Abstract
Streptococcus pyogenes is a human-specific pathogen that relies on its host for metabolic substrates. Rgg-like proteins constitute a family of transcriptional regulators present in several gram-positive bacteria. In S. pyogenes, Rgg influences the expression of several virulence-associated proteins localized to the cell wall and extracellular environment. Secreted enzymes may degrade host macromolecules, thereby liberating metabolic substrates. To determine if Rgg regulation of exoprotein expression is associated with altered metabolism, the catabolic activities of S. pyogenes strain NZ131 (serotype M49) and an isogenic rgg mutant strain were analyzed during growth with complex and defined media. As expected, the wild-type strain preferentially used glucose and produced lactic acid during the exponential phase of growth. In contrast, the rgg mutant fermented arginine in the exponential phase of growth, even in the presence of glucose. Arginine degradation was associated with a neutral culture pH and excretion of NH3 and ornithine. Arginine, serine, and asparagine were depleted from mutant cultures during growth. The addition of arginine and serine to culture media increased the growth yield and NH3 production of mutant but not wild-type cultures. Addition of asparagine had no effect on the growth yield of either strain. Altered metabolism of arginine and serine in the mutant was associated with increased transcript levels of genes encoding arginine deiminase and a putative serine dehydratase. Thus, Rgg coordinates virulence factor synthesis and catabolic activity and may be important in the pathogen's adaptation to changes in the availability of metabolic substrates.
Streptococcus pyogenes is a human-specific pathogen that relies on its host for catabolic and anabolic substrates. The host-restricted parasitism is reflected in a small genome (1.8 Mb) that lacks genes associated with several metabolic pathways. For example, S. pyogenes generates energy by substrate-level phosphorylation and does not encode a complete tricarboxylic acid cycle or respiratory system (2, 12, 31). In addition, S. pyogenes is auxotrophic for most amino acids (30). The acquisition of metabolic substrates in vivo may involve the degradation of host macromolecules, which is likely to contribute to disease.
Rgg proteins constitute a family of gram-positive transcriptional regulators. These include Rgg of Streptococcus gordonii, which is required for extracellular glycosyltransferase G expression (32, 33), GadR of Lactococcus lactis, which is required for glutamate-dependent acid tolerance (26), MutR, which is required for expression of the mutacin lantibiotic (MutA) of Streptococcus mutans (24), and the plasmid-encoded LasX of Lactobacillus sakei, which induces lasA-W transcription and represses transcription of lasXY (25). The lasA-W genes code for the synthesis, immunity, and transport of the lantibiotic lactocin S (25, 28); lasY encodes a putative ABC transporter (29). Rgg of S. pyogenes, also known as RopB, is required for the expression of the secreted cysteine protease SPE B (3, 16). Subsequent genome scale analyses revealed that Rgg influenced the expression of additional secreted proteins associated with virulence, including the M49 protein, and mitogenic factors 1 and 3 (6, 7). In addition, rgg inactivation altered the expression of known and putative transcriptional regulators, including several two-component regulatory systems, which are important in the transcriptional response to changing environmental conditions (6).
SPE B degrades host proteins, thereby liberating peptides, which are potential catabolic substrates (5). To test the hypothesis that rgg inactivation alters metabolism, we analyzed the catabolic activity of the rgg mutant strain in complex and defined media. The results show significant differences in catabolic substrate preference during growth of the mutant compared to the isogenic wild-type strain. The mutant ferments arginine and degrades serine, even in the presence of high concentrations of glucose. Thus, Rgg coordinates the expression of virulence factors and metabolic enzymes, which may be critical for the pathogen's adaptation to changes in the availability of metabolic substrates.
MATERIALS AND METHODS
Strains and growth media.
S. pyogenes strain NZ131 (serotype M49) and mutant derivatives NZ131 speB::ermAB and NZ131 rgg::ermAB have been previously described (3, 4). S. pyogenes was grown at 37°C with Trypticase soy agar containing 5% sheep blood (Becton Dickinson, Cockeysville, Md.), with 10 ml of Todd-Hewitt broth containing 0.2% (wt/vol) yeast extract (THY; Difco Laboratories, Detroit, Mich.) in 15-ml tubes, or with 10 ml of chemically defined medium (CDM) obtained from JR Biosciences (Denver, Pa.). CDM was prepared without arginine, serine, and glucose, which were added separately at the indicated concentrations. The supplemental compounds were obtained from Sigma Chemical Co. (St. Louis, Mo.).
RNA isolation.
S. pyogenes was grown in 10 ml of THY broth in 15-ml tubes (Corning, New York, N.Y.) for 2 to 3 h (A600 = 0.2) at 37°C. Cultures were centrifuged, and the bacteria were suspended in 200 μl of diethyl pyrocarbonate (Sigma Chemical Co.)-treated water and frozen in liquid nitrogen. RNA was isolated with a FastPrep Instrument (Qbiogene, Carlsbad, Calif.) and a FastPrep Blue kit (Qbiogene), as previously described (10).
Quantitative reverse transcription (RT)-PCR.
Oligonucleotide primers and probes were designed with Primer Express 1.0 software (ABI Prism, PE Biosystems, Framingham, Mass.) and purchased from either MegaBases Inc. (Evanston, Ill.) or PE Biosystems. The following primers (forward and reverse) and TaqMan probes were used: PsagP, 5′-CAATTACCCATTTGCCATCGA-3′, 5′-CACCTGTACCGATAGTTGCGAAT-3′, and 6FAM-TGCCAAACCTTTATTTCACACGGGACC-TAMRA; PsdhA, 5-CTGTTGCCGGCCTAGTAGAAGT-3′, 5′-CTGCGACAAGGGCGAAACT-3′, and 6FAM-CCTTGTGTCAAACGCAATGCCCTAGG-TAMRA; PgryA, 5′-CGACTTGTCTGAACGCCAAA-3′, 5′-TTATCACGTTCCAAACCAGTCAA-3′, and 6FAM-CGACGCAAACGCATATCCAAATAGCTTG-TAMRA. Amplification and detection were done with the ABI Prism 7700 Sequence Detection System (PE Applied Biosystems) by using TaqMan One-Step RT-PCR Master Mix reagents (PE Biosystems), as recommended by the manufacturer. Each assay was done in triplicate with at least two independently isolated RNA samples. Amplification and analysis were done as previously described (10). The quantity of cDNA for each experimental gene was normalized to the quantity of gyrA cDNA in each sample, and the mean ± standard error of the mean of independently isolated RNA samples was determined. The unpaired Student's t test was used to compare the means between wild-type and mutant transcript levels.
Metabolite analysis.
The concentration of glucose in sterile culture supernatant fluids was determined with a glucose-oxidase kit, as previously described (5). The concentrations of ammonia, lactate, citrate, acetate, formate, glutamate, and ethanol were determined with kits purchased from R-Biopharm, Inc. (Marshall, Mich.), as described by the manufacturer. The concentrations of free amino acids were determined with a Beckman amino acid analyzer by the Scientific Resource Consortium Inc. (Minneapolis, Minn.).
RESULTS
Growth of S. pyogenes in THY.
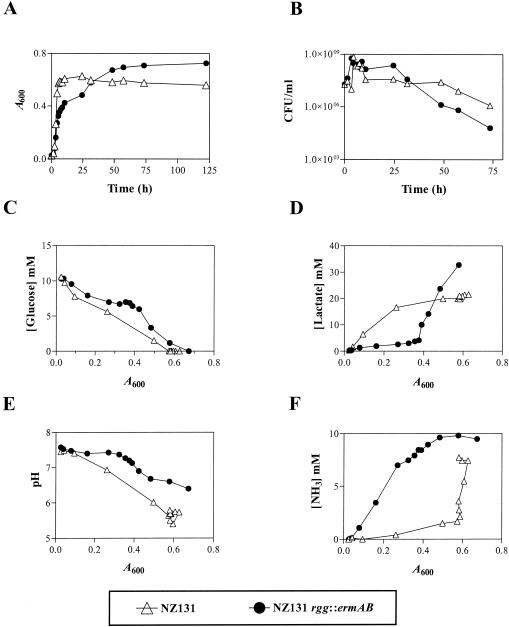
S. pyogenes strain NZ131 and an rgg mutant strain designated NZ131 rgg::ermAB were cultured with THY broth at 37°C with 5% CO2 and no agitation. Aliquots were removed at regular intervals, and the A600 and number of viable bacteria were determined. The isogenic strains grew at similar rates for approximately 8 h. After 8 h the growth rate of the mutant decreased; however, the mutant culture attained a higher growth yield after approximately 25 h of culture (A600 = 0.677 ± 0.005) than did the wild-type strain (A600 = 0.580 ± 0.002; P = 0.002) (Fig. 1A). The viability of NZ131 rgg::ermAB was greater than that of the wild-type strain after 24 h of growth but less after prolonged incubation (75 h) (Fig. 1B). The differences in growth suggested that rgg inactivation altered cellular metabolism.
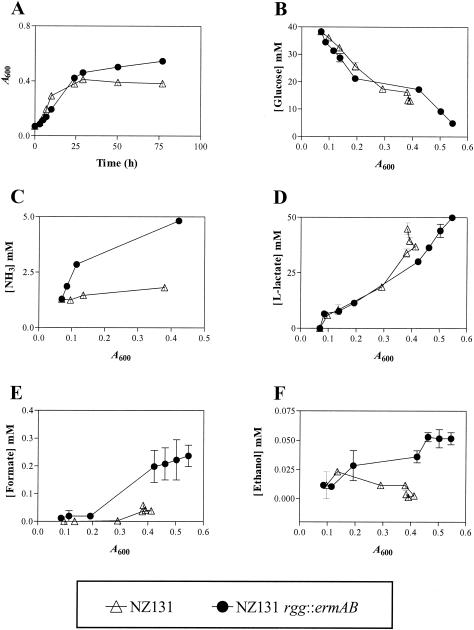
FIG. 1.
Growth of S. pyogenes NZ131 and NZ131 rgg::ermAB with THY broth. (A) At the indicated times, aliquots were removed and the optical density was determined. (B) Aliquots were serially diluted to determine viability. Sterile culture filtrates were prepared, and the concentrations of glucose (C), lactate (D), hydrogen ions (pH) (E), and ammonia (F) were determined. The results shown are representative of at least two independent experiments.
Altered metabolism is associated with rgg inactivation.
Wild-type strain NZ131 preferentially ferments glucose to lactate during exponential growth with THY (5). Entry into the stationary phase coincides with the depletion of glucose from the medium (5). To determine if the rgg mutant fermented glucose, the concentrations of glucose and lactate were determined during growth of NZ131 wild-type and NZ131 rgg::ermAB with THY broth. As expected, the wild-type strain degraded glucose during the exponential phase of growth and glucose depletion coincided with entry into the stationary phase at an A600 of ≈0.55 (Fig. 1C). The concentrations of glucose in rgg mutant cultures decreased at a slightly lower rate than did those of wild-type cultures until the culture reached an A600 of 0.4 (Fig. 1C). At higher cell densities, the rate of glucose depletion in mutant cultures was similar to that of the wild-type strain (Fig. 1C). Lactate accumulated in the wild-type culture during the exponential phase of growth (Fig. 1D) and correlated with decreased concentrations of glucose (compare Fig. 1C and D). Lactate accumulation ended concomitantly with glucose depletion (compare Fig. 1C and D) and entry into the stationary phase at an A600 of 0.6. In contrast, lactate was not produced by rgg mutant cultures in significant amounts during the initial exponential phase of growth (Fig. 1D). However, lactate accumulated in the mutant culture media at A600 between 0.4 and 0.6 (Fig. 1D). Thus, the rgg mutant does not preferentially ferment glucose to produce lactate, in marked contrast to the isogenic wild-type strain. Several anaerobic bacteria ferment citrate and glutamate. In addition, the genome sequences of S. pyogenes encode a putative citrate lyase, which is the first step in the fermentation of citrate. To determine if the mutant was fermenting citrate or glutamate, the concentration of each was determined during growth of the wild-type and mutant strains. No change in the concentration of glutamate or citrate was detected in wild-type or mutant culture media during growth, indicating that the compounds were not fermented (data not shown). To assess if acidic end products other than lactate were excreted by the mutant, the pHs of the culture media were determined during growth. The mutant culture remained neutral throughout overnight incubation (Fig. 1E) and became only slightly acidic at cell densities with an A600 greater than 0.4, which correlated with the accumulation of lactate in the media (Fig. 1D). In contrast, growth of the wild-type strain was associated with rapid acidification of the media and the pH reached a final value of 5.3 (Fig. 1E).
The rgg mutant ferments nitrogen-containing compounds in the exponential phase of growth.
The slight increase in the pH of rgg mutant cultures during exponential growth suggested that the mutant fermented nitrogen (N)-containing compounds, which typically results in NH3 excretion. To test this hypothesis, the concentrations of NH3 in wild-type and rgg mutant cultures were determined. NH3 accumulated in the mutant culture during the early exponential phase of growth (Fig. 1F), suggesting that N-containing compounds, such as amino acids, were being degraded. In contrast, the wild-type strain produced NH3 only in the stationary phase, subsequent to the depletion of glucose (Fig. 1F). Taken together, the results supported the idea that NZ131 rgg::ermAB ferments N-containing compounds during exponential growth in the presence of glucose.
The rgg mutant ferments amino acids in the presence of glucose.
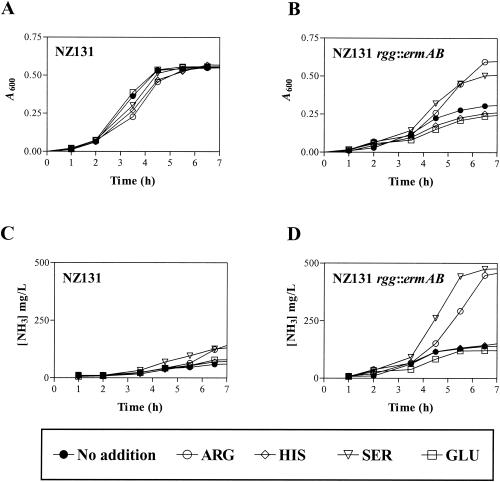
One explanation for ammonia accumulation in mutant cultures was that the mutant fermented amino acids during the initial phase of growth. If this is correct, supplementation of media with an appropriate amino acid will extend the initial exponential phase of growth and increase the growth yield of mutant cultures compared to unsupplemented cultures or cultures supplemented with amino acids that were not catabolized. In addition, increased growth yield will be associated with increased NH3 excretion. To test the hypothesis, a 2 mM concentration of each amino acid was added individually to 10 ml of THY and inoculated with either NZ131 or NZ131 rgg::ermAB. Aliquots were removed during growth, and the A600 and concentrations of NH3 in sterile filtrates were determined. Supplementation with arginine or serine significantly enhanced the growth yield of the rgg mutant after 6.5 h of culture (A600 = 0.595 or 0.504, respectively), compared to the unsupplemented control (A600 = 0.305). Supplementation did not affect the growth yield of the wild-type strain (compare Fig. 2A and B). Increased growth yield correlated with increased NH3 excretion (Fig. 2D), indicating that the rgg mutant degrades arginine and serine, even in the presence of glucose. Supplementation with other amino acids affected neither growth nor ammonia accumulation (data not shown).
FIG. 2.
Growth and ammonia production of NZ131 and NZ131 rgg::ermAB cultured with THY media supplemented with amino acids. Strains were cultured with THY (•) or THY supplemented with arginine (○), serine (▿), glutamic acid (□), or histidine (⋄). At regular intervals, aliquots were removed and the A600 (A and B) and the concentrations of ammonia (C and D) in sterile filtrates were determined. Results shown are representative of two independent experiments.
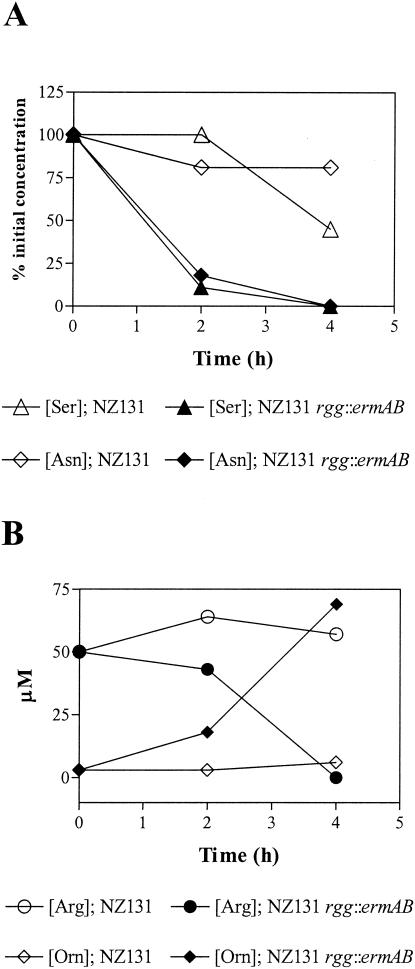
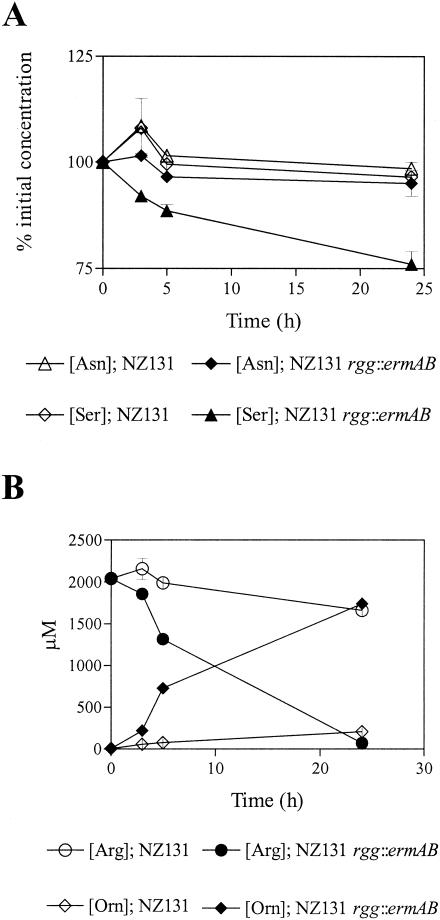
S. pyogenes ferments arginine via the arginine deiminase pathway (ADI), which is present in a variety of bacteria (1, 9, 13, 14, 17). The first enzyme in the pathway is arginine deiminase (ArcA/SagP), which catalyzes the deamination of arginine to NH3 and citrulline. Ornithine carbamoyltransferase converts citrulline to CO2, ornithine, and carbamoylphosphate. ATP is generated by carbamate kinase-mediated conversion of carbamoylphosphate to NH3 and CO2. In addition, the genome sequences of S. pyogenes encode the putative α and β subunits of serine dehydratase. Serine dehydratase catalyzes the transformation of l-serine to pyruvate and NH3. Pyruvate can subsequently be degraded by a variety of fermentation pathways to generate ATP. To determine if amino acids including arginine and serine were depleted from mutant culture media, wild-type and mutant strains were grown in THY and the concentrations of free amino acids were determined. Serine, arginine, and asparagine were depleted from the mutant growth media after approximately 4 h of growth (Fig. 3A and B). Arginine degradation was associated with the excretion of ornithine, an end product of the ADI pathway (Fig. 3B). No significant changes in the concentrations of other free amino acids were detected in mutant cultures (data not shown). No significant changes in the concentrations of amino acids were detected in wild-type cultures (Fig. 3A and B). Together the results indicate that the mutant degrades arginine and serine during the initial period of growth in THY, despite the presence of glucose. Moreover, increased growth yields of the mutant with arginine- and serine-supplemented media and excretion of ornithine and NH3 indicated that the amino acids were fermented.
FIG. 3.
Concentrations of free amino acids during growth of NZ131 (open symbols) and NZ131 rgg::ermAB (closed symbols) with THY. (A) Percentage of the initial concentrations of serine (Ser) and asparagine (Asn) present in sterile culture filtrates. (B) Concentrations of arginine (Arg) and ornithine (Orn) in sterile culture filtrates.
Transcripts associated with amino acid catabolism are more abundant in the rgg mutant.
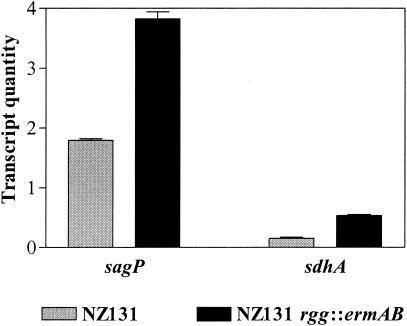
Rgg and Rgg-like proteins are transcriptional regulatory proteins. To determine if altered catabolism in the rgg mutant strain was associated with changes in the expression of genes associated with amino acid catabolism, real-time RT-PCR was used to quantitate transcripts encoding the putative α subunit of serine dehydratase (sdhA) and arginine deiminase (sagP) in the exponential phase of growth (A600 = 0.2). Transcripts encoding SagP and SdhA were 2.1- and 3.6-fold higher, respectively, in the rgg mutant than in the isogenic wild-type strain (P < 0.005) (Fig. 4). Together the results indicate that inactivation of the rgg gene relieves the repression of enzymes responsible for the degradation of arginine and serine.
FIG. 4.
Increased transcription of amino acid catabolism genes in NZ131 rgg::ermAB. The quantities of transcripts encoding arginine deiminase (sagP) and the putative α subunit of serine dehydratase (sdhA) were determined with quantitative RT-PCR. The quantity of cDNA for each gene was normalized to the quantity of gyrA cDNA in each RNA sample. The values shown are the means ± standard errors of the means from two independently isolated RNA preparations analyzed in triplicate.
Growth characteristics of NZ131 and NZ131 rgg::ermAB in CDM.
THY medium is rich in peptides. To determine if similar changes in metabolism occur in the absence of peptides, the isogenic strains were cultured with CDM and catabolic activity was assessed by measuring the concentrations of glucose, lactate, NH3, and free amino acids during growth. The CDM did not contain peptides and consisted of 0.1 g of each amino acid/liter, vitamins, purine and pyrimidine precursors, ferrous sulfate, ferric nitrate, salts, and dextrose. Similar to results obtained with complex media, the growth yield of the mutant (A600 = 0.545 ± 0.001) was higher than that of the wild-type strain (A600 = 0.383 ± 0.001) after 50 h of culture (P = 0.0004) (Fig. 5A). Glucose was depleted from cultures at similar rates until the cultures reached an A600 of ≈0.2. At an A600 of approximately 0.25, the rate of glucose depletion decreased in both cultures and the concentration of glucose in the wild-type culture remained static (Fig. 5B). In contrast, the mutant strain continued to deplete glucose, which correlated with an increase in bacterial density (compare Fig. 5A and B). In contrast to growth with THY, entry into the stationary phase of growth was not associated with glucose depletion. Ammonia was excreted by the rgg mutant but not the wild-type strain when cultured with CDM (Fig. 5C). Lactate was produced during the exponential growth phase for both strains (Fig. 5D). It is unclear if lactate was formed in the mutant culture via glycolysis and reduction of pyruvate or if increased expression of the putative serine dehydratase converted serine to pyruvate, which was then reduced to lactate.
FIG. 5.
Growth of S. pyogenes NZ131(Δ) and NZ131 rgg::ermAB (•) with CDM. At the indicated times, aliquots were removed and the optical density (A) and concentrations of glucose (B), NH3 (C), lactate (D), formate (E), and ethanol (F) in sterile filtrates were determined. The data shown are the means ± standard errors of the means obtained from two independent experiments.
To detect addition changes in metabolite excretion, the concentrations of acetate, formate, and ethanol in sterile filtrates obtained from wild-type and rgg mutant cultures during growth with CDM were determined. No significant differences in acetate formation between the isogenic strains were detected (data not shown). However, minor levels of formate and ethanol were detected in filtrates from stationary phase cultures of the rgg mutant (Fig. 5E and F). In contrast, the compounds were not detected during growth of wild-type strain NZ131 (Fig. 5E and F).
The concentrations of free amino acids were determined during culture of the isogenic strains with CDM. In contrast to the results obtained with THY, no significant change was detected in the concentration of asparagine, even after prolonged incubation (Fig. 6A). Similar to results obtained with complex media, the mutant degraded serine (Fig. 6A) and arginine and excreted ornithine (Fig. 6B). Neither arginine nor serine was depleted from wild-type cultures (Fig. 6A and B). The results obtained with CDM support the idea that arginine and serine catabolism is derepressed in the rgg mutant strain.
FIG. 6.
Concentrations of free amino acids during culture of NZ131 (open symbols) and NZ131 rgg::ermAB (closed symbols) with CDM. (A) Percentage of the initial concentrations of serine (Ser) and asparagine (Asn) in culture filtrates. (B) Concentrations of arginine (Arg) and ornithine (Orn) in culture filtrates. The data shown are the means ± standard errors of the means obtained from two independent experiments.
DISCUSSION
Rgg and Rgg-like proteins constitute a family of transcriptional regulatory proteins encoded in the genomes of several gram-positive bacteria including Streptococcus, Lactococcus, Lactobacillus, and Listeria; homologues have not been identified in gram-negative bacteria. Rgg of S. pyogenes is an important regulator of virulence-associated gene products localized to both the cell wall and the extracellular environment (3, 6, 16). Inactivation of rgg in strain NZ131 altered the growth of the mutant, suggesting that Rgg also regulates the expression of metabolic genes. Comparative analyses of mutant and wild-type strains cultured with complex and defined media showed that the rgg mutant fermented arginine in the presence of glucose. In contrast, the wild-type strain preferentially fermented glucose and amino acid catabolism was repressed. In addition, inactivation of rgg was associated with altered metabolism of serine. Consistent with these phenotypes, transcripts encoding enzymes associated with arginine and serine catabolism were more abundant in the mutant strain. The results show that rgg inactivation relieves repression of enzymes involved in the metabolism of arginine and serine.
Amino acid fermentation.
A variety of bacteria ferment amino acids during anaerobic growth. Serine metabolism in S. pyogenes is poorly understood; however, the genome encodes proteins with similarity to the subunits of serine dehydratase (sdhA, sdhB), a heterodimeric enzyme that catalyzes the deamination of serine to pyruvate and NH3. Derepression of the putative serine dehydratase in NZ131 rgg::ermAB was associated with increased growth yield in serine-supplemented media and increased NH3 production. In addition, serine was depleted from complex and defined media during exponential growth of the mutant but not during growth of the wild-type strain. SdhAB converts serine to pyruvate, which can be reduced to lactate. The higher yield of lactate in mutant cultures (Fig. 1D) is probably due to serine metabolism. Together the results indicate that rgg inactivation is associated with altered metabolism of serine.
Arginine degradation by S. pyogenes was first reported in 1940 (14), and supplementation of media with arginine has previously been shown to increase the growth yields of several serotypes of S. pyogenes (30). Arginine deiminase of S. pyogenes is also known as the antitumor compound streptococcal acid glycoprotein (SAGP). SAGP has arginine deiminase activity and is encoded in an operon that encodes other enzymes of the pathway, and directed mutants of sagP lack arginine deiminase activity (9, 10). One mole of ornithine is excreted (8) for each mole of arginine degraded by the ADI pathway, which is consistent with our results (Fig. 3B and 6B). Excretion of ornithine by the rgg mutant shows that the pathway is not functioning in a biosynthetic direction to generate citrulline or for pyrimidine biosynthesis. Excretion in streptococci is facilitated by an arginine-ornithine antiporter, and transport does not consume energy (11, 23). Therefore, one mole of ATP is generated per mole of arginine metabolized.
We conclude that Rgg represses arginine fermentation based on the following results: (i) arginine supplementation increased the growth yield of the rgg mutant, which was associated with increased NH3 production; (ii) arginine was depleted from mutant cultures, and ornithine was excreted; and (iii) transcript levels of sagP were elevated in the rgg mutant. In general, expression of the ADI pathway is induced by arginine and repressed by glucose, which is mediated, at least in part, by carbon catabolite repression. In addition, the activity of the antiporter is inhibited by glucose (8, 27). Thus, altered glycolytic activity associated with rgg inactivation may relieve carbon catabolite repression, resulting in expression of the ADI pathway. Additional information is needed to determine if Rgg acts directly or indirectly to control ADI expression.
The excretion of NH3 associated with amino acid fermentation enhances acid tolerance in several bacterial species. For example, inactivation of sagP (arc) in S. pyogenes serotype M5 decreased the viability of bacteria exposed to low pH (9). Although the primary function of amino acid fermentation is probably to generate energy, since the expression of ADI is regulated by the availability of metabolic substrates and not pH, neutralization of acidic microenvironments may also be important in sustaining infection.
Rgg regulation is complex.
The mechanism of Rgg regulation remains unclear but appears to be complex. For example, transcripts of rgg are detected during both the exponential and the stationary phases of growth (7); however, speB is expressed primarily in the stationary phase of growth (5, 34). Moreover, Rgg acts both to repress amino acid catabolic activity, based on the results described here, and to activate speB expression (3, 16). It remains to be determined if the disparate effects on transcription are due to secondary or “downstream” effects, such as perturbations of other regulatory networks, or if Rgg has bifunctional regulatory activity. In this regard, the Rgg-like protein LasX is both an enhancer and repressor of transcription in L. sakei (25).
Regulatory link between amino acid catabolism and speB expression.
Coordinately regulated genes often encode proteins with related functions. Rgg is required for speB expression, an extracellular cysteine protease (3, 16). One putative function of bacterial proteases secreted into the environment is to degrade proteins and liberate amino acids and peptides, which could be used as catabolic substrates. In addition, degradation of host proteins may facilitate the invasion of nutrient-rich environments. Results from several studies indicate that potential catabolic substrates including glucose, amino acids, and peptides influence speB expression. (i) SPE B is produced primarily during the stationary phase of growth concomitant with glucose depletion, when strain NZ131 is cultured with THY (10). SPE B production is inhibited by the addition of glucose during the exponential phase of growth, even as the culture enters the stationary phase (10). (ii) speB transcription is inhibited when cultured with amino-acid depleted CDM; transcription is subsequently induced by the addition of peptides (22). (iii) Inactivation of genes encoding components of the oligopeptide and dipeptide transport systems diminish speB transcript levels, indicating a link between peptide availability and expression of speB (20, 21). (iv) Amino acid starvation inhibited speB expression in a relA-independent manner, even though rgg transcription was slightly enhanced (31a). Here, we show that in addition to regulating speB expression, Rgg represses amino acid catabolism, which provides further evidence of a regulatory, and perhaps functional, link between amino acid catabolism and expression of the secreted SPE B protease. Rgg may both activate and repress transcription, in a fashion similar to that of the bifunctional regulatory activity of LasX (25). In this regard, rgg expression does not correlate with speB expression. For example, rgg was expressed in both the exponential and stationary phases of growth in wild-type strain NZ131, although speB was expressed only in the stationary phase of growth. One possibility is that during growth with glucose, Rgg represses alternate catabolic pathways including genes encoding the enzymes arginine deiminase and serine dehydratase. Following the depletion of glucose, Rgg may activate transcription. The result would be derepression of genes associated with amino acid catabolism and expression of SPE B, which could facilitate amino acid catabolism following the degradation of host proteins. Clearly, additional information is necessary to distinguish between direct effects of Rgg on transcription and changes in expression associated with perturbation of other regulatory circuits.
Catabolic activity in vivo.
S. pyogenes ferments carbohydrates via the Embden-Meyerhof-Parnas pathway to form pyruvate, which is reduced to lactic acid. As with many bacteria, glucose is the preferred catabolic substrate and glucose represses the expression of alternative catabolic pathways, including those associated with amino acid catabolism (18, 19). Gene expression and the corresponding phenotype of S. pyogenes are typically characterized during growth with media that contain levels of glucose significantly higher than those likely to be available in vivo. Because S. pyogenes preferentially ferments glucose, little information regarding the phenotype of S. pyogenes during catabolism of amino acids is available. Although it is difficult to estimate the concentration of amino acids at the various anatomical sites colonized by S. pyogenes, amino acids are available, since the bacterium is auxotrophic for most amino acids. Thus, the acquisition of amino acids is necessary for growth, even if glucose or another carbohydrate is fermented. Clearly, host proteins are abundant at sites of colonization. The SPE B protease cleaves a variety of human extracellular matrix proteins, including fibronectin and vitronectin (15). Coordinate regulation of speB and sagP expression by Rgg suggests a possible functional relationship between the gene products. We speculate that the phenotype of S. pyogenes during amino acid fermentation may be more representative of the phenotype in necrotic lesions in which degradation of host proteins may provide important metabolic substrates.
Acknowledgments
The project described was supported by NIH grant number P20 RR16479-02 from the BRIN Program of the National Center for Research Resources and by a grant to L.R. (MCB-0077904) from the National Science Foundation.
REFERENCES
- 1.Bauchop, T., and S. R. Elsden. 1960. The growth of microorganisms in relation to their energy supply. J. Gen. Microbiol. 23:457-469. [DOI] [PubMed] [Google Scholar]
- 2.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaussee, M. S., D. Ajdic, and J. J. Ferretti. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect. Immun. 67:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaussee, M. S., D. Gerlach, C.-E. Yu, and J. J. Ferretti. 1993. Inactivation of the streptococcal erythrogenic toxin B gene (speB) in Streptococcus pyogenes. Infect. Immun. 61:3719-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaussee, M. S., E. R. Phillips, and J. J. Ferretti. 1997. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect. Immun. 65:1956-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crow, V. L., and T. D. Thomas. 1982. Arginine metabolism in lactic streptococci. J. Bacteriol. 150:1024-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degnan, B. A., M. C. Fontaine, A. H. Doebereiner, J. J. Lee, P. Mastroeni, G. Dougan, J. A. Goodacre, and M. A. Kehoe. 2000. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 68:2441-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degnan, B. A., J. M. Palmer, T. Robson, C. E. Jones, M. Fischer, M. Glanville, G. D. Mellor, A. G. Diamond, M. A. Kehoe, and J. A. Goodacre. 1998. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extract is associated with arginine deiminase activity. Infect. Immun. 66:3050-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driessen, A. J., B. Poolman, R. Kiewiet, and W. N. Konings. 1987. Arginine transport in Streptococcus lactis is catalyzed by a cationic exchanger. Proc. Natl. Acad. Sci. USA 17:6093-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunsalus, I. C., and C. W. Shuster. 1961. Energy-yielding metabolism in bacteria, p. 1-58. In R. Y. Stanier (ed.), The Bacteria, vol. II. Academic Press, Inc., New York, N.Y.
- 14.Hills, G. M. 1940. Ammonia production by pathogenic bacteria. J. Biochem. 34:1057-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapur, V., S. Topouzis, M. W. Majesky, L.-L. Li, M. R. Hamrick, R. J. Hamill, J. M. Patti, and J. M. Musser. 1993. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb. Pathog. 15:327-346. [DOI] [PubMed] [Google Scholar]
- 16.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierce, W. A., and A. G. C. White. 1955. Arginine and glucose metabolism in a strain of Streptococcus pyogenes. J. Bacteriol. 69:230-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pine, L., and M. W. Reeves. 1972. Correlation of M protein production with those factors found to influence growth and substrate utilization of Streptococcus pyogenes. Infect. Immun. 5:668-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pine, L., and M. W. Reeves. 1978. Regulation of the synthesis of M protein by sugars, Todd Hewitt broth, and horse serum, in growing cells of Streptococcus pyogenes. Microbios 21:185-212. [PubMed] [Google Scholar]
- 20.Podbielski, A., and B. A. B. Leonard. 1998. The group A streptococcal dipeptide permease (Dpp) is involved in the uptake of essential amino acids and affects the expression of cysteine protease. Mol. Microbiol. 28:1323-1334. [DOI] [PubMed] [Google Scholar]
- 21.Podbielski, A., B. Pohl, M. Woischnik, C. Körner, K.-H. Schmidt, E. Rozdzinski, and B. A. B. Leonard. 1996. Molecular characterization of group A streptococcal (GAS) oligopeptide permease (Opp) and its effect on cysteine protease production. Mol. Microbiol. 21:1087-1099. [DOI] [PubMed] [Google Scholar]
- 22.Podbielski, A., M. Woischnik, B. Kreikemeyer, K. Bettenbrock, and B. A. Buttaro. 1999. Cysteine protease SpeB expression in group A streptococci is influenced by the nutritional environment but SpeB does not contribute to obtaining essential nutrients. Med. Microbiol. Immunol. (Berlin) 188:99-109. [DOI] [PubMed] [Google Scholar]
- 23.Poolman, B., A. J. M. Driessen, and W. N. Konings. 1987. Regulation of arginine-ornithine exchange and the arginine deiminase pathway in Streptococcus lactis. J. Bacteriol. 169:5597-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi, F., P. Chen, and P. W. Caufield. 1999. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl. Environ. Microbiol. 65:652-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawlinson, E. L., I. F. Nes, and M. Skaugen. 2002. LasX, a transcriptional regulator of the lactocin S biosynthetic genes in Lactobacillus sakei L45, acts both as an activator and a repressor. Biochimie 84:559-567. [DOI] [PubMed] [Google Scholar]
- 26.Sanders, J. W., K. Leenhouts, J. Burghoorn, J. R. Brands, G. Venema, and J. Kok. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 27.Simon, J. P., B. Wargnies, and V. Stalon. 1982. Control of enzyme synthesis in the arginine deiminase pathway of Streptococcus faecalis. J. Bacteriol. 150:1085-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skaugen, M., C. I. Abildgaard, and I. F. Nes. 1997. Organization and expression of a gene cluster involved in the biosynthesis of the lantibiotic lactocin S. Mol. Gen. Genet. 253:674-686. [DOI] [PubMed] [Google Scholar]
- 29.Skaugen, M., E. L. Andersen, V. H. Christie, and I. F. Nes. 2002. Identification, characterization, and expression of a second, bicistronic, operon involved in the production of lactocin S in Lactobacillus sakei L45. Appl. Environ. Microbiol. 68:720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slade, H. D. 1954. The metabolism of amino acids by streptococci, p. 65-86. In M. McCarty (ed.), Streptococcal infections. Columbia University Press, New York, N.Y.
- 31.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Steiner, K., and H. Malke. 2000. Life in protein-rich environments: the relA-independent response of streptococcus pyogenes to amino acid starvation. Mol. Microbiol. 38:1004-1016. [DOI] [PubMed] [Google Scholar]
- 32.Sulavik, M. C., and D. B. Clewell. 1996. Rgg is a positive transcriptional regulator of the Streptococcus gordonii gtfG gene. J. Bacteriol. 178:5826-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sulavik, M. C., G. Tardif, and D. B. Clewell. 1992. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J. Bacteriol. 174:3577-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unnikrishnan, M., J. Cohen, and S. Sriskandan. 1999. Growth-phase-dependent expression of virulence factors in an M1T1 clinical isolate of Streptococcus pyogenes. Infect. Immun. 67:5495-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]