Abstract
The multicellular developmental cycle of Myxococcus xanthus requires large-scale changes in gene transcription, and recent findings indicate that NtrC-like activators play a prominent role in regulating these changes. In this study, we made insertions in 28 uncharacterized ntrC-like activator (nla) genes and found that eight of these insertions cause developmental defects. Hence, these results are consistent with the idea that M. xanthus uses a series of different NtrC-like activators during fruiting body development. Four of the eight developmental mutants we identified have motility defects. The nla1, nla19, and nla23 mutants show S-motility defects, while the nla24 mutant shows defects in both S-motility and A-motility. During development, aggregation of the nla1, nla19, and nla23 mutants is delayed slightly and the nla24 mutant shows no signs of aggregation or sporulation. The nla4, nla6, nla18, and nla28 mutants have no appreciable loss in motility, but they fail to aggregate and to sporulate normally. The nla18 mutant belongs to a special class of developmental mutants whose defects can be rescued when they are codeveloped with wild-type cells, suggesting that nla18 fails to produce a cell-cell signal required for development. The three remaining activator mutants, nla4, nla6, and nla28, appear to have complex developmental phenotypes that include deficiencies in cell-cell developmental signals.
The social lifestyle of the gram-negative soil bacterium Myxococcus xanthus is rather unusual in the prokaryotic world. In nature, large groups of M. xanthus cells feed on prey bacteria to obtain their nutrients. When this nutrient supply is depleted, M. xanthus cells initiate a complex developmental program that culminates in the formation of a multicellular fruiting body. Once the fruiting body is molded into its final form, individual rod-shaped cells within the fruiting body differentiate into a specialized and dormant cell (a spherical spore) that is resistant to many forms of environmental stress (for reviews, see references 8 and 53).
All of the morphological events that occur during M. xanthus development are accompanied by large-scale changes in gene expression, and the products of many of these genes are absolutely required for normal development (14, 35, 36, 43, 47, 57, 60). The temporal and spatial expression of developmental genes is coordinated by a series of cell-cell signals; each signal is required for the expression of different sets of genes (7, 21, 26, 32, 34, 39, 41, 42). For example, A-signal is required for gene expression during all stages of M. xanthus development, while C-signal is only required for gene expression during the later stages of development.
Recent findings indicate that M. xanthus uses σ54-like promoters to drive expression of many developmentally regulated genes; the hallmarks of σ54 promoters are recognition sequences located around −12 and −24 bp upstream of the transcriptional start site (13, 15, 20, 30, 50, 64, 66; J. S. Jakobsen, E. Licking, and D. Kaiser, personal communication). Moreover, expression studies suggest that σ54 promoters are activated during all stages of development, although they appear to be used more frequently in the early stages of development than in the later stages (for late promoters, see references 4, 11, and 12). In addition to being used frequently in developing cells, the products of many genes under the control of σ54 promoters play critical roles in M. xanthus development (13, 14, 16, 17, 18, 36). Taken together, these results indicate that σ54 promoters play an important role in modulating gene expression during M. xanthus development.
Transcription from σ54 promoter elements has been studied in a variety of bacterial systems in recent years (46, 66). Transcription is dependent on the σ54 protein, which directs RNA polymerase to recognition sequences in the −12- and −24-bp regions of the σ54 promoter. Expression also requires a NtrC-like activator, which is a DNA binding protein that allows σ54-loaded RNA polymerase to form a transcriptionally active, open promoter complex. Each NtrC activator has a specific DNA recognition sequence (called an enhancer) that is typically located 70 to 150 bp upstream of the −12- and −24-bp regions of the σ54 promoters. Presumably, the enhancer binding sequence allows each NtrC-like activator to specify which set of genes will be transcribed by σ54-RNA polymerase. In many cases, NtrC-like proteins function as response regulators in two-component regulatory circuits. Hence, the activity of an NtrC-like protein is often modulated via phosphorylation by a histidine kinase partner, which is a sensor protein that detects changes in the internal or external environment of a cell (see reference 48).
The preponderance of developmental promoters with σ54 hallmarks led to the suggestion that NtrC-like activators are key components of the transcriptional machinery that coordinates gene expression during M. xanthus development (16, 30). In the past 5 years, four NtrC-like activators (ActB, MrpB, Mxa287, and PilR) that are required for normal development have been identified and characterized (16, 17, 18, 19, 57, 58, 64). Two systems control M. xanthus gliding motility: the adventurous (A) system, which helps control individual cell movement, and the social (S) system, which helps control movement in multicellular groups (24, 25). The product of the pilR gene has been implicated in M. xanthus S-motility, which is required for normal aggregation into multicellular fruiting bodies. Cells carrying mutations in actB, mxa287, or mrpB have normal motility, but they are still unable to complete development. Based on the expression and morphological data, it appears that Mxa287 and MrpB are required prior to aggregation while ActB is required as aggregation begins. Recent work also suggests that ActB plays a direct or indirect role in the production of C-signal.
The work presented here focuses on NtrC-like transcriptional activators, proteins that appear to be important components in the regulatory machinery that controls the M. xanthus developmental cycle. Using the M. xanthus genome sequence, we have identified 37 genes that are likely to code for members of the NtrC family of proteins. In this study, we systematically inactivated the 28 uncharacterized activator (nla) genes. Preliminary analysis of the 28 nla mutants showed that 8 are defective for development. Four of these nla mutants have been classified as motility mutants, while the remaining four nla mutants have relatively normal motility, but they are defective for aggregation and sporulation. These results are consistent with the idea that a series of NtrC-like activators are used during fruiting body development in M. xanthus.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Plasmids were propagated in Escherichia coli strain Top10F (Invitrogen). M. xanthus strain DK1622 (27) is wild type for fruiting body development, sporulation, A- and S-motility, and growth. M. xanthus strains AG301-AG328 are derivatives of DK1622. Each of these strains carries a pNBC plasmid insertion in an ntrC-like activator (nla) gene. M. xanthus strains AG367-AG374 are derivatives of DK1622. Each of these strains carries a pNBC plasmid insertion immediately downstream of an nla gene. Plasmid pCR2.1-TOPO (Invitrogen) was used for cloning PCR-generated fragments of nla genes. The pCR2.1-TOPO plasmids carrying nla gene fragments were designated pNBC1-pNBC28, and these plasmids were used to create strains AG301-AG328. The pCR2.1-TOPO plasmid was also used to clone PCR fragments carrying the 3′ ends of nla genes and 10 to 600 bp of downstream DNA. The pCR2.1-TOPO plasmids carrying these PCR fragments were designated pNBC33-pNBC40, and these plasmids were used to create strains AG367-AG374. Strains DK1253 (25) and DK1218 (25) are DK1622 derivatives that are wild type for growth, but they have a defect in S-motility and A-motility, respectively. DK2161 (25) is a derivative of strain DK1622 that is wild type for growth, but it has defects in S- and A-motility. Strains AG329-AG332 are derivatives of DK1253 that carry pNBC plasmid insertions in nla genes, and strains AG333-AG336 are derivatives of DK1218 that carry pNBC plasmid insertions in nla genes. Strains AG341-AG344 are derivatives of DK1622 that carry insertions of pNBC29-pNBC32. Plasmids pNBC29-pNBC32 were constructed by introducing activator gene fragments into pSWU22, which confers resistance to oxytetracycline (S. S. Wu and D. Kaiser, unpublished results). Strains DK5057 (38) and DK5208 (34) are derivatives of DK1622 that carry the Tn5-132 Ω4560 asgA476 and Tn5-132::csgA mutations, respectively.
Identifying and cloning regions of the M. xanthus nla loci.
Using the genome sequence (95% complete) provided by the Monsanto Company, R. D. Welch, J. S. Jakobsen, and D. Kaiser (personal communication) have identified approximately 7,000 putative genes in the M. xanthus chromosome. The results of BLAST searches indicate that 37 of these M. xanthus genes are likely to encode NtrC-like transcriptional activators; the highly conserved central domain that is characteristic of members of the NtrC family of proteins was found in each case (46). Of the 37 ntrC-like activator genes that were identified in the M. xanthus genome sequence, 28 were uncharacterized. Internal fragments of each of the 28 activator genes were generated using PCR as described in the Invitrogen TOPO TA cloning kit (thermostable DNA polymerase and gene-specific primers were used in the PCRs). The sequences of the primers used are available elsewhere (http://www.wsu.edu/∼aggarza/nla-primers.htm). Each PCR fragment was cloned (using the procedure specified by the manufacturer [Invitrogen]) into a pCR2.1-TOPO plasmid, generating plasmids pNBC1-pNBC28. Plasmids pNBC33-pNBC40 were generated using a similar strategy. The nla4, nla6, nla18, and nla28 fragments were also introduced into pSWU22 (Wu and Kaiser, unpublished), generating plasmids pNBC29-pNBC32. The compositions of pNBC1-pNBC40 were confirmed by digesting plasmid DNA with the appropriate endonucleases.
Plasmid transfer to M. xanthus.
Plasmids containing fragments of nla loci (pNBC1-pNBC40) were electroporated into M. xanthus cells (using the technique of Plamann et al. [49]). Following electroporation, cells were placed into flasks containing 1.5 ml of CTTYE (see below) and incubated at 32°C for 12 to 24 h with vigorous agitation. Aliquots (500 μl) of these cultures were added to 5.0 ml of CTT soft agar and poured onto CTTYE plates containing kanamycin or tetracycline. Chromosomal DNA was isolated from Kanr and Tetr colonies and used for Southern blot analysis (51). Briefly, chromosomal DNA from each Kanr or Tetr colony was digested with a restriction enzyme that cuts once within the multicloning site of the pNBC plasmid and a second restriction enzyme that cuts either upstream or downstream of the plasmid insertion site. Two probes were used for Southern blots, pCR2.1-TOPO (Kanr) and pSWU22 (Tetr). Transformants that carry a single copy of a pNBC plasmid integrated into the target nla locus by homologous recombination were identified by a single band of the appropriate size. After Southern blot analysis was used to confirm that the transformants carried the appropriate insertions, they were scored for development, motility, and auxotrophy as needed.
Media used for growth, motility assays, and developmental assays.
M. xanthus strains were grown at 32°C in CTTYE broth containing 1.0% Casitone (Difco Laboratories), 0.5% yeast extract (Difco Laboratories), 10.0 mM Tris-HCl (pH 8.0), 1.0 mM KH2PO4, and 8.0 mM MgSO4 or on plates containing CTTYE broth and 1.5% Difco Bacto Agar. Motility of M. xanthus strains was assayed at 32°C on CTTYE plates containing 1.5 or 0.4% Difco Bacto Agar. CTTYE broth and plates were supplemented with 40 μg of kanamycin sulfate (Sigma)/ml or 10 μg of oxytetracycline/ml as needed. CTT soft agar contains 1.0% Casitone (Difco Laboratories), 10.0 mM Tris-HCl (pH 8.0), 1.0 mM KH2PO4, 8.0 mM MgSO4, and 0.7% Difco Bacto Agar. E. coli Top10F was grown at 37°C in Luria broth (LB) containing 1.0% tryptone (Difco), 0.5% yeast extract (Difco), and 0.5% NaCl or in plates containing LB and 1.5% Difco Bacto Agar. LB and LB plates were supplemented with 40 μg of kanamycial sulfate (Sigma)/ml or 10 μg of oxytetracycline/ml as needed. Fruiting body development was carried out at 32°C on plates containing TPM buffer (10.0 mM Tris-HCl [pH 8.0], 1.0 mM KH2PO4, and 8.0 mM MgSO4) and 1.5% Difco Bacto Agar. Sporulation in CTTYE broth was induced by adding glycerol to achieve a final concentration of 0.5 M.
Nutritional requirements.
To assay M. xanthus strains carrying nla gene insertions for auxotrophy, wild-type (DK1622) and mutant cells were inoculated onto plates containing A1 minimal medium (5) and agarose and plates were incubated at 32°C for 5 to 10 days. Following the incubation period, the growth of activator insertion mutants was compared to the growth of parental strain DK1622.
M. xanthus development.
M. xanthus strains were inoculated into flasks containing CTTYE broth, and the cultures were incubated at 32°C with vigorous swirling. After each culture reached a density of 5 × 108 cells/ml, the cells were pelleted, the supernatant was removed, and the cells were resuspended in TPM buffer to a density of 5 × 109 cells/ml. Aliquots (20 μl) of this cell suspension were spotted onto TPM agar plates and incubated at 32°C. The progress of fruiting body development was monitored visually using a Nikon Eclipse TE 2000-U inverted phase-contrast microscope. Images were captured with a digital camera (Photometrics COOLSNAP HQ) and analyzed using Metavue software 5.0 (Universal Imaging Corporation).
To determine the sporulation efficiency of each M. xanthus strain, developing cells were harvested from TPM agar plates after 5 days as described previously (35). The cells were resuspended in 400 μl of TPM buffer, the cell suspension was sonicated, and the sonicated cells were incubated at 50°C for 2 h. The number of heat- and sonication-resistant spores that germinated into colonies was determined as described by Thöny-Meyer and Kaiser (60). Glycerol sporulation was induced and analyzed as described by Licking et al. (43).
For extracellular complementation of nla mutants by wild-type cells, nla cells were grown in CTTYE broth, concentrated to a density of 5 × 109 cells/ml, and mixed at a 1:1 ratio with wild-type cells and 20-μl aliquots of the cell mixtures were spotted onto TPM agar plates. For extracellular complementation of asg or csg cells by nla mutants, cells were also mixed at a ratio of 1:1. The sporulation efficiency for each strain in the mixed-culture fruiting body was determined as described above. Colonies derived from activator insertion mutants were distinguished from the wild-type, asg, or csg colonies on the basis of their resistance to kanamycin or oxytetracycline.
Motility assays.
The swarm expansion assay described by Kaiser and Crosby (29) and Shi and Zusman (52) was used to examine the motility of wild-type M. xanthus cells and cells carrying nla gene insertions. Cells were grown to a density of 5 × 108 cells/ml in CTTYE broth and pelleted by centrifugation, the supernatant was removed, and the cells were resuspended in CTTYE broth to a density of 5 × 109 cells/ml. Aliquots (3 μl) of the concentrated cells were spotted onto CTTYE plates containing 1.5 or 0.4% agar, the spots were allowed to dry, and the plates were placed at 32°C. After the plates were incubated for 3 to 5 days, five swarms of each strain were measured and their mean diameter was normalized to the mean diameter of five swarms formed by wild-type strain DK1622. Activator (nla) mutants whose mean swarm diameter on 1.5 or 0.4% agar plates was less than 80.0% of that of the wild-type strain were classified as motility mutants; only this class of nla mutants had noticeable defects in their colony edge morphologies, which is characteristic of M. xanthus motility mutants. During the 3- to 5-day incubation period on CTTYE plates, the edges of swarms were monitored using a Nikon Eclipse TE 2000-U inverted phase-contrast microscope. Images were captured and analyzed as described above for developmental assays.
Nucleotide sequence accession numbers.
The DNA sequences of the 28 nla genes analyzed in this study have been deposited in GenBank. The accession numbers are AY337488-AY337515.
RESULTS
Making insertions in ntrC-like activator (nla) genes in M. xanthus.
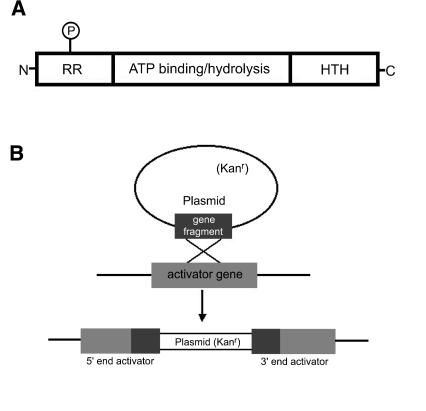
Members of the NtrC family of proteins function as transcriptional activators, modulating gene expression from σ54 promoter elements. Typically, NtrC-like activators contain three functional domains: an N-terminal region that modulates their activity in response to regulatory signals, a C-terminal DNA binding domain that allows each activator to recognize specific DNA sequences, and a conserved central domain that couples the energy of ATP hydrolysis to σ54-RNA polymerase activation (Fig. 1A).
FIG. 1.
(A) Common structure of NtrC-like activators. The N-terminal domain is the least conserved of the domains indicated, and its length ranges from about 12 to 400 amino acids. Many activators serve as response regulators in two-component regulatory systems, as indicated by the letters RR. In the two-component paradigm, a histidine kinase sensor modulates the activity of a response regulator partner by phosphorylation of a conserved aspartate residue (P). The C-terminal domain of activators is about 65 to 130 amino acids. This region contains a helix-turn-helix (HTH) motif, which is characteristic of many DNA binding proteins. The central domain of activators is the most highly conserved region, and it consists of approximately 240 amino acids. This central domain is required for ATP binding and hydrolysis, which helps σ54-bound RNA polymerase become transcriptionally active. The central region of the activators may also be involved in contacting σ54-RNA polymerase. Data for this figure are taken from work by Morett and Segovia (46) and Xu and Hoover (66). (B) Disruption of ntrC-like activator (nla) genes by homologous recombination. Internal fragments of activator genes were cloned into a plasmid vector that confers resistance to kanamycin. After electroporation of the plasmid clones into wild-type M. xanthus cells, a single homologous crossover produces a tandem duplication of the internal fragment and incorporation of the vector into the chromosomal copy of the gene. The likely result of the crossover is an inactivated (knockout) copy of the activator gene.
The distinguishing characteristic of the NtrC-like proteins is the highly conserved central domain (46). Hence, we used this conserved domain to probe the M. xanthus chromosomal sequence (Monsanto Company) for genes that encode NtrC-like activators. Using this strategy, we identified 37 M. xanthus genes that are likely to encode NtrC-like activators. Twenty-eight of the putative ntrC-like genes that we identified (designated nla1-nla28 for ntrC-like activator genes 1 to 28) were uncharacterized. Based on the DNA sequence information, it appears that each of the 28 Nla proteins contains the three characteristic modules of NtrC-like activators. In addition, the deduced amino sequence in the N-terminal region of the Nla proteins suggests that they may function as response regulators in two-component signal transduction circuits.
To identify Nla proteins that play roles in M. xanthus development, we systematically inactivated each of the 28 nla genes. To do this, a 300- to 600-bp partial fragment of the conserved central region of each nla gene was generated using PCR, the PCR fragments were cloned into a plasmid that confers kanamycin resistance (Kanr), and plasmid DNA was electroporated into wild-type M. xanthus cells. Although the plasmids we introduced into M. xanthus are incapable of autonomous replication, they can integrate into the chromosomal copies of nla genes by homologous recombination of the cloned PCR fragments. A single crossover yields Kanr electroporants with two incomplete copies of the nla gene separated by vector DNA (Fig. 1B). Moreover, each plasmid insertion creates two truncated copies of the conserved central region of the target nla gene, a region that is essential for activator protein function. Hence, the plasmid insertions are likely to inactivate the nla genes and their corresponding protein products. Phenotypic characterization of nla1-nla28 mutants is described below.
Development.
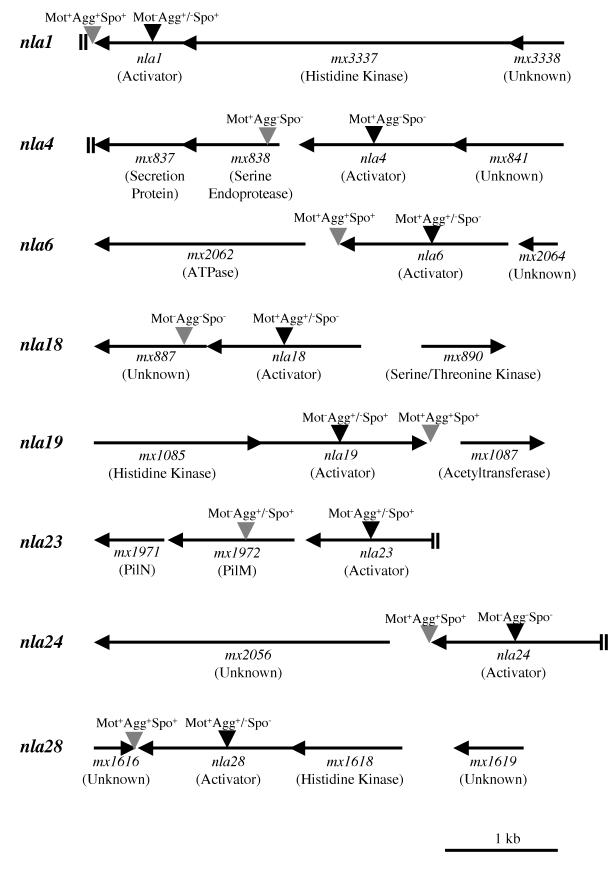
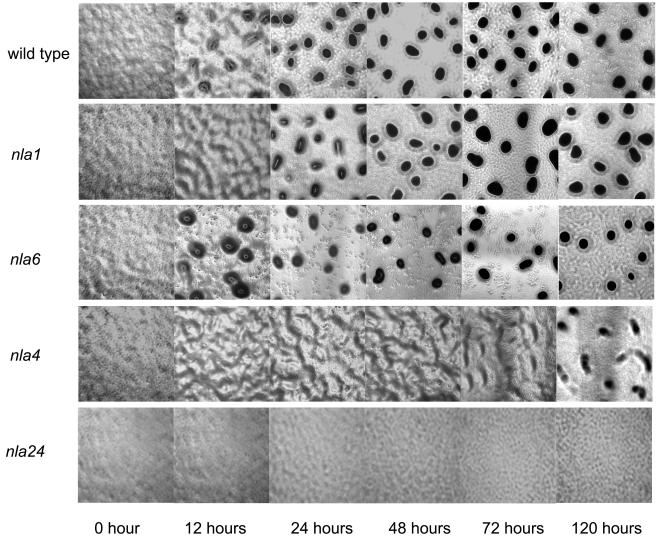
To determine which of the 28 nla insertions produce defects in fruiting body development, nla mutant cells and wild-type DK1622 cells were placed on TPM starvation agar. The large majority of nla mutants (20 out of 28) showed no obvious defects in aggregation or sporulation efficiency relative to wild-type M. xanthus cells. In contrast, eight of the nla mutants showed developmental defects that were detectable using our assay conditions (Table 1 and Fig. 2). Based on their developmental phenotypes, these nla mutants were grouped into four categories (see Fig. 3 for representative examples). Aggregation of the nla1, nla19, and nla23 group of activator mutants was delayed 12 to 24 h compared to that of wild-type cells, but after this short delay, the progress of aggregation was similar to that of the wild type. When we performed spore assays on the nla1, nla19, and nla23 mutants, we found sporulation efficiencies similar to those of wild-type DK1622 cells. The nla6, nla18, and nla28 group of mutants also showed a short delay in aggregation. However, the sporulation efficiencies of nla28 and nla6 mutants were reduced about 50- to 500-fold compared to that of the wild type, while the nla18 mutant produced no detectable spores. Aggregation of the nla4 mutant cells appeared to be delayed and incomplete compared to that of wild-type M. xanthus cells. During the first 12 to 72 h, nla4 cells showed the early signs of aggregation; they formed branch-like elevations that rest on top of an otherwise continuous mat of cells. After 5 days, the nla4 mutant formed loose aggregates, but these aggregates failed to compact after an additional 3 days of development (data not shown). In addition to its aggregation defect, the sporulation efficiency of the nla4 mutant was reduced about 500-fold compared to that of the wild type. Of the eight nla mutants that failed to develop normally, nla24 appears to have the most severe aggregation defect. After 5 days of development on TPM starvation agar, nla24 mutant cells showed no signs of aggregation, and no improvement was observed when this mutant was given an additional 3 days to develop (data not shown). When we examined the sporulation efficiency of the nla24 mutant, no spores were detected.
TABLE 1.
Developmental phenotypes of wild-type and nla mutant strainsa
| Strain | Aggre- gationb | Fruiting body spores (% of wild type)c | Glycerol spores (% of wild type)d |
|---|---|---|---|
| DK1622 (wild type) | + | 100.0 ± 9.5 | 100.0 ± 16.2 |
| AG301 (nla1) | +/− | 89.4 ± 9.6 | 83.5 ± 3.8 |
| AG304 (nla4) | − | 0.2 ± 0.2 | 2.4 ± 2.3 |
| AG306 (nla6) | +/− | 0.2 ± 0.2 | 5.1 ± 0.7 |
| AG318 (nla18) | +/− | <0.0002 | 0.1 ± 0.1 |
| AG319 (nla19) | +/− | 107.7 ± 14.6 | 78.8 ± 5.8 |
| AG323 (nla23) | +/− | 129.4 ± 17.6 | 122.0 ± 9.2 |
| AG324 (nla24) | − | <0.0002 | 8.9 ± 2.7 |
| AG328 (nla28) | +/− | 2.1 ± 1.5 | 76.8 ± 4.5 |
Cells were placed on TPM agar and allowed to develop for 5 days. Development was monitored visually using phase-contrast microscopy. Spore assays were performed three times for each strain. The mean values (± standard deviations) for the spore assays are shown as percentages of DK 1622 (wild type). The number of spores produced by wild-type cells ranged from 3 × 106 to 6 × 106.
+, produced normal-looking fruiting bodies; −, failed to produce normal-looking fruiting bodies; +/−, produced normal-looking fruiting bodies but aggregation was delayed.
Values were determined by transferring sonication- and heat-resistant spores to CTTYE agar plates, incubating the plates for 5 days, and counting the number of colonies that arose from the spores.
Values were determined by counting the number of sonication- and heat-resistant spores by using a Petroff-Hausser chamber and phase-contrast microscopy.
FIG.2.
Physical map of nla loci implicated in development. Gene designations were taken from the work of R. D. Welch, J. S. Jakobsen, and D. Kaiser (personal communication). The potential functions of some of the gene products are indicated in parentheses below the gene designations. “Unknown” indicates that in BLAST searches, no potential function for a gene product was obtained. Black inverted triangles represent insertions within nla genes, and gray inverted triangles represent insertions downstream of nla genes. The motility (Mot), aggregation (Agg), and sporulation (Spo) phenotypes produced by each insertion are indicated above the insertions. The data were taken from the results shown in Table 1, Table 2, and Table 4. Double bars (||) at the end of a line indicate that the upstream or downstream sequence was not available.
FIG. 3.
Behavior of representative nla mutants during development on TPM agar plates. Cells were spotted on TPM starvation agar and monitored visually as described in Materials and Methods. Development of the indicated nla mutants and wild-type strain DK1622 was observed for 5 days using a phase-contrast microscope. The aggregation phenotypes of nla19 and nla23 mutant cells are similar to those of nla1 mutant cells, and the aggregation phenotypes of nla18 and nla28 mutant cells are similar those of nla6 mutant cells. Photographs were taken after 12, 24, 48, 72, and 120 h using a total magnification of ×40.
Normally, M. xanthus sporulation takes place inside a multicellular fruiting body. However, Dworkin and Gibson (10) showed that rod-shaped vegetative cells undergo a rapid and synchronous conversion into stress-resistant, spherical spores when glycerol is added to a nutrient broth culture. Presumably, glycerol-induced sporulation bypasses many of the early events required for production of fruiting body spores (e.g., starvation and aggregation) by activating the sporulation program directly. Therefore, many mutants that are blocked in the early stages of fruiting body development are capable of forming stress-resistant glycerol spores (17, 41). To determine whether our collection of eight nla developmental mutants can form glycerol spores, glycerol was added to wild-type and nla mutant cells growing in CTTYE nutrient broth. The efficiency of glycerol-induced sporulation for each of the nla mutants relative to that of wild-type M. xanthus cells is shown in Table 1. The efficiencies of glycerol-induced sporulation in the nla1, nla19, and nla 23 mutants, which produced normal levels of fruiting body spores, were similar to that in wild-type cells. Similarly, four of the five nla mutants that showed defects in fruiting body sporulation also showed defects in glycerol-induced sporulation, although the defects in glycerol-induced sporulation were less severe. The nla28 mutant was the one case in which defects in fruiting body sporulation did not translate into defects in glycerol-induced sporulation.
Auxotrophy.
Previous studies have shown that auxotrophic mutations can produce defects in M. xanthus development (for example, see reference 36). Given that the nla strains were originally isolated and propagated under nutrient-rich conditions, we wanted to determine whether any of these mutants are auxotrophs by examining their growth on A1 minimal plates (35). Wild-type strain DK1622 showed confluent growth on A1 minimal plates after 7 days of incubation at 32°C. Similarly, all of the nla mutants, including mutants with developmental defects, showed confluent growth on A1 minimal plates after 7 to 10 days of incubation at 32°C. Based on these results, it appears that none of the 28 nla mutants we identified are auxotrophs.
Motility.
M. xanthus cells must be motile to aggregate into a multicellular fruiting body, and mutants that are defective for motility display a variety of developmental phenotypes (23). Two motility systems control M. xanthus swarming (or gliding) motility on a solid surface, the A and S systems (24, 25). Mutants that are defective for either A-motility (A− S+ cells) or S-motility (A+ S− cells) swarm at a reduced rate, while mutants that are defective for both types of motility (A− S− cells) have a nonswarming phenotype.
The motility of the eight nla mutants that are defective for development was examined using swarm expansion assays (29). For these assays, nla cells and wild-type cells were grown in CTTYE nutrient broth to a density of 5 × 108 cells/ml, the cells were concentrated 10-fold, and 3-μl aliquots were placed on CTTYE plates containing 0.4 or 1.5% agar. The rationale for using 1.5 and 0.4% agar plates for our swarm expansion assays is based on the findings of Shi and Zusman (52): A-motility appears to be favored on relatively firm and dry surfaces (1.5% agar plates), while S-motility appears to be favored on soft and wet surfaces (0.4% agar plates). Hence, a particular motility defect may be more evident with one agar concentration than with the other. After the plates were incubated for 3 to 5 days at 32°C, the diameters of nla mutant colonies were compared to the diameters of wild-type colonies (Table 2). To be classified a motility mutant, the mean diameters of nla colonies on either 0.4 or 1.5% agar had to be less than 80% of those of wild-type colonies (see Materials and Methods for the rationale). Four of the nla mutants that we tested met this criterion. The nla24 mutant colonies were significantly smaller than wild-type colonies on both 1.5 and 0.4% agar plates, with mean diameters ranging from about 34 to 41% of those of the wild type. For comparison, the mean diameters of colonies from the A− S− mutant DK2161 (nonswarming) ranged from about 37 to 38% of that of the wild type under our assay conditions (Table 2). Moreover, when we used phase-contrast microscopy to examine nla24 mutant colonies, their edges appeared to be smooth (Fig. 4), a characteristic associated with nonswarming A− S− mutants. Taken together, these findings suggest that the nla24 mutant is defective for both A- and S-motility, which is consistent with the finding that mutant shows no signs of aggregation during development. The nla23 mutant also produced smaller colonies on both agar surfaces; the mean diameters of nla23 mutant colonies on 0.4 and 1.5% agar plates were about 51 and 76% of those of wild-type colonies, respectively. The edges of nla23 mutant colonies seemed to be lacking S-motile flares, but they were not smooth, indicating that the nla23 mutant retains some motility (Fig. 4). Finally, the mean diameters of nla1 and nla19 mutant colonies were similar to the wild type on 1.5% agar plates, which favor A-motility, but their mean colony diameters were significantly smaller than the wild type (approximately 65 to 69%) on 0.4% agar plates, which favor S-motility. Perhaps the developmental delay observed for the nla1, nla19, and nla23 mutants is due to their reduced ability to swarm.
TABLE 2.
Swarm diameters of wild-type and nla strains on 0.4 and 1.5% agara
| Strain | Mean swarm diameter (% of wild type)
|
|
|---|---|---|
| Soft agar (0.4%) | Hard agar (1.5%) | |
| DK 1622 (wild type) | 100 ± 3 | 100 ± 2 |
| DK 2161 (A− S−) | 38 ± 1 | 37 ± 1 |
| AG301 (nla1) | 69 ± 5 | 92 ± 4 |
| AG304 (nla4) | 83 ± 5 | 86 ± 2 |
| AG306 (nla6) | 100 ± 1 | 108 ± 3 |
| AG318 (nla18) | 86 ± 2 | 88 ± 5 |
| AG319 (nla19) | 65 ± 2 | 99 ± 3 |
| AG323 (nla23) | 51 ± 2 | 76 ± 2 |
| AG324 (nla24) | 41 ± 1 | 34 ± 1 |
| AG328 (nla28) | 90 ± 2 | 94 ± 4 |
Cells were grown to a density of 5 × 108 cells/ml in CTTYE broth and concentrated 10-fold. A total of 3 μl of concentrated cells was spotted on CTTYE containing 0.4 or 1.5% agar, and the cells were incubated for 5 days at 32°C. The mean diameter (± standard deviation) of five swarms produced by each nla strain was determined and normalized to the mean diameter of five swarms produced by wild-type strain DK1622.
FIG. 4.
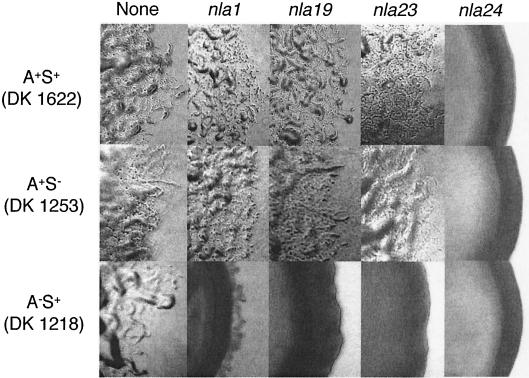
Colony edge morphologies produced by nla insertions. The nla1, nla19, nla23, and nla24 insertions were transferred into A+ S+ (DK1622), A− S+ (DK1218), and A+ S− (DK1253) backgrounds, and colony edges were observed using phase-contrast microscopy (×40 magnification). Photographs were taken after 5 days on CTTYE plates.
To determine whether the nla1, nla19, and nla23 insertions cause defects in either the A- or S-motility system, they were introduced into A− S+ (DK1218) and A+ S− (DK1253) mutant strains by electroporation and the colony edges of the resulting double mutants were examined using phase-contrast microscopy (Fig. 4). When the nla1, nla19, and nla23 insertion mutations were introduced into the DK1253 (A+ S−) recipient, colony edge morphologies were similar to the morphologies of wild-type cells carrying the same insertion mutations. However, when the nla1, nla19, and nla23 insertion mutations were introduced into the DK1218 (A− S+) background, colony edges were smooth, similar to the colony edge of a nonswarming A− S− double mutant. Based on these findings, we believe that the nla1, nla19, and nla23 insertions cause defects in S-motility. It is important that after extended incubation, DK1218 cells that carried the nla1 insertion produced a few S-motile flares. Our interpretation of this result is that nla1 is a leaky S-motility mutation. The nla24 insertion, which produced a nonswarming phenotype in an otherwise wild-type background, produced nonswarming phenotypes when introduced into strains DK1253 and DK1218 (Fig. 4). These findings are consistent with the idea that nla24 causes a defect in both the A- and S-motility systems.
Cell-cell signaling.
Several cell-cell signaling mutants have been isolated in previous analyses of the M. xanthus developmental cycle (6, 7, 14, 21, 41). Although these mutants are unable to produce a particular cell-cell signal required for development, their ability to respond to the signal is fully intact. Hence, their developmental defects can be transiently overcome when they are mixed with wild-type M. xanthus cells; wild-type cells provide the signal that the mutants lack (no genetic exchange occurs). Of the cell-cell signals identified in these studies, A- and C-signals have been studied the most extensively. A-signal is a diffusible cell density signal that is required prior to the onset of aggregation, while C-signal is a contact-stimulated signal that is required early in the aggregation phase of development (31, 32, 33, 34, 39, 40).
In recent studies, the NtrC-like activators ActB and MrpB have been implicated in the production of cell-cell signals required for normal development in M. xanthus (17, 18, 58). In light of these findings, we decided to examine A- and C-signal production in the nla mutants that are defective for development (nla mutants classified as motility mutants were excluded). In these experiments, nla cells were mixed with the A-signal-deficient mutant DK5057 (asgA) and the C-signal-deficient mutant DK5208 (csgA). Subsequently, the cell mixtures were codeveloped on TPM starvation agar for 5 days and the sporulation efficiencies of the A-signal-deficient and the C-signal-deficient mutants were determined (Table 3). For comparison, the sporulation efficiencies of these signaling mutants after 5 days of codevelopment with wild-type DK1622 cells are also shown in Table 3. When they were mixed with wild-type cells, sporulation in the A-signal-deficient and C-signal-deficient mutants was almost completely rescued. In contrast, sporulation in these signaling mutants was improved when they were mixed with nla4, nla6, and nla28 cells but the rescue was incomplete. Sporulation in C-signal-deficient cells was restored to wild-type levels when they were mixed and codeveloped with nla18 mutant cells. However, nla18 mutant cells were unable to restore sporulation in the A-signal-deficient mutant as efficiently as wild-type cells. Taken together, these results suggest that C-signal and/or A-signal production in the nla4, nla6, nla18, and nla28 mutants is reduced.
TABLE 3.
Extracellular complementation of asgA, csgA, and nla mutantsa
| Strain mixture
|
Sporulation of test strain (% of wild type) | |
|---|---|---|
| Test strain | Coculture strain | |
| DK1622 (wild type) | 100 ± 12.2 | |
| DK5057 (asgA) | <0.001 | |
| DK5208 (csgA) | <0.001 | |
| AG341 (nla4) | 0.3 ± 0.2 | |
| AG342 (nla6) | 0.2 ± 0.1 | |
| AG343 (nla18) | <0.002 | |
| AG344 (nla28) | 4.0 ± 2.6 | |
| AG341 (nla4) | DK1622 (wild type) | 0.3 ± 0.2 |
| AG342 (nla6) | DK1622 (wild type) | 0.2 ± 0.2 |
| AG343 (nla18) | DK1622 (wild type) | 28.4 ± 6.4 |
| AG344 (nla28) | DK1622 (wild type) | 5.4 ± 3.4 |
| DK5057 (asgA) | DK1622 (wild type) | 88.2 ± 8.9 |
| DK5208 (csgA) | DK1622 (wild type) | 83.8 ± 11.4 |
| DK5057 (asgA) | AG341 (nla4) | 1.2 ± 0.8 |
| DK5208 (csgA) | AG341 (nla4) | 0.2 ± 0.03 |
| DK5057 (asgA) | AG342 (nla6) | 12.7 ± 3.4 |
| DK5208 (csgA) | AG342 (nla6) | 8.3 ± 2.6 |
| DK5057 (asgA) | AG343 (nla18) | 9.6 ± 1.9 |
| DK5208 (csgA) | AG343 (nla18) | 79.3 ± 11.5 |
| DK5057 (asgA) | AG344 (nla28) | 15.9 ± 5.9 |
| DK5208 (csgA) | AG344 (nla28) | 8.3 ± 3.4 |
The indicated strains were mixed at a 1:1 ratio and spotted onto TPM agar plates as described in Materials and Methods. The sporulation efficiencies of test strains were determined after 5 days of development as described in Table 1. The mean values for the spore assays were determined from three independent experiments and normalized to the mean value for strain DK1622 alone (wild type for sporulation). The number of spores produced by DK1622 cells ranged from 3 × 106 to 5 × 106. Standard deviations are shown next to the means.
The results presented above suggest that nla4, nla6, nla18, and nla28 may be defective for production of at least one cell-cell signal. If the primary reason for the developmental phenotypes of these nla mutants is a defect in C-signal and/or A-signal production, then wild-type cells should be able to provide the signal(s) that the mutants lack, rescuing sporulation in nla cells. To examine this possibility, nla mutant cells were mixed with wild-type DK1622 cells and spotted onto TPM starvation agar. After 5 days of codevelopment with wild-type cells, the sporulation efficiencies of the nla mutants were determined. Codevelopment with wild-type cells made no improvement in the sporulation efficiencies of nla4, nla6, or nla28 cells (Table 3). Apparently, the developmental defects of these three nla mutants are more complex than a simple lack of signal production (see Discussion). When a pure culture of nla18 mutant cells was allowed to develop on TPM agar for 5 days, no nla18 spores were detected (Table 3). In mixtures with wild-type cells, sporulation in nla18 mutant improved more than 30,000-fold, which is a significant and almost complete rescue of the nla18 sporulation defect (Table 3). The fact that nla18 spores produced colonies that retained their developmental defects indicates that no permanent genetic exchange occurred between wild-type cells and nla18 mutant cells (data not shown). These results are consistent with the idea that the nla18 mutant strain is unable to produce a cell-cell signal required for M. xanthus development but retains the ability to respond to the signal when it is provided by wild-type cells.
Making insertions downstream of the nla genes.
Because the nla mutations were created by plasmid insertions, they have the potential to block transcription of genes located downstream of their respective insertion sites via polar effects. To examine the issue of polarity by nla gene insertions, we made plasmid insertions 10 to 600 bp downstream of the eight nla genes implicated in M. xanthus development and examined their effects on motility, aggregation, and sporulation (Fig. 2 and Table 4). If the defects caused by an nla insertion are simply due to a polar effect on a downstream gene(s), then an insertion immediately flanking the 3′ end of the nla gene should yield defects similar to the insertion in the nla gene itself. Motility and development of strains carrying insertions downstream of nla1, nla6, nla19, nla24, and nla28 are similar to those of the wild type. Thus, it appears that the defects caused by insertions in these five nla genes are not due to polar effects on downstream genes. The insertion downstream of nla18 yielded cells with a nonswarming phenotype similar to that of DK2161 cells, which are defective for A-motility and S-motility. Presumably, this motility defect led to the observed developmental phenotypes; cells carrying an insertion downstream of nla18 showed no signs of aggregation and produced no spores. Given that the insertion within the nla18 gene yielded cells that are motile and capable of aggregating into fruiting bodies, we believe that it is unlikely that the defects caused by the nla18 insertion are simply due to a polar effect. An additional finding that is consistent with this proposal is that the sporulation defect of cells carrying the nla18 insertion was corrected when they were codeveloped with wild-type cells, while the sporulation defect of cells carrying the insertion downstream of nla18 was not corrected when they were mixed and codeveloped with wild-type cells (data not shown). In the case of nla4 and nla23, the downstream insertions yielded motility and/or developmental defects similar to those of original nla gene insertions. DNA sequence analysis places these insertions in genes that are located 164 and 114 bp downstream of nla4 and nla23, respectively. These data suggest that the downstream genes may not be part of the same operons as nla4 and nla23. However, given the uncertainty about the nla4 and nla23 operon structures, we conclude that either nla4 and nla23 and their respective downstream genes are important for M. xanthus development or the defects produced by insertions in nla4 and nla23 are due to polar effects on these downstream genes.
TABLE 4.
Phenotypes produced by insertions in nla locia
| Insertion | Motilityb | Aggregationc | Sporulationd |
|---|---|---|---|
| AG301 (nla1) | − | +/− | + |
| AG367 (nla1de) | + | + | + |
| AG304 (nla4) | + | − | − |
| AG368 (nla4d) | + | − | − |
| AG306 (nla6) | + | +/− | − |
| AG369 (nla6d) | + | + | + |
| AG318 (nla18) | + | +/− | − |
| AG370 (nla18d) | − | − | − |
| AG319 (nla19) | − | +/− | + |
| AG371 (nla19d) | + | + | + |
| AG323 (nla23) | − | +/− | + |
| AG372 (nla23d) | − | +/− | + |
| AG324 (nla24) | − | − | − |
| AG373 (nla24d) | + | + | + |
| AG328 (nla28) | + | +/− | − |
| AG374 (nla28d) | + | + | + |
+, Swarm diameters were 80 to 100% of that of the wild-type cells; −, swarm diameters were less than 80% of that of the wild-type cells.
+, Produced normal-looking fruiting bodies; −, failed to produce normal-looking fruiting bodies; +/−, produced normal-looking fruiting bodies but aggregation was delayed.
+, Sporulation was 50 to 100% of that of the wild-type cells; −, sporulation was less than 50% of that of the wild-type cells.
d, Insertion downstream of the indicated nla gene.
DISCUSSION
During M. xanthus development, cells undergo an elaborate series of morphological changes that culminate in the formation of a multicellular, spore-filled fruiting body. All of the morphological events that occur during this developmental cycle are accompanied by large-scale changes in gene expression (34, 35, 39). How does M. xanthus orchestrate programmed changes in gene transcription during its developmental cycle? It has been proposed that M. xanthus uses a series of NtrC-like activators to turn on specific sets of genes at specific times in development (16, 30). Hence, this model speculates that M. xanthus uses a cascade of NtrC activators during fruiting body development in much the same way as B. subtilis uses a cascade of sigma factors during sporulation (37).
Given that NtrC activators seem to play an important role in the M. xanthus developmental cycle, we made insertions in 28 uncharacterized ntrC-like activator (nla) genes. Of the 28 nla mutants that we analyzed, 8 had obvious defects in development. Three of the nla mutants (nla1, nla19, and nla23) that failed to develop normally have S-motility defects. In earlier studies, Hodgkin and Kaiser (25) showed that many of their S-motility mutants are defective for aggregation into multicellular fruiting bodies. The results of developmental assays with the nla1, nla19, and nla23 mutants are consistent with these findings; aggregation of each of these mutants is delayed compared to that of wild-type cells. The first NtrC-like activator linked to S-motility was PilR, and like our three S-motility mutants, pilR strains show a delay in aggregation. Work by Wu and Kaiser (64) indicates that PilR is required for transcription of the pilA gene, which codes for the external structural subunit of type IV pili. Extension and retraction of polar type IV pili are thought to provide the power for S-motility in M. xanthus, suggesting that S-motility is similar to twitching motility in Neisseria and Pseudomonas (28, 45, 54, 59, 63). Because the location of nla23 places it near a cluster of pilus genes on the M. xanthus chromosome (63, 65), it is tempting to speculate that the Nla23 protein plays a role in pilus biogenesis. In addition to type IV pili, S-motility requires peritrichous fibrils, which are extracellular appendages composed of proteins and carbohydrates (1, 2, 3, 9, 61, 67, 68). It appears that the function of fibrils is to help promote cohesion between neighboring cells and cell cohesion to a solid surface. Future work will be needed to determine whether Nla1, Nla19, and Nla23 are involved in the biogenesis of type IV pili and/or extracellular fibrils.
The insertion in nla24 produces a nonswarming phenotype, suggesting that the nla24 mutant is defective for A- and S-motility. Unlike S-motility, little is known about the motor that drives A-motility, although recent studies have linked A-motility to slime extrusion at the M. xanthus cell poles (62). Given that the product of the nla24 gene has similarity to NtrC-like transcriptional activators, it seems plausible that the Nla24 protein functions as a global regulator of swarming motility in M. xanthus. Our recent expression studies support this idea; the nla24 mutant fails to express genes required for A- and S-motility (data not shown). To date, the only other locus that has been implicated in both A- and S-motility is mglA (55, 56). The product of the mglA gene has similarity to GTP binding proteins (22). The results of MacNeil et al. (44) indicate that MglA is involved in modulating the activity of A- and S-motility components rather than functioning as a regulator of motility gene transcription, which is the likely role of Nla24.
Presumably, the motility defects of the nla24 mutant play an important role in its inability to aggregate and to sporulate when placed on TPM starvation agar. However, it should be noted that glycerol-induced sporulation, which bypasses the need for aggregation, is defective in the nla24 mutant. Our interpretation of this result is that the defect in fruiting body sporulation observed for the nla24 mutant is not simply due to a loss of motility. Perhaps Nla24 plays a dual role in motility and the sporulation process in M. xanthus.
Four of the nla mutants (nla4, nla6, nla18, and nla28) show no appreciable loss in swarming motility, but they are defective for fruiting body development. The aggregation phenotypes of the nla6, nla18, and nla28 mutants are similar; aggregation of these mutants is delayed for about 12 to 24 h. In contrast, aggregation of the nla4 mutant was delayed and incomplete compared to that of the wild type. In addition to these aggregation defects, sporulation in all four of these nla mutants is either reduced or abolished. What could cause the developmental phenotypes of these nla mutants? Based on the results of mixing experiments with A-signal-deficient and C-signal-deficient cells, the nla4, nla6, nla18, and nla28 mutants may be defective for production of cell-cell signals required for fruiting body development in M. xanthus. However, the fact that the sporulation defects of nla4, nla6, and nla28 mutants are not overcome when mixed with wild-type cells indicates that a lack of signal production is not the only developmental process that has been altered in these mutants. One possible explanation for these observations is that nla4, nla6, and nla28 mutants are defective for production of cell-cell signals and for the subsequent response to their production. When mixed and codeveloped, nla18 cells rescue the sporulation defect of C-signal-deficient cells but they fail to rescue the sporulation defect of A-signal-deficient cells completely. In addition, the sporulation efficiency of the nla18 mutant is improved about 30,000-fold when mixed with wild-type M. xanthus cells. Thus, it seems that nla18 belongs to a special class of mutants that fail to produce cell-cell developmental signals, but they retain the ability to respond to the signals when wild-type cells provide them. Another interesting finding from our studies is that glycerol-induced sporulation in nla4, nla6, and nla18 cells is reduced compared to that of the wild type. A defect in glycerol-induced sporulation is a rare occurrence among the many developmental mutants that have been isolated with M. xanthus. One notable exception is the Ω7536 mutant, which is unable to complete a stable conversion to a spherical spore after glycerol induction (43).
Research with a variety of bacterial systems has shown that NtrC-like proteins activate transcription of genes with σ54 promoter elements. Presumably, NtrC-like proteins play a similar role in regulating gene expression in M. xanthus cells. Based on our findings and those in previous studies, 12 NtrC-like activators have now been implicated in M. xanthus development. To date, the M. xanthus genome sequence is 95% complete, suggesting that additional activators required for normal development may be uncovered. Phenotypic characterization of activator mutants indicates that NtrC-like proteins take part in a variety of processes required for normal development, including A-motility, S-motility, and production of cell-cell signals. Hence, the current data support the proposal that a series of NtrC-like proteins are used during the multicellular developmental cycle of M. xanthus. Further analysis will be needed to determine which σ54-dependent genes each NtrC-like protein regulates, how they recognize the promoter elements of these genes, and whether their activity and/or expression is modulated in developing cells.
Acknowledgments
We thank Steve Slater and the Monsanto Company for giving us access to the M. xanthus genome sequence prior to publication and Barry Goldman, Dale Kaiser, Roy Welch, and Jimmy Jakobsen for providing a preliminary annotation of the M. xanthus genome sequence. We also thank Mike Konkel for the use of optical equipment and David Dutton for critical reading of the manuscript.
This work was supported in part by a National Science Foundation grant (0212052) to A.G.G.
REFERENCES
- 1.Arnold, J. W., and L. J. Shimkets. 1988. Inhibition of cell-cell interactions in Myxococcus xanthus by Congo red. J. Bacteriol. 170:5765-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, J. W., and L. J. Shimkets. 1988. Cell surface properties correlated with cohesion in Myxococcus xanthus. J. Bacteriol. 170:5771-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behmlander, R. M., and M. Dworkin. 1994. Biochemical and structural analyses of the extracellular matrix fibrils of Myxococcus xanthus. J. Bacteriol. 176:6295-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandner, J. P., and L. Kroos. 1998. Identification of the Ω4400 regulatory region, a developmental promoter of Myxococcus xanthus. J. Bacteriol. 180:1995-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bretscher, A. P., and D. Kaiser. 1978. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J. Bacteriol. 133:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, K., and D. R. Zusman. 1999. AsgD, a new two-component regulator required for A-signaling and nutrient sensing during early development of Myxococcus xanthus. Mol. Microbiol. 34:268-281. [DOI] [PubMed] [Google Scholar]
- 7.Downard, J., S. V. Ramaswamy, and K.-S. Kil. 1993. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus xanthus development. J. Bacteriol. 175:7762-7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dworkin, M. 1996. Recent advances in the social and developmental biology of the myxobacteria. Microbiol. Rev. 60:70-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dworkin, M. 1999. Fibrils as extracellular appendages of bacteria: their role in contact-mediated cell-cell interactions in Myxococcus xanthus. Bioessays 21:590-595. [DOI] [PubMed] [Google Scholar]
- 10.Dworkin, M., and S. M. Gibson. 1964. A system for studying microbial morphogenesis: rapid formation of microcysts in Myxococcus xanthus. Science 146:243-244. [DOI] [PubMed] [Google Scholar]
- 11.Fisseha, M., M. Gloudemans, R. E. Gill, and L. Kroos. 1996. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J. Bacteriol. 178:2539-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisseha, M., D. Biran, and L. Kroos. 1999. Identification of the Ω4499 regulatory region controlling developmental expression of a Myxococcus xanthus cytochrome P-450 system. J. Bacteriol. 181:5467-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garza, A. G., J. S. Pollack, B. Z. Harris, A. Lee, I. M. Keseler, E. F. Licking, and M. H. Singer. 1998. SdeK is required for early fruiting body development in Myxococcus xanthus. J. Bacteriol. 180:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garza, A. G., B. Z. Harris, J. S. Pollack, and M. H. Singer. 2000. The asgE locus is required for cell-cell signaling during Myxococcus xanthus development. Mol. Microbiol. 35:812-824. [DOI] [PubMed] [Google Scholar]
- 15.Garza, A. G., B. Z. Harris, B. M. Greenberg, and M. H. Singer. 2000. Control of asgE expression during growth and development in Myxococcus xanthus. J. Bacteriol. 182:6622-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorski, L., and D. Kaiser. 1998. Targeted mutagenesis of σ54 activator proteins in Myxococcus xanthus. J. Bacteriol. 180:5896-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorski, L., T. Gronewold, and D. Kaiser. 2000. A σ54 activator protein necessary for spore differentiation within the fruiting body of Myxococcus xanthus. J. Bacteriol. 182:2438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gronewold, T. M., and D. Kaiser. 2001. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol. Microbiol. 40:744-756. [DOI] [PubMed] [Google Scholar]
- 19.Gronewold, T. M., and D. Kaiser. 2002. act operon control of developmental gene expression in Myxococcus xanthus. J. Bacteriol. 184:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gulati, P., D. Xu, and H. B. Kaplan. 1995. Identification of the minimum regulatory region of a Myxococcus xanthus A-signal-dependent developmental gene. J. Bacteriol. 177:4645-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagen, D. C., A. P. Bretcher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 22.Hartzell, P. L., and D. Kaiser. 1991. Function of MglA, a 22-kilodalton protein essential for gliding in Myxococcus xanthus. J. Bacteriol. 173:7625-7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartzell, P. L., and P. Youderian. 1995. Genetics of gliding motility and development in Myxococcus xanthus. Arch. Microbiol. 164:309-323. [DOI] [PubMed] [Google Scholar]
- 24.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171:167-176. [Google Scholar]
- 25.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus: two gene systems control movement. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 26.Julien, B., D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiser, D. 2000. Bacterial motility: how do pili pull? Curr. Biol. 10:R777-R780. [DOI] [PubMed] [Google Scholar]
- 29.Kaiser, D., and C. Crosby. 1983. Cell movement and its coordination in swarms of Myxococcus xanthus. Cell Motil. 3:227-245. [Google Scholar]
- 30.Keseler, I. M., and D. Kaiser. 1995. An early A-signal-dependent gene in Myxococcus xanthus has a σ54-like promoter. J. Bacteriol. 177:4638-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, S. K., and D. Kaiser. 1990. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates the fruiting body morphogenesis of Myxococcus xanthus. Genes Dev. 4:896-905. [DOI] [PubMed] [Google Scholar]
- 32.Kim, S. K., and D. Kaiser. 1990. Cell alignment required in differentiation of Myxococcus xanthus. Science 249:926-928. [DOI] [PubMed] [Google Scholar]
- 33.Kim, S. K., and D. Kaiser. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 61:19-26. [DOI] [PubMed] [Google Scholar]
- 34.Kroos, L., and D. Kaiser. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1:840-854. [DOI] [PubMed] [Google Scholar]
- 35.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 36.Kroos, L., A. Kuspa, and D. Kaiser. 1990. Defects in fruiting body development caused by Tn5-lac insertions in Myxococcus xanthus. J. Bacteriol. 172:484-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kroos, L., B. Zhang, H. Ichikawa, and Y. T. Yu. 1999. Control of sigma factor activity during Bacillus subtilis sporulation. Mol. Microbiol. 31:1285-1294. [DOI] [PubMed] [Google Scholar]
- 38.Kuspa, A., and D. Kaiser. 1989. Genes required for developmental signaling in Myxococcus xanthus: three asg loci. J. Bacteriol. 171:2762-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuspa, A., L. Kroos, and D. Kaiser. 1986. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev. Biol. 117:267-276. [DOI] [PubMed] [Google Scholar]
- 40.Kuspa, A., L. Plamann, and D. Kaiser. 1992. A-signaling and cell density requirement for Myxococcus xanthus development. J. Bacteriol. 174:7360-7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaRossa, R., J. Kuner, D. Hagen, C. Manoil, and D. Kaiser. 1983. Developmental cell interactions of Myxococcus xanthus: analysis of mutants. J. Bacteriol. 153:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, S., B. U. Lee, and L. J. Shimkets. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 43.Licking, E., L. Gorski, and D. Kaiser. 2000. A common step for changing cell shape in fruiting body and starvation-independent sporulation of Myxococcus xanthus. J. Bacteriol. 182:3553-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacNeil, S. D., A. Mouzeyan, and P. L. Hartzell. 1994. Genes required for gliding motility and development in Myxococcus xanthus. Mol. Microbiol. 14:785-795. [DOI] [PubMed] [Google Scholar]
- 45.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 46.Morett, C., and L. Segovia. 1993. The σ54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 175:6067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogawa, M., S. Fujitani, X. Mao, S. Inouye, and T. Komano. 1996. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol. Microbiol. 22:757-767. [DOI] [PubMed] [Google Scholar]
- 48.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 49.Plamann, L., J. M. Davis, B. Cantwell, and J. Mayor. 1994. Evidence that asgB encodes a DNA-binding protein essential for growth and development of Myxococcus xanthus. J. Bacteriol. 176:2013-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romeo, J. M., and D. R. Zusman. 1991. Transcription of the myxobacterial hemagglutinin gene is mediated by a σ54-like promoter and a cis-acting upstream regulatory region of DNA. J. Bacteriol. 173:2969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Shi, W., and D. Zusman. 1993. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. USA 90:3378-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimkets, L. J. 1990. Social and developmental biology of the myxobacteria. Microbiol. Rev. 54:473-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stephens, K., and D. Kaiser. 1987. Genetics of gliding motility in Myxococcus xanthus: molecular cloning of the mgl locus. Mol. Gen. Genet. 207:256-266. [Google Scholar]
- 56.Stephens, K., P. Hartzell, and D. Kaiser. 1989. Gliding motility in Myxococcus xanthus: mgl locus, RNA, and predicted protein products. J. Bacteriol. 171:819-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun, H., and W. Shi. 2001. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. J. Bacteriol. 183:4786-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun, H., and W. Shi. 2001. Analyses of mrp genes during development. J. Bacteriol. 183:6733-6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun, H., D. R. Zusman, and W. Shi. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143-1146. [DOI] [PubMed] [Google Scholar]
- 60.Thöny-Meyer, L., and D. Kaiser. 1993. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J. Bacteriol. 175:7450-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weimer, R. M., C. Creighton, A. Stassinopoulos, P. Youderian, and P. L. Hartzell. 1998. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J. Bacteriol. 180:5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolgemuth, C., E. Hoiczyk, D. Kaiser, and G. Oster. 2002. How myxobacteria glide. Curr. Biol. 12:369-377. [DOI] [PubMed] [Google Scholar]
- 63.Wu, S. S., and D. Kaiser. 1995. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18:547-558. [DOI] [PubMed] [Google Scholar]
- 64.Wu, S. S., and D. Kaiser. 1997. Regulation of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, S. S., J. Wu, Y. L. Cheng, and D. Kaiser. 1998. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol. Microbiol. 29:1249-1261. [DOI] [PubMed] [Google Scholar]
- 66.Xu, H., and T. R. Hoover. 2001. Transcriptional regulation at a distance in bacteria. Curr. Opin. Microbiol. 4:138-144. [DOI] [PubMed] [Google Scholar]
- 67.Yang, Z., Y. Geng, D. Xu, H. B. Kaplan, and W. Shi. 1998. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol. Microbiol. 30:1123-1130. [DOI] [PubMed] [Google Scholar]
- 68.Yang, Z., X. Ma, L. Tong, H. B. Kaplan, L. J. Shimkets, and W. Shi. 2000. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J. Bacteriol. 182:5793-5808. [DOI] [PMC free article] [PubMed] [Google Scholar]