. For SDS extracts, bacterial cells were washed once in phosphate-buffered saline (PBS) or TM buffer, suspended in 2× sample buffer to a volume equivalent to a calculated OD600 of 5, and boiled for 5 min. Bacteria were pelleted by centrifugation, and soluble SDS-extracted proteins were recovered in the supernatant. Secreted proteins were prepared by adding 5% trichloroacetic acid to bacterial supernatants. Precipitates were washed with acetone, solubilized in 2× sample buffer to a volume equivalent to a calculated OD600 of 5, and boiled for 5 min.
Metabolic labeling and immunoprecipitation experiments.
For metabolic labeling of intracellularly grown bacteria, the mouse macrophage-like cell line J774 was used, and the experiment was performed as described previously with some modifications (27). Macrophages were seeded in 35-mm-diameter tissue culture dishes and infected with L. monocytogenes at a multiplicity of infection (MOI) of 5 to 10 bacteria per cell. Infected cells were washed with PBS 30 min postinfection, and gentamicin (10 μg/ml) was added at 1 h postinfection. At 3.5 h postinfection, J774 cells were starved for methionine and cysteine, and cytochalasin D (0.25 μg ml−1) was added to stop bacterial cell-to-cell spread. At 4 h postinfection, host protein synthesis inhibitors (cycloheximide, 22.5 μg ml−1; anisomycin, 30 μg ml−1) and proteasome inhibitor LLnL (50 μM) were added. Ten minutes later, infected cells were pulse-labeled for 5 min with 90 μCi of [35S]Met (Express protein labeling from NEN Life Science) and chased for 5 min in pulsing medium supplemented with chloramphenicol (20 μg ml−1), tetracycline (10 μg ml−1), methionine (5 mM), and cysteine (0.5 mM). Where indicated, samples were chased for an additional 10 min in buffer (133 mM KCl, 1 mM MgCl2, 15 mM Mes, 15 mM HEPES) adjusted to pH 7.3 or 6.5 and supplemented with nigericin (10 μM) to equilibrate the intracellular pH of the host. This buffer also contained all of the supplements mentioned above for the pulsing and chasing medium. Samples were washed three times with ice-cold PBS and lysed on ice in 500 μl of NP-40 buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 1% NP-40) supplemented with protease inhibitors. Lysates were passed several times through a 26-gauge needle to shear the DNA. Bacteria (L. monocytogenes does not lyse in this buffer) and insoluble material were pelleted by centrifugation at 20,000 × g for 20 min at 4°C. Supernatants were transferred to new tubes, and 500 μl of radioimmunoprecipitation assay (RIPA) buffer (NP-40 buffer with 0.1% SDS, 0.5% deoxycholate, 1 mM EDTA, cocktail of protease inhibitors) containing the appropriate antibodies was added to each sample for detection of bacterially secreted proteins. Pellets were suspended in 100 μl of protoplast buffer supplemented with phage endolysin, protease inhibitors, and Na azide. Protoplast formation was monitored by microscopy. Four hundred microliters of NP-40 buffer and 500 μl of RIPA buffer containing the appropriate antibodies were added to each sample. The samples were incubated overnight at 4°C and cleared by centrifugation at 20,000 × g for 20 min at 4°C. Preswollen protein A-Sepharose beads in RIPA buffer were added to each sample, and this mixture was incubated at 4°C for 2 h. Protein A beads were washed four times with ice-cold RIPA buffer, and immunoprecipitates were recovered in 2× sample buffer. Samples were boiled for 5 min, protein A beads were pelleted by centrifugation, and supernatants were loaded on 11% bisacrylamide gels. Immunoprecipitates were detected by autoradiography with a BioMax Transcreen LE (Kodak) and by phosphorimaging. A STORM 860 scanner (Molecular Dynamics) was used to scan the phosphor screen, and results were analyzed with ImageQuant software (Molecular Dynamics).
Western immunoblot.
After completion of SDS-polyacrylamide gel electrophoresis (PAGE), the gels were equilibrated in transfer buffer (48 mM Tris-39 mM glycine-10% methanol). The proteins were electrotransferred to polyvinylidene difluoride membrane (Bio-Rad) by using a semidry transfer apparatus. The membranes were blocked with 2% bovine serum albumin in TBST buffer (10 mM Tris-HCl [pH 8.0]-150 mM NaCl-0.05% Tween 20) for 2 to 4 h. Antibodies were diluted in the same buffer and incubated overnight at 4°C: 1/1,600 for InlA, 1/500 for Mpl, 1/100 for PC-PLC, 1/50,000 for PI-PLC, and 1/12,000 for PrfA. Membranes were washed three times in TBST and reacted for 1 to 2 h with alkaline phosphatase-conjugated goat anti-rabbit (1/25,000) or anti-mouse (1/10,000) secondary antibodies (Molecular Probes). After several washes, nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate were added as colorimetric substrates for alkaline phosphatase. The reaction was stopped by rinsing the membrane in water.
RESULTS
Detection of bacteria-associated PC-PLC and Mpl from broth-grown bacteria.
The amino acid sequences of PC-PLC and Mpl reveal the presence of N-terminal signal sequences, and, accordingly, both proteins can be detected in the supernatant of bacterial cultures (11, 16, 17, 27, 31, 47). PC-PLC is secreted as a proenzyme with a predicted molecular mass of 30 kDa, and activation requires cleavage of a 24-amino-acid N-terminal prodomain. By SDS-PAGE, the pro and active forms of the enzyme migrate as approximately 32- and 28-kDa proteins, respectively (27). Mpl is secreted as a proenzyme of 55 kDa (11). The pro- and catalytic domains of Mpl are predicted to be 21 and 34 kDa, respectively (31). Recently, we reported that intracellular bacteria carry a pool of PC-PLC that is rapidly secreted in response to a decrease in host intracellular pH (28). Moreover, this phenomenon is Mpl dependent, suggesting the existence of bacterially associated Mpl.
The localization of PC-PLC from broth-grown bacteria was investigated. Bacteria were grown in LB-MOPS-G1P or LB-MOPS-glucose to an OD600 of 1.0 to 1.2, and the cultures were separated in three fractions: bacterially associated proteins (bacterial lysates), surface extractable proteins (SDS-βME extracts), and secreted proteins (supernatants). Control proteins included InlA, a nonextractable surface protein that is bound to the peptidoglycan in a covalent manner (10, 13); PI-PLC, a secreted protein (18); and PrfA, a cytoplasmic protein (23). InlA and PrfA were found exclusively in the bacterial cell lysate as expected (Fig. 1). PI-PLC was found in all three fractions, although bacterially associated PI-PLC was completely extractable with SDS-βME, suggesting that it was in the process of being translocated across the cell wall. PC-PLC was detected in the bacterial lysate and supernatant, and bacterially associated PC-PLC was not extractable with SDS-βME. There was no difference in the migration patterns of bacterially associated PC-PLC and secreted PC-PLC, suggesting that bacterially associated PC-PLC had been translocated across the bacterial membrane and that the signal sequence had been processed.
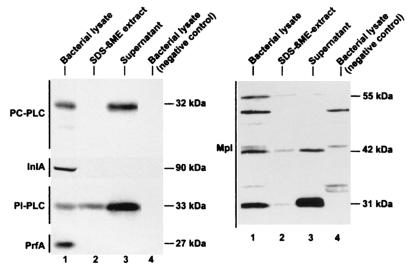
FIG. 1.
Immunodetection of PC-PLC and Mpl from broth-grown bacteria. Bacteria were grown in LB-MOPS-G1P or LB-MOPS-glucose (DP-L1075). Bacterial lysates, SDS-βME extracts, and secreted proteins were prepared as described in Materials and Methods. Equivalent amounts of culture OD units were loaded per lane. Proteins were detected by Western immunoblotting. The various polypeptides migrated as indicated on the figure. Lanes 1 to 3 (both panels), 10403S wild-type strain. Negative controls in lane 4 (left panels from the top) include DP-L1935 (ΔplcB), EJ-L12 (ΔinlA), DP-L1552 (ΔplcA), and DP-L1075 (Tn917 insertion in prfA). The negative control in lane 4 (right panel) is DP-L2296 (Δmpl).
The localization of Mpl from broth-grown bacteria was also investigated. Four Mpl-specific species were detected by Western immunoblotting and migrated as polypeptides of 57 (presumably the preproenzyme, which was not consistently detectable), 55, 42, and 31 kDa (Fig. 1). It is unclear which one of the 42- and 31-kDa species represents the active form of Mpl. The proenzyme (55 kDa) was found predominantly in the bacterial lysate. The 42-kDa protein was found in equal amounts in the bacterial lysate and supernatant, whereas the 31-kDa protein was found predominantly in the supernatant fraction. Minute amounts of all three Mpl species were extractable with SDS-βME, suggesting that most of bacterially associated Mpl was not in the process of being translocated across the cell wall.
Localization of bacterially associated PC-PLC and Mpl.
Bacterial fractionation was performed to determine the localization of bacterially associated PC-PLC and Mpl. Broth-grown bacterial cells were stripped of their cell wall with a cell wall endopeptidase and separated from the protoplasts by centrifugation. The purity of the fractions was determined by detection of control protein markers: InlA, a surface protein that is covalently anchored to the cell wall (10, 13); and PrfA, a cytoplasmic protein (23). PC-PLC, Mpl, and the control markers were detected by Western immunoblotting. The results show that InlA fractionated specifically with the bacterial cell wall, whereas PrfA fractionated with the protoplasts (Fig. 2 and 3). PC-PLC consistently fractionated with the bacterial cell wall (Fig. 2). However, the distribution of all three species of Mpl varied from being cell wall associated (H. Marquis, unpublished data) to being dispersed in both cell wall and protoplast fractions (Fig. 3). The migration behaviors of cell wall- and protoplast-associated Mpl were undistinguishable, suggesting that the cell wall-associated fraction of Mpl was not a cleaved product of the protoplast-associated fraction.
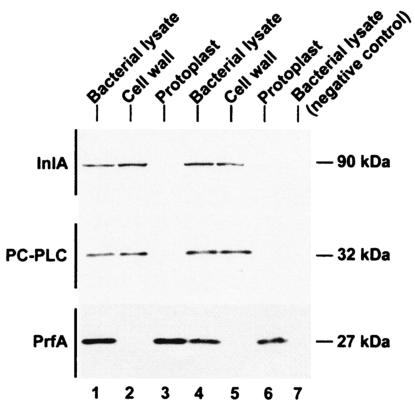
FIG. 2.
Detection of PC-PLC in the cell wall fraction of broth-grown bacteria. Bacteria were grown in LB-MOPS-G1P or LB-MOPS-glucose (DP-L1075). Bacterial lysates, cell walls, and protoplasts were prepared as described in Materials and Methods. Equivalent amounts of culture OD units were loaded per lane. Proteins were detected by Western immunoblotting. The various polypeptides migrated as indicated on the figure. Lanes: 1 to 3,10403S wild-type strain; 4 to 6, DP-L2296 (Δmpl); 7 (top to bottom), EJ-L12 (ΔinlA), DP-L1935 (ΔplcB), and DP-L1075 (Tn917 insertion in prfA).
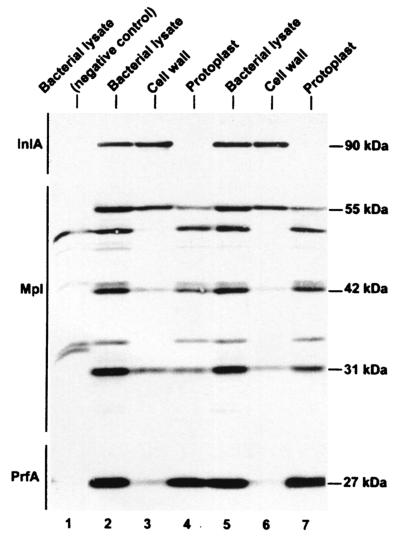
FIG. 3.
Detection of Mpl in the cell wall and protoplast fractions of broth-grown bacteria. Bacteria were grown in LB-MOPS-G1P or LB-MOPS-glucose (DP-L1075). Bacterial lysates, cell walls, and protoplasts were prepared as described in Materials and Methods. Equivalent amounts of culture OD units were loaded per lane. Proteins were detected by Western immunoblotting. The various polypeptides migrated as indicated on the figure. Lanes: 2 to 4,10403S wild-type strain; 5 to 7, DP-L1935 (ΔplcB); 1 (from top to bottom), EJ-L12 (ΔinlA), DP-L2296 (Δmpl), and DP-L1075 (Tn917 insertion in prfA).
Rapid secretion of PC-PLC in response to a decrease in intracellular pH is Mpl dependent. We investigated whether the bacterial association of PC-PLC and Mpl was dependent on the presence of the other protein. The localization of PC-PLC in an Mpl-negative background and of Mpl in a PC-PLC-negative background was determined. The results indicated that neither PC-PLC nor Mpl localization was influenced by the presence of the other protein (Fig. 2, lanes 4 to 6; and Fig. 3, lanes 5 to 7).
In gram-positive bacteria, proteins targeted to the bacterial cell wall contain a sorting signal (LPXTG) followed by a stretch of hydrophobic amino acids (37). Sortase A catalyzes the covalent anchoring of these proteins to the cell wall (2, 29). In Staphylococus aureus, a second sortase enzyme was shown to recognize an NPQTN motif (30). Neither PC-PLC nor Mpl has characteristics of cell wall-anchored proteins. Nevertheless, we tested whether sortase A was involved in the bacterial localization of PC-PLC or Mpl. Immunodetection of PC-PLC and Mpl from bacterial lysates, SDS-βME extracts, and supernatants did not reveal any differences between the sortase A-negative mutant (Bug 1768) and the two wild-type strains tested, EGD and 10403S (H. Marquis, unpublished data). This result indicates that PC-PLC and Mpl association with the bacterial cell wall was independent of sortase A.
Translocation of PC-PLC across the bacterial cell wall.
We investigated the efficacy of secretion of de novo-made PC-PLC and PI-PLC in intracellularly grown bacteria. Infected J774 cells were pulse-labeled for 5 min and chased for 5 min, and the phospholipases were immunoprecipitated from host cell lysates and from bacterial lysates. The ratios of bacterially associated versus secreted PC-PLC and PI-PLC were approximately 3:1 and 1:3, respectively (Fig. 4, lanes 1, 2, 9, and 10), as determined by image analysis of the phosphor scan. Bacterially associated PC-PLC and PI-PLC comigrated with the secreted species, indicating that the protein signal sequences had been processed. These results suggest that translocation of PC-PLC across the bacterial cell wall is slower than that of PI-PLC.
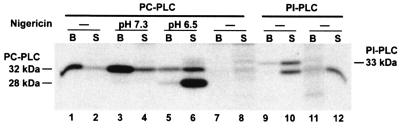
FIG. 4.
Detection of de novo-made PC-PLC and PI-PLC from intracellular bacteria by autoradiography. Bacteria grown in J774 cells were pulse-labeled for 5 min and chased for 5 min at 37°C as described in Materials and Methods. When indicated, samples were treated for an additional 10 min with nigericin in buffer at pH 7.3 or 6.5. PC-PLC and PI-PLC were immunoprecipitated from bacterial lysates (B [bacterially associated]) and host cell lysates (S [secreted in the host]), fractionated by SDS-PAGE, and detected by autoradiography. Lanes: 1 to 8, immunoprecipitation of PC-PLC; 9 to 12, immunoprecipitation of PI-PLC. Lanes 1 to 6, 9, and 10 show results from cells infected with the wild-type strain, 10403S. Negative controls are in lanes 7 and 8 (ΔplcB) and 11 and 12 (ΔplcA). Proform and processed forms of PC-PLC migrated as indicated on the left side of the figure. PI-PLC migrated as indicated on the right side of the figure. In lanes 10 and 12, the faster-migrating band is nonspecific, because it is present in the deletion mutant.
The efficacy of PC-PLC translocation across the cell wall in response to a decrease in host intracellular pH was also investigated. Pulse-labeled infected cells were perfused with buffer at pH 7.3 or 6.5 in presence of nigericin for a period of 10 min. PC-PLC was immunoprecipitated from host cell lysates and bacterial lysates. The ratios of bacterially associated versus secreted PC-PLC were 4:1 at pH 7.3 and 1:6 at pH 6.5 (Fig. 4, lanes 3, 4, 5, and 6). PC-PLC was detected in its proform at pH 7.3, whereas the processed form of PC-PLC was predominant at pH 6.5. These results confirm that pH contributes to the regulation of PC-PLC activation and translocation across the bacterial cell wall. However, this phenomenon was not observed when broth-grown bacteria were subjected to a rapid decrease in pH (H. Marquis, unpublished data), suggesting that intracellular factors other than pH contribute to the regulation of PC-PLC activation and secretion.
The total amount of PC-PLC detected appears to be significantly higher after the chase at pH 6.5 than after the pulse or after the chase at pH 7.3. Perhaps, bacterially associated PC-PLC is degraded by bacterially associated proteases during protoplast formation. Alternatively, intracellular bacteria may be more resistant to protoplast formation influencing the apparent amount of bacterially associated protein detected. In either case, the results would underestimate the ratio of PC-PLC that remains bacterially associated during intracellular bacterial growth.
DISCUSSION
This study shows that PC-PLC and Mpl fractionate with secreted proteins and with the bacterial lysate. Extraction of bacterially associated PC-PLC and Mpl requires cell wall hydrolysis, even though there is no evidence that the proteins are anchored to the cell wall. Pulse-chase labeling of de novo-made proteins indicates that the rate of synthesis of PC-PLC exceeds its rate of translocation across the bacterial cell wall. This slow rate of translocation across the cell wall is not a feature of all secreted L. monocytogenes proteins, because PI-PLC, a protein similar in size to PC-PLC, is secreted efficiently during the same time frame, and bacterially associated PI-PLC is extractable without digestion of the bacterial cell wall. Efficient translocation of PC-PLC across the bacterial cell wall occurs upon a decrease in host intracellular pH, which also coincides with the proteolytic activation of PC-PLC. These results suggest that translocation of PC-PLC across the bacterial cell wall is rate limiting, leading to its accumulation presumably at the membrane-cell wall interface. Colocalization of PC-PLC and Mpl at the cell wall-membrane interface would allow for rapid activation and translocation of PC-PLC across the bacterial cell wall upon formation of a nascent vacuole during cell-to-cell spread.
In gram-positive bacteria, secretion can be divided in two major steps: translocation across the cytoplasmic membrane and translocation across the cell wall. There are subsets of proteins that translocate across the cytoplasmic membrane, yet remain bacterially associated. Among those are transmembrane proteins, lipoproteins, and cell wall-anchored proteins that can either be covalently or noncovalently anchored to the cell wall (5, 32, 42). Each subset of proteins can be identified by a signature motif within their amino acid sequence. Analysis of the amino acid sequences of PC-PLC and Mpl does not suggest that the protein could remain bacterially associated after translocation across the cytoplasmic membrane. Accordingly, both proteins were found in the bacterial supernatant. However, both proteins were also found to be bacterially associated, and all of the bacterially associated PC-PLC and Mpl species comigrated with the secreted species by SDS-PAGE, indicating that they had been translocated across the cytoplasmic membrane and that the signal sequence had been processed. Moreover, cell wall hydrolysis was a prerequisite for their extraction even when tested with a sortase A-negative mutant. Interestingly, Tweten and Iandolo (45) made a similar observation for staphylococcal enterotoxin B (SEB), a secreted protein of S. aureus. SEB transiently associates with the cell wall and can only be extracted by treating bacteria with a cell wall hydrolase. Bacterially associated SEB is resistant to extracellular proteases and not extractable under high- and low-salt conditions. Conceivably, the mechanism regulating translocation of SEB and PC-PLC across the cell wall may reflect a common strategy for delayed secretion of virulence factors in gram-positive bacterial pathogens.
Translocation of proteins across the cell wall of gram-positive bacteria is the rate-limiting step for many secreted proteins. The cell wall of gram-positive bacteria is thick (25 to 50 nm) and highly cross-linked (1), and it is not well understood how proteins translocate across this dense mesh-like structure, although several factors are known to influence the efficacy of this process. Levels of d-alanylation of teichoic and lipoteichoic acids influence the net charge of cell wall and the associated levels of cations (19, 20, 43). A negatively charged cell wall can bind high concentrations of cations, assisting folding of some proteins (8, 24). In the absence of cations or d-alanylation, the cell wall is highly negatively charged, which may interfere with translocation of proteins (41). Chaperone-like proteins and thiol-disulfide oxidoreductases act on the trans side of cytoplasmic membrane, assisting protein folding (3, 22, 48), which is required for protein translocation across the cell wall and for protection against proteolysis. Perhaps protein folding is a factor influencing PC-PLC translocation across the cell wall. The propeptide of PC-PLC is likely to influence protein folding, and the proform of the enzyme may adopt a conformation that is not compatible with rapid protein translocation across the cell wall. Cleavage of PC-PLC propeptide could result in a conformational change that is more compatible with protein translocation across the bacterial cell wall. Accordingly, it is the processed form of PC-PLC that is primarily translocated upon a decrease in host intracellular pH. Alternatively, an increase in cell wall porosity may be necessary for translocation of PC-PLC across the cell wall. Bacterially associated PC-PLC is not detectable by immunofluorescence microscopy of intact bacteria, except at the site of bacterial division (27), suggesting that PC-PLC can diffuse more efficiently at this site, presumably because of an increase in cell wall porosity. The bacterial cell wall contains several autolysins implicated in cell wall growth and turnover (6, 21, 39). The environmental conditions that prevail in the host cytosol and in the vacuole may influence autolysin activities and, concomitantly, protein translocation across the cell wall.
Proteases are present at the membrane-cell wall interface of gram-positive bacteria and are responsible for degradation of unfolded or misfolded proteins that may accumulate in excessive amounts and potentially be harmful for bacterial survival (21, 40). In this study, pulse-labeling experiments indicated that the rate of synthesis of PC-PLC exceeds its rate of translocation across the cell wall. However, by Western immunoblotting, we observed that larger amounts of PC-PLC are present in the bacterial supernatant than in the bacterial lysate from bacteria grown in broth for 5 h. A possible explanation for this discrepancy would be that proteases control the total amount of PC-PLC that accumulate at the membrane-cell wall interface.
In conclusion, this study indicates that bacterial secretion of PC-PLC is limited by a slow rate of translocation across the cell wall leading to the accumulation of a pool of bacterially associated PC-PLC, presumably at the membrane-cell wall interface. Colocalization of PC-PLC and Mpl at the membrane-cell wall interface is consistent with Mpl being involved with the activation and rapid translocation of PC-PLC in response to a decrease in intracellular pH. An immediate bacterial response to an environmental change, such as vacuolar acidification, would increase the bacterial ability to survive in its host. Perhaps, regulation of protein translocation across the cell wall is a mechanism of great importance for the virulence of gram-positive bacterial pathogens.
Acknowledgments
We thank J. F. Miller from UCLA for the L. monocytogenes inlA deletion mutant strain and Darren Higgins and Angelika Grundling from Harvard Medical School for critical reading of the manuscript.
This work was supported by Public Health Service grant AI-42800 (to H.M.).
REFERENCES
- 1.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis, and turnover, p. 381-410. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 2.Bierne, H., S. K. Mazmanian, M. Trost, M. G. Pucciarelli, G. Liu, P. Dehoux, L. Jansch, F. G. Portillo, O. Schneewind, and P. Cossart. 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43:869-881. [DOI] [PubMed] [Google Scholar]
- 3.Bolhuis, A., G. Venema, W. J. Quax, S. Bron, and J. M. van Dijl. 1999. Functional analysis of paralogous thiol-disulfide oxidoreductases in Bacillus subtilis. J. Biol. Chem. 274:24531-24538. [DOI] [PubMed] [Google Scholar]
- 4.Brundage, R. A., G. A. Smith, A. Camilli, J. A. Theriot, and D. A. Portnoy. 1993. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl. Acad. Sci. USA 90:11890-11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabanes, D., P. Dehoux, O. Dussurget, L. Frangeul, and P. Cossart. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10:238-245. [DOI] [PubMed] [Google Scholar]
- 6.Calamita, H. G., and R. J. Doyle. 2002. Regulation of autolysins in teichuronic acid-containing Bacillus subtilis cells. Mol. Microbiol. 44:601-606. [DOI] [PubMed] [Google Scholar]
- 7.Camilli, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambert, R., and M. F. Petit-Glatron. 1999. Anionic polymers of Bacillus subtilis cell wall modulate the folding rate of secreted proteins. FEMS Microbiol. Lett. 179:43-47. [DOI] [PubMed] [Google Scholar]
- 9.Dancz, C. E., A. Haraga, D. A. Portnoy, and D. E. Higgins. 2002. Inducible control of virulence gene expression in Listeria monocytogenes: temporal requirement of listeriolysin O during intracellular infection. J. Bacteriol. 184:5935-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhar, G., K. F. Faull, and O. Schneewind. 2000. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry 39:3725-3733. [DOI] [PubMed] [Google Scholar]
- 11.Domann, E., M. Leimeister-Wächter, W. Goebel, and T. Chakraborty. 1991. Molecular cloning, sequencing, and identification of a metalloprotease gene from Listeria monocytogenes that is species specific and physically linked to the listeriolysin gene. Infect. Immun. 59:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freitag, N. E., L. Rong, and D. A. Portnoy. 1993. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 61:2537-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaillard, J.-L., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from Gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 14.Gaillard, J.-L., P. Berche, J. Mounier, S. Richard, and P. Sansonetti. 1987. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect. Immun. 55:2822-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gedde, M. M., D. E. Higgins, L. G. Tilney, and D. A. Portnoy. 2000. Role of listeriolysin O in cell-to-cell spread of Listeria monocytogenes. Infect. Immun. 68:999-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geoffroy, C., J. Raveneau, J.-L. Beretti, A. Lecroisey, J.-A. Vazquez-Boland, J. E. Alouf, and P. Berche. 1991. Purification and characterization of an extracellular 29-kilodalton phospholipase C from Listeria monocytogenes. Infect. Immun. 59:2382-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldfine, H., N. C. Johnston, and C. Knob. 1993. Nonspecific phospholipase C of Listeria monocytogenes: activity on phospholipids in Triton X-100-mixed micelles and in biological membranes. J. Bacteriol. 175:4298-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldfine, H., and C. Knob. 1992. Purification and characterization of Listeria monocytogenes phosphatidylinositol-specific phospholipase C. Infect. Immun. 60:4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heptinstall, S., A. R. Archibald, and J. Baddiley. 1970. Teichoic acids and membrane function in bacteria. Nature 225:519-521. [DOI] [PubMed] [Google Scholar]
- 20.Hyyrylainen, H. L., M. Vitikainen, J. Thwaite, H. Wu, M. Sarvas, C. R. Harwood, V. P. Kontinen, and K. Stephenson. 2000. d-Alanine substitution of teichoic acids as a modulator of protein folding and stability at the cytoplasmic membrane/cell wall interface of Bacillus subtilis. J. Biol. Chem. 275:26696-26703. [DOI] [PubMed] [Google Scholar]
- 21.Jolliffe, L. K., R. J. Doyle, and U. N. Streips. 1980. Extracellular proteases modify cell wall turnover in Bacillus subtilis. J. Bacteriol. 141:1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kontinen, V. P., P. Saris, and M. Sarvas. 1991. A gene (prsA) of Bacillus subtilis involved in a novel, late stage of protein export. Mol. Microbiol. 5:1273-1283. [DOI] [PubMed] [Google Scholar]
- 23.Leimeister-Wächter, M., C. Haffner, E. Domann, W. Goebel, and T. Chakraborty. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 87:8336-8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leloup, L., A. el Haddaoui, R. Chambert, and M. F. Petit-Glatron. 1997. Characterization of the rate-limiting step of the secretion of Bacillus subtilis alpha-amylase overproduced during the exponential phase of growth. Microbiology 143:3295-3303. [DOI] [PubMed] [Google Scholar]
- 25.Loessner, M. J., A. Schneider, and S. Scherer. 1996. Modified Listeria bacteriophage lysin genes (ply) allow efficient overexpression and one-step purification of biochemically active fusion proteins. Appl. Environ. Microbiol. 62:3057-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquis, H., V. Doshi, and D. A. Portnoy. 1995. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect. Immun. 63:4531-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marquis, H., H. Goldfine, and D. A. Portnoy. 1997. Proteolytic pathways of activation and degradation of a bacterial phospholipase C during intracellular infection by Listeria monocytogenes. J. Cell Biol. 137:1381-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marquis, H., and E. J. Hager. 2000. pH-regulated activation and release of a bacteria-associated phospholipase C during intracellular infection by Listeria monocytogenes. Mol. Microbiol. 35:289-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 30.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mengaud, J., C. Geoffroy, and P. Cossart. 1991. Identification of a new operon involved in Listeria monocytogenes virulence: its first gene encodes a protein homologous to bacterial metalloproteases. Infect. Immun. 59:1043-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niebuhr, K., T. Chakraborty, P. Köllner, and J. Wehland. 1993. Production of monoclonal antibodies to the phosphatidylcholine-specific phospholipase C of Listeria monocytogenes, a virulence factor for this species. Med. Microbiol. Lett. 2:9-16. [Google Scholar]
- 34.Portnoy, D. A., P. S. Jacks, and D. J. Hinrichs. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167:1459-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raveneau, J., C. Geoffroy, J.-L. Beretti, J.-L. Gaillard, J. E. Alouf, and P. Berche. 1992. Reduced virulence of a Listeria monocytogenes phospholipase-deficient mutant obtained by transposon insertion into the zinc metalloprotease gene. Infect. Immun. 60:916-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ripio, M.-T., K. Brehm, M. Lara, M. Suarez, and J. A. Vazquez-Boland. 1997. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J. Bacteriol. 179:7174-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 38.Smith, G. A., H. Marquis, S. Jones, N. C. Johnston, D. A. Portnoy, and H. Goldfine. 1995. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 63:4231-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 40.Stephenson, K., and C. R. Harwood. 1998. Influence of a cell-wall-associated protease on production of α-amylase by Bacillus subtilis. Appl. Environ. Microbiol. 64:2875-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephenson, K., C. L. Jensen, S. T. Jorgensen, J. H. Lakey, and C. R. Harwood. 2000. The influence of secretory-protein charge on late stages of secretion from the Gram-positive bacterium Bacillus subtilis. Biochem. J. 350:31-39. [PMC free article] [PubMed] [Google Scholar]
- 42.Sutcliffe, I. C., and R. R. B. Russell. 1995. Lipoproteins of gram-positive bacteria. J. Bacteriol. 177:1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thwaite, J. E., L. W. J. Baillie, N. M. Carter, K. Stephenson, M. Rees, C. R. Harwood, and P. T. Emmerson. 2002. Optimization of the cell wall microenvironment allows increased production of recombinant Bacillus anthracis protective antigen from B. subtilis. Appl. Environ. Microbiol. 68:227-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tilney, L. G., and D. A. Portnoy. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109:1597-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tweten, R. K., and J. J. Iandolo. 1983. Transport and processing of staphylococcal enterotoxin B. J. Bacteriol. 153:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Wart, H. E., and H. Birkedal-Hansen. 1990. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl. Acad. Sci. USA 87:5578-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vazquez-Boland, J.-A., C. Kocks, S. Dramsi, H. Ohayon, C. Geoffroy, J. Mengaud, and P. Cossart. 1992. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect. Immun. 60:219-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vitikainen, M., T. Pummi, U. Airaksinen, E. Wahlstrom, H. Wu, M. Sarvas, and V. P. Kontinen. 2001. Quantitation of the capacity of the secretion apparatus and requirement for PrsA in growth and secretion of α-amylase in Bacillus subtilis. J. Bacteriol. 183:1881-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]