Abstract
Strains of Thermus thermophilus accumulate primarily trehalose and smaller amounts of mannosylglycerate in response to salt stress in yeast extract-containing media (O. C. Nunes, C. M. Manaia, M. S. da Costa, and H. Santos, Appl. Environ. Microbiol. 61:2351-2357, 1995). A 2.4-kbp DNA fragment from T. thermophilus strain RQ-1 carrying otsA (encoding trehalose-phosphate synthase [TPS]), otsB (encoding trehalose-phosphate phosphatase [TPP]), and a short sequence of the 5′ end of treS (trehalose synthase [TreS]) was cloned from a gene library. The sequences of the three genes (including treS) were amplified by PCR and sequenced, revealing that the genes were structurally linked. To understand the role of trehalose during salt stress in T. thermophilus RQ-1, we constructed a mutant, designated RQ-1M6, in which TPS (otsA) and TPP (otsB) genes were disrupted by gene replacement. Mutant RQ-1M6 accumulated trehalose and mannosylglycerate in a medium containing yeast extract and NaCl. However, growth in a defined medium (without yeast extract, known to contain trehalose) containing NaCl led to the accumulation of mannosylglycerate but not trehalose. The deletion of otsA and otsB reduced the ability to grow in defined salt-containing medium, with the maximum salinity being 5% NaCl for RQ-1 and 3% NaCl for RQ-1M6. The lower salt tolerance observed in the mutant was relieved by the addition of trehalose to the growth media. In contrast to trehalose, the addition of glycine betaine, mannosylglycerate, maltose, and glucose to the growth medium did not allow the mutant to grow at higher salinities. The results presented here provide crucial evidence for the importance of the TPS/TPP pathway for the synthesis and accumulation of trehalose and the decisive contribution of this disaccharide to osmotic adaptation in T. thermophilus RQ-1.
Thermophilic organisms, like the vast majority of other microorganisms, accumulate compatible solutes in response to water stress imposed by salt. However, the compatible solutes of thermophilic and hyperthermophilic prokaryotes are generally different from those of their mesophilic counterparts (34), and some compatible solutes, namely, di-myo-inositol-phosphate, di-mannosyl-di-myo-inositol-phosphate, diglycerol phosphate, and mannosylglyceramide, are confined to organisms that grow at extremely high temperatures. Mannosylglycerate is also a common compatible solute of thermophiles and hyperthermophiles (21, 28, 35). Despite this association with organisms that grow at extremely high temperatures, mannosylglycerate was initially identified in red algae of the order Ceramiales (4, 15). Trehalose, a canonical compatible solute of mesophiles, also accumulates in a few thermophilic and hyperthermophilic organisms, where it appears to serve as a compatible solute (18, 21, 35). This nonreducing disaccharide has also been implicated in several stress responses in prokaryotes and eukaryotic microorganisms (10, 32, 36, 37) and also serves as an intermediate in the synthesis of glycolipids, sulfolipids, and lipooligosaccharides in mycobacteria (3).
The most common pathway for the synthesis of trehalose in bacteria involves trehalose-phosphate synthase (TPS), encoded by the gene otsA, which converts UDP-glucose and glucose-6-phosphate to trehalose-6-phosphate. This intermediate is subsequently dephosphorylated to yield trehalose via a specific trehalose-6-phosphate phosphatase (TPP), encoded by otsB (12). Another pathway converts maltose to trehalose via a trehalose synthase encoded by treS. Homologues of treS are known to occur in a Pimelobacter sp. (38), Mycobacterium tuberculosis (8), Thermus thermophilus AT-62 and GK24 (16, 39), and Deinococcus radiodurans (40). A third pathway, found in several bacteria, such as Arthrobacter and Rhizobium spp. (22-24), M. tuberculosis (8), and the hyperthermophilic archaeon Sulfolobus acidocaldarius (25), converts the terminal α(1→4)-linked unit of a glucose polymer to an α(1→1) linkage by maltooligosyltrehalose synthase encoded by treY. The terminal disaccharide is then cleaved by a maltooligosyltrehalose trehalohydrolase, encoded by treZ, to yield free trehalose. Organisms may possess one, two, or even all three of these pathways (8).
The species of the genus Thermus have an optimum growth temperature of 70 to 75°C, and the majority, being exclusively isolated from continental hydrothermal springs venting fresh water, do not grow in media containing more than 1% (wt/vol) NaCl. Strains of T. thermophilus, however, are commonly isolated from marine hot springs and are capable of growing in media containing up to about 5% NaCl (7). Trehalose is the major compatible solute of the strains of T. thermophilus grown in yeast extract-tryptone-NaCl medium although these organisms also accumulate lower levels of mannosylglycerate (28). The accumulation of trehalose, during osmotic stress, by T. thermophilus in yeast extract-containing medium is, as in many other organisms, most likely due to its uptake from the medium (18, 27). The role of compatible solutes in the osmotic adjustment of thermophilic and hyperthermophilic organisms remains largely unknown because we lack genetic tools for the manipulation of the organisms and defined media to examine the events leading to the accumulation of compatible solutes under conditions of osmotic stress. However, genetic tools have been developed for T. thermophilus strains that can aid our understanding of osmotic adjustment in this organism (11, 19). In this study we identified and sequenced the complete otsA, otsB, and treS genes of T. thermophilus RQ-1, deleted the otsA and otsB genes for the synthesis of trehalose in strain RQ-1, developed a defined medium lacking yeast extract, examined the effect of these deletions on the osmotic adjustment of the mutant, and examined the effect of the addition of organic solutes on the growth and accumulation of compatible solutes during salt stress.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
RQ-1 (DSM 9247) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). T. thermophilus strains were grown in Thermus medium, containing tryptone (1.0 g/liter) and yeast extract (1.0 g/liter) as source of growth factors, carbon, and nitrogen (41). Escherichia coli DH5α (2) and E. coli XL1-Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA2 lac [F′ proAB lacIqZΔM15 Tn10(Tetr)]) were used as hosts for the cloning vectors. E. coli was grown in YT medium containing, per liter, 10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl. Ampicillin was added to the medium at a final concentration of 100 μg/ml, and kanamycin was added at a final concentration of 30 μg/ml for selection of plasmids. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside and isopropyl-thio-β-d-galactoside (IPTG) were added to the medium at final concentrations of 80 μg/ml and 0.5 mM, respectively. Plasmid pKT1 (19) was kindly provided by José Berenguer (Centro de Biología Molecular, Universidad Autónoma de Madrid, Madrid, Spain).
Construction of a T. thermophilus gene library.
The following degenerate oligonucleotides were designed based on conserved regions of several mesophilic trehalose-phosphate synthase genes available from public databases: forward primer 5′-TGG(A/G)TCCA(C/T)GACTA(C/T)CAC-3′ and reverse primer 5′-AC(G/C)A(A/G)GTTCAT(G/C)CCGTC-3′. A 0.720-kbp PCR fragment was amplified from T. thermophilus RQ-1 with these primers. This fragment was subsequently used as a probe for the isolation of the complete gene from a genomic library.
Total DNA was isolated from T. thermophilus RQ-1 as described by Marmur (20). A partial genomic library from RQ-1, containing DNA fragments selected by Southern blot analysis of genomic DNA with the 0.720-kbp fragment probe, was obtained by complete digestion of total DNA with restriction enzyme HindIII, followed by ligation of purified fragments of around 2.5 kbp into the HindIII site of pUC18 (Invitrogen) and subsequent transformation of E. coli XL1-Blue with the ligation mixture. Positive clones were detected by colony hybridization with a digoxigenin-labeled probe (Roche Molecular Biochemicals). Nucleotide sequences were determined by AGOWA (Berlin, Germany). Inserts of positive pUC18 clones and pGEM-T Easy (Promega) clones were sequenced in both orientations by using vector- and insert-specific oligodeoxynucleotide primers. Nucleic acid and protein sequence analyses were performed with programs of the Wisconsin Genetics Computer Group software package (9). The European Bioinformatics Institute databases (www.ebi.ac.uk) and functionally annotated genomes in the Kyoto Encyclopedia of genes and genomes (www.genome.ad.jp) were screened for homology by using the (T) FASTA (29, 30) and (T) BLAST (1) algorithms.
DNA amplification and analysis.
DNA manipulation techniques used in this study were carried out as described by Sambrook et al. (33). For sequencing, recombinant plasmid DNAs were prepared with plasmid kits (Roche Molecular Biochemicals). PCR amplifications were carried out in a Perkin-Elmer Gene Amp PCR system 2400 in reaction mixtures (50 μl) containing 100 ng of genomic T. thermophilus DNA, 200 ng of each primer, 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, 50 mM KCl, 0.5 U of Taq or Pfu DNA polymerase, and 0.2 mM concentrations of each deoxynucleoside triphosphate (Amersham Biosciences). The mixture was preincubated for 5 min at 94°C and then subjected to 25 cycles of denaturation at 94°C for 1 min. Annealing was performed at 48°C for 1 min, and primer extension was at 72°C for 1 min. The extension reaction in the last cycle was prolonged for 7 min. Amplification products were purified from agarose gels for use as hybridization probes and for cloning into the pGEM-T Easy vector (Promega). Southern blots of restriction endonuclease-cleaved genomic DNA and colony blots were hybridized with DNA probes labeled with a digoxigenin DNA labeling and detection kit (Roche Molecular Biochemicals).
Amplification of treS from RQ-1 genomic DNA was performed with a GC-rich PCR system (Roche), with primers based on the AT-62 treS gene sequence. Two separate amplifications were necessary to obtain the complete gene. The first PCR product was a 1.482-kbp fragment and was obtained using the forward primer 5′-GTGGACCCCCTCTGGTACAAG-3′ and the reverse primer 5′-GGCCTGGGTGTAGCG-3′. The second PCR product was a 1.683-kbp fragment obtained by amplification with the forward primer 5′-GCCTTCTCCCGCGCC-3′ and the reverse primer 5′-CTAGGCTTTTCCGGCCTTGGCC-3′. Both fragments were cloned into pGEM-T Easy vector and were sequenced in both orientations, as described above.
Construction of T. thermophilus otsA/otsB deletion mutants.
A 691-bp fragment corresponding to the 5′ region of otsA (otsA′) was amplified from the chromosomal DNA of strain RQ-1, using the forward primer 5′-GCGGGTACCATGGGGCTAATCATCG-3′ and the reverse primer 5′-GCGAAGCTTGCGCCCCGTGTCTATCC-3′. Restriction sites (underlined) for KpnI and HindIII, respectively, were added to the primers to clone this fragment, immediately upstream of the pslA-kat cassette, into the pKT1 vector (19), yielding pAKT1. Using a similar strategy, a 492-bp fragment corresponding to the 3′ region of otsB (′otsB) was amplified. The forward primer used was 5′-GCGGAATTCGGACAAGGGCTTCG-3′ and the reverse primer was 5′-GCGGCGGCCGCCTAAAGGCTAGTGGGTCG-3′. Restriction sites (underlined) for EcoRI and NotI, respectively, were introduced. The PCR-amplified 3′ region of otsB (′otsB) was cloned into EcoRI/NotI restriction sites of the TOPO 2.1 cloning vector (Invitrogen), prior to cloning into pAKT1. ′otsB was removed from the TOPO vector by EcoRI/NotI digestion followed by digestion of vector DNA with RsaI and BspMI, to eliminate possible contaminations with TOPO cloning vector in the subsequent steps. Then, ′otsB was cloned into pAKT1, to yield pAKBT1. Vector pAKBT1 was used to transform T. thermophilus RQ-1. The method used for plasmid transformation was essentially that of Koyama et al. (17). For this purpose, the recipient strain was grown at 65°C in a transformation medium which contained, per liter of water, 4 g of tryptone (Difco), 2 g of yeast extract (Difco), 1.5 g NaCl, 1 mM MgCl2, and 0.5 mM CaCl2 (pH 7.5). Samples (1 ml) of the culture with a turbidity of 0.3 (610 nm) were transferred to 5-ml sterile tubes, and 20 μg of DNA was added. After 1.5 h of incubation at 65°C under strong aeration, cells were directly plated onto each of three agar Thermus agar plates containing 30 μg of kanamycin per ml and incubated at 55, 60, and 65°C.
Development of a defined medium lacking yeast extract and growth of the organisms.
A defined medium lacking yeast extract (which contains trehalose) was developed to examine growth parameters in the presence or absence of putative exogenous compatible solutes. The defined medium (TD medium) was composed of tryptone (2 g/liter) and a vitamin solution containing thiamine, riboflavin, pyridoxine, biotin, folic acid, inositol, nicotinic acid, pantothenic acid, p-aminobenzoic acid, and cyanocobalamin (Sigma-Aldrich) at a final concentration of 40 μg/liter. This medium was supplemented with NaCl to a final concentration of 1 to 6% (wt/vol) to test the effect of salinity on growth and on the accumulation of organic solutes. Growth of the organisms was performed in 1-liter metal-capped Erlenmeyer flasks containing 200 ml of medium in a reciprocal-water bath shaker (120 rpm) at 70°C. For the cultivation of mutants, the culture medium did not contain kanamycin, but their ability to grow in the presence and absence of kanamycin was always tested on solid medium during or after cultivation. The effect of organic solutes on growth and compatible solute accumulation was examined by the addition of filter-sterilized (Gelman type GN-6; pore size, 0.45 μm; diameter, 47 mm) trehalose, maltose, glucose, glycine betaine, and potassium mannosylglycerate to the culture medium at a final concentration of 0.26 mM.
Overexpression of otsA and otsB genes in E. coli.
The otsA gene was amplified by PCR with the forward primer 5′-GCGGAATTCATGGGGCTCATCATCG-3′ and the reverse primer 5′-GCGCTGCAGTCATCCCTCCTCCAAGGAGGC-3′. The otsB gene was amplified with the forward primer 5′-GCGGAATTCATGAGGGCGGAAAACCCCG-3′ and the reverse primer 5′-GCGCTGCAGCTAAAGGCTAGTGGGTCG-3′. The forward primers contained an additional EcoRI sequence (underlined), immediately upstream of the ATG codon, and the reverse primers had an additional PstI recognition site (underlined), directly behind the TGA stop codon. The PCR products were purified after digestion with EcoRI and PstI and ligated into the corresponding sites into the pKK223-3 expression vector (Amersham Biosciences) to obtain pKKTPS and pKKTPP, respectively. Each construction was transformed into E. coli XL1-Blue cells. Host cells containing pKKTPS or pKKTPP were grown to mid-exponential growth phase, induced with IPTG, and grown further for 6 to 8 h.
Preparation of cell extracts.
T. thermophilus RQ-1, T. thermophilus RQ-1M6, and transformed E. coli cells were harvested by centrifugation (7,000 × g, 10 min, 4°C), and the cell pellet was resuspended in 20 mM Tris-HCl (pH 7.6) containing (per milliliter of suspension) MgCl2 (25 mM), DNase I (10 μg), and the protease inhibitors phenylmethylsulfonyl fluoride (80 μg), leupeptin (20 μg), and antipain (20 μg). Cells were disrupted with a sonicator (Labsonic U; B. Braun, Melsungen, Germany), followed by centrifugation (13,000 × g, 1 h, 4°C). The supernatant was dialyzed against 20 mM Tris-HCl (pH 7.6) to remove endogenous low-molecular-weight compounds (including trehalose) prior to examination of cell extracts for enzyme activity.
Enzymatic assays.
Cell extracts of E. coli XL1-Blue expressing otsA from pKKTPS or otsB from pKKTPP were denatured at 70°C for 20 min and centrifuged (13,000 × g, 2 min) to remove precipitated proteins. The supernatant solutions (10 μl) were incubated with the TPS substrates UDP-glucose (Sigma-Aldrich) and glucose-6-phosphate (Sigma-Aldrich) or the TPP substrate trehalose-6-phosphate (Sigma-Aldrich), at a final concentration of 2 mM, in 20 mM Tris-HCl (pH 7.6) with 25 mM MgCl2. The reaction mixtures were incubated at 65°C for 1 h. The product of the TPS reaction was incubated for an additional hour with 2 U of alkaline phosphatase (Roche) at 37°C to form trehalose. T. thermophilus RQ-1 and RQ-1M6 cell extracts were directly incubated with UDP-glucose and glucose-6-phosphate at 65°C for 1 h. To visualize trehalose, samples were spotted onto thin-layer chromatography (TLC) plates.
TreS activity was assessed in crude cell extracts obtained from cultures of RQ-1 and RQ-1M6 grown in Thermus medium until late exponential phase of growth (turbidity of 0.6 at 610 nm). The cell extracts obtained by sonication were dialyzed against 25 mM MOPS (morpholinepropanesulfonic acid), pH 7.6. Samples (50 μl) of these extracts were incubated with [14C]maltose or [14C]trehalose at a final concentration of 1.35 μM. Aliquots (10 μl) were removed at different times (0.5, 1.0, 1.5, 2.0, and 10.0 min) and extracted with 5 μl of 12% trichloroacetic acid. After incubation on ice (10 min) and centrifugation (2 min, 13,000 × g), the supernatants were applied to TLC plates as described above. The solvent system was composed of butanol-pyridine-water (7:3:1 [vol/vol/vol]). After drying, the TLC plates were autoradiographed for 4 days. [14C]maltose and [14C]glucose were obtained from Amersham Biosciences, and [14C]trehalose was prepared as described previously (13)
Extraction and determination of intracellular solutes.
Wild-type (RQ-1) and mutant (RQ-1M6) cells were harvested by centrifugation (7,000 × g, 10 min, 4°C) during mid-exponential-phase growth and washed twice with a NaCl solution identical in concentration to that of the growth medium. Cell pellets were extracted twice with boiling 80% ethanol as described previously (31). The protein content of the cells was determined by the Bradford assay (5), after sonication of cells, from an aliquot of the suspension before extraction of compatible solutes.
TLC.
TLC was performed on Silica Gel 60 plates (Merck) with a solvent system composed of butanol-ethanol-water (5:3:2 [vol/vol/vol]). The sugars and sugar derivatives were visualized by spraying with α-naphtholsulfuric acid solution followed by charring at 120°C (14). Authentic standards of trehalose, UDP-glucose, and glucose-6-phosphate were used for comparative purposes.
Quantification of organic solute pools by NMR spectroscopy.
Freeze-dried extracts were dissolved in D2O and analyzed by nuclear magnetic resonance (NMR). NMR spectra were acquired in a Bruker AMX300 spectrometer using a 5-mm inverse detection probe head at 25°C, with presaturation of the water signal, a 60° flip angle, and a repetition delay of 60 s. A known amount of sodium formate was added and used as a concentration standard.
Nucleotide sequence accession number.
The sequences of otsA, otsB, and treS reported in this paper have been deposited in GenBank under accession number AY275558.
RESULTS
Sequence analysis of trehalose-synthesizing genes.
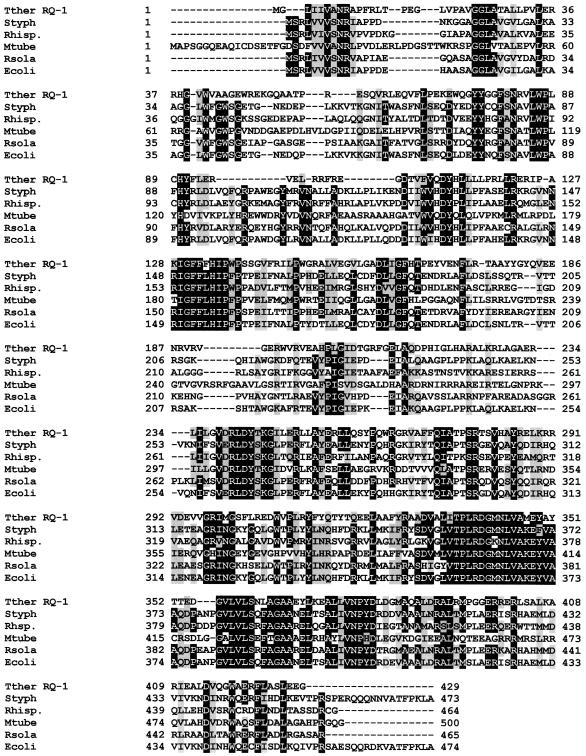
A 2.4-kbp fragment carrying the T. thermophilus RQ-1 otsA (encoding TPS) and otsB (encoding TPP) genes was cloned from a gene library, and its complete sequence was determined. The deduced amino acid sequence of TPS had a calculated molecular mass of 52.64 kDa and had significant similarities with several TPS proteins from other organisms, including 35% identity with E. coli TPS (GenPept AAC74966) (Fig. 1). The deduced amino acid sequence of TPP from strain RQ-1, with a calculated molecular mass of 26.91 kDa, also had significant similarities to several TPP proteins. The otsB sequence from strain RQ-1 was identical to the plasmid-borne homologue from T. thermophilus strain GK24 (AF135796). Immediately downstream of the otsB gene we found a sequence for treS (trehalose synthase). The product of treS, a 965-amino-acid protein, had 98 and 99% amino acid identity to the corresponding proteins from T. thermophilus AT-62 and GK24, respectively (16, 39). The activity of the recombinant TPS or TPP of strain RQ-1, overexpressed in E. coli, was confirmed by TLC using UDP-glucose and glucose-6-phosphate as substrates for TPS and trehalose-phosphate for TPP (results not shown).
FIG. 1.
Alignment of TPS of T. thermophilus RQ-1. The deduced amino acid sequence of otsA of RQ-1 was aligned with those of homologous proteins: Styph, Salmonella enterica serovar Typhimurium (GenPept AF213176); Rhisp., Rhizobium sp. (GenPept AAB91813); Mtube, M. tuberculosis (GenPept CAB08713); Rsola, Ralstonia solanacearum (GenPept CAD17882); Ecoli, E. coli (GenPept AAC74966). Black shading represents identity, and gray shading represents conservative changes.
Construction and isolation of T. thermophilus RQ-1 otsA/otsB deletion mutants.
T. thermophilus mutants, derived from the parental strain RQ-1, were constructed by homologous recombination using a plasmid-encoded deletion between otsA′ and ′otsB (Fig. 2). Sixty kanamycin-resistant colonies were obtained after transformation at 55°C in Thermus medium plates with yeast extract (a source of trehalose) without the addition of NaCl.
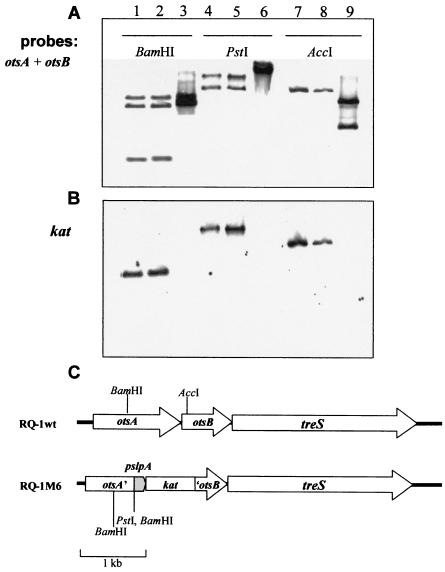
FIG. 2.
Verification of the internal deletion in otsA and otsB. Southern blots of genomic DNA from T. thermophilus RQ-1 and two kanamycin-resistant clones (RQ-1M6 and RQ-1M8) are shown. Bands were identified by hybridization with a 2.05-kbp fragment corresponding to otsA and otsB (A) or kat (B). Lanes 1, 4, and 7, RQ-1M6; lanes 2, 5, and 8, RQ-1M8; lanes 3, 6, and 9, RQ-1. (C) Genetic organization of trehalose-synthesizing genes in the chromosomes of the wild type and an RQ-1 deletion mutant.
All colonies were plated on Thermus medium plates with kanamycin and NaCl up to 4% for 48 h. None of the colonies grew on the plates containing 4% NaCl and 12 strains did not grow in medium with 3% NaCl. Three mutants that did not grow in medium with 3% NaCl, designated RQ-1M3, RQ-1M6, and RQ-1M8, were randomly chosen. Southern blotting experiments performed with genomic DNA of these kanamycin-resistant mutants that was digested with BamHI, PstI, and AccI confirmed that the construction was properly inserted into the chromosome. A 2.05-kbp fragment corresponding to otsA and otsB genes and a 0.78-kbp fragment corresponding to kat gene were used as probes to distinguish between the normal wild-type and insertional forms of the genes in the mutants (Fig. 2).
Digestion of the wild-type strain DNA with BamHI followed by Southern blot hybridization with an otsA/otsB probe produced two bands, while the mutant strains produced three bands that confirm the complete insertion of the kat gene. These results show that an extra BamHI restriction site, 273 bp downstream of the original wild-type restriction site, was responsible for the additional band in the mutants (Fig. 2A). These results were confirmed with AccI and PstI. The restriction site present near the amino-terminal end of the otsB gene in the wild-type strain, demonstrated by the two-band pattern (Fig. 2A), was not present in the mutants, demonstrating that the required 213 bp of this region of otsB gene was removed and replaced by the pslpA-kat cassette. PstI sites are not present in otsA or otsB genes of the wild-type strain; however, the correct insertion of the pslpA-kat cassette led to the introduction of a PstI restriction site shown by the two-band pattern in the mutants. These results show that the replacement of 659 bp at the C-terminal end of otsA and 213 bp at the N-terminal end of otsB by the pslpA-kat cassette occurred via double-recombination events. Probing with the kat gene indicated its presence in the mutant strains alone (Fig. 2B).
In contrast to the wild-type RQ-1 strain, the extracts of the partial otsA and otsB deletion mutants did not, as assessed by TLC, produce trehalose from UDP-glucose and glucose-6-phosphate. The conversion of maltose to trehalose, indicative of TreS activity, could not be detected in crude dialyzed extracts of RQ-1 and RQ-1M6. The reverse reaction could also not be detected (results not shown). Southern blot analysis of the transformants and preliminary salt tolerance tests showed that all three strains (RQ-1M3, RQ-1M6, and RQ-1M8) were identical. RQ-1M6 was chosen for further examination.
Growth of strains RQ-1 and RQ-1M6 in defined medium with different NaCl levels.
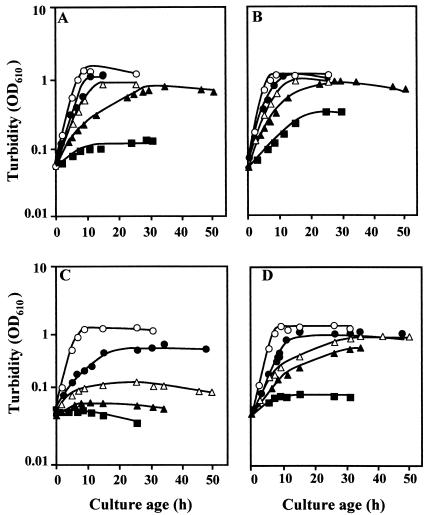
Growth of RQ-1 and RQ-1M6 was similar in the defined TD medium (without an external source of trehalose) without NaCl or containing 1% (wt/vol) NaCl (Fig. 3A and C). The growth rate of RQ-1M6 decreased considerably in the medium containing 2% NaCl and was insignificant in TD medium containing 3% NaCl. The growth rate of RQ-1M6 was 7.3 times lower at this salinity than that of the wild-type strain, and the cell yield was about half that of the parental strain. RQ-1 grew consistently in TD medium containing 5% NaCl but did not grow in the medium containing 6% NaCl (Fig. 3A).
FIG. 3.
Growth of wild-type and mutant T. thermophilus strains in TD medium with 0% (○), 3% (•), 4% (▵), 5% (▴), and 6% (▪) NaCl. RQ-1 (A and B) and RQ-1M6 (C and D) were grown in TD medium in the absence (A and C) or presence (B and D) of exogenous trehalose. OD610, optical density at 610 nm.
Effect of exogenous trehalose on growth.
The addition of trehalose to the defined medium without NaCl had no effect on the growth rate or the cell yield of the wild type or the mutant. However, the addition of trehalose favored the growth of the wild-type strain and the mutant under salt stress conditions (Fig. 3B and D). Exogenous trehalose caused an increase in the growth rate of the wild-type strain in TD medium containing 5% NaCl from 0.06 to 0.12 h−1 and allowed growth in medium with 6% NaCl. We observed no alteration in the final yield after growth in TD medium containing 3 and 4% NaCl and a slight increase of 0.19 and 0.22 optical density unit in medium containing 5 and 6% NaCl, respectively (Fig. 3B).
The effect of exogenous trehalose on the growth of RQ-1M6 was more pronounced. The maximum NaCl concentration for growth of the mutant in TD medium increased from 3 to 5% with the addition of trehalose. The growth rate of the mutant was lower than that of the wild type in trehalose-containing defined medium with 3 and 4% NaCl, but the final yield was similar. The mutant also grew in TD medium containing 5% NaCl, although the growth rate and the final yield were lower than those of the wild type. However, the mutant did not grow, even in the presence of trehalose, in medium containing 6% NaCl (Fig. 3D).
Effect of exogenously provided mannosylglycerate, glycine betaine, maltose, and glucose on growth.
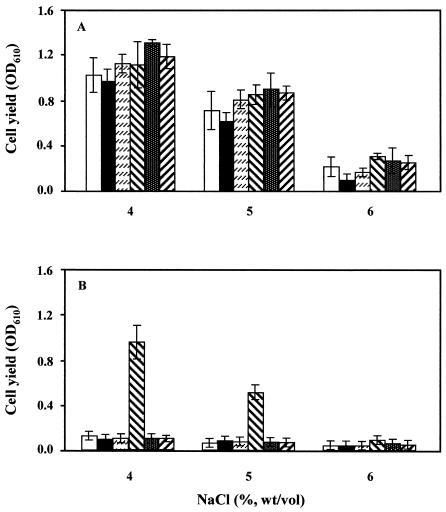
The addition of the solutes mannosylglycerate, glycine betaine, maltose, and glucose to medium containing 4, 5, and 6% NaCl had no effect on the growth of the mutant. We observed, however, that maltose and glucose, like trehalose, increased the growth rate of the wild-type strain, and glycine betaine caused a slight increase in the growth rate, while mannosylglycerate did not (results not shown). Maltose and glucose also increased the cell yield of the wild-type strain, but betaine had only a very slight effect on the cell yield of this strain, while mannosylglycerate had no effect (Fig. 4).
FIG. 4.
Effect of the presence of exogenous organic solutes on the cell yield of RQ-1 and RQ-1M6. RQ-1 (A) and mutant RQ-1M6 (B) were grown in TD medium containing 4, 5, and 6% NaCl. Within each group, bars represent (from left to right) TD medium without exogenous solutes and TD medium supplemented with mannosylglycerate, glycine betaine, trehalose, maltose, and glucose. OD610, optical density at 610 nm.
Accumulation of organic osmolytes by RQ-1 and RQ-1M6.
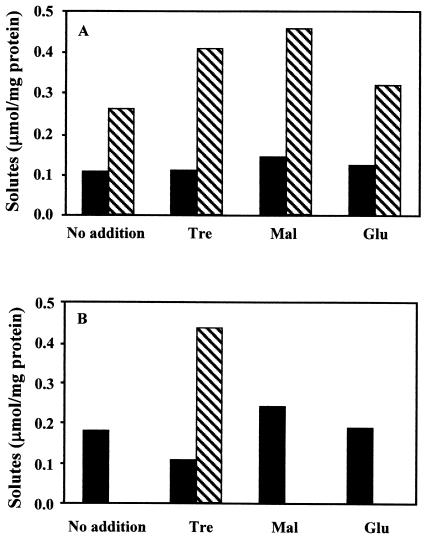
Growth of the wild-type strain in defined TD medium containing 3% NaCl led to the accumulation of trehalose and low levels of mannosylglycerate. The addition of trehalose or maltose led to a slight increase in the levels of intracellular trehalose. There was, however, no increase in the intracellular levels of mannosylglycerate; glucose and maltose were never detected (Fig. 5).
FIG. 5.
Accumulation of compatible solutes by RQ-1 and RQ-1M6. RQ-1 (A) and RQ-1M6 (B) were grown in TD medium containing 3% NaCl without the addition of exogenous organic solutes (no addition) or with trehalose (Tre), maltose (Mal), or glucose (Glu). Bars represent intracellular concentrations of mannosylglycerate (▪) and trehalose (▧).
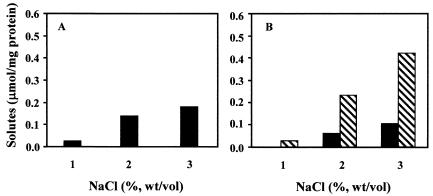
Growth of the mutant in TD medium led to the progressive accumulation of mannosylglycerate in medium containing 1 to 3% NaCl, but trehalose was not detected. On the other hand, growth of the mutant in TD medium containing 1 to 3% NaCl with exogenous trehalose led to the accumulation of progressively larger amounts of the disaccharide and smaller amounts of mannosylglycerate. At the same salt concentration, the level of mannosylglycerate in these cells was always lower than in TD medium without trehalose (Fig. 6). The addition of maltose or glucose to TD medium containing 3% NaCl did not lead to their intracellular accumulation or the conversion of these sugars into trehalose. Glycine betaine was never detected in cell extracts (Fig. 5).
FIG. 6.
Effect of salinity on the accumulation of solutes by RQ-1M6. RQ-1M6 was grown in TD medium without (A) or with (B) exogenous trehalose. Bars represent intracellular concentrations of mannosylglycerate (▪) and trehalose (▧).
DISCUSSION
Trehalose is a very common solute of bacteria, yeasts, fungi, and archaea that appears to play several roles related to stress protection (6, 8). The protecting role of trehalose in osmotic adjustment, heat stress, and dehydration has been amply demonstrated, although under some experimental conditions, the precise role of this sugar is unclear because it may have several roles in the same organism (36, 37).
T. thermophilus strains generally grow in media containing NaCl at concentrations as high as 5 to 6%. In media containing yeast extract, the major compatible solute is trehalose, while mannosylglycerate is found in smaller amounts (28). Under these conditions, trehalose could be synthesized de novo or could be taken up from the growth media, because the disaccharide is present in the yeast extract (42).
Here we show that strain RQ-1 possesses the pathway for the synthesis of trehalose that proceeds via trehalose-6-phosphate. Cloning and functional overexpression of otsA and otsB from T. thermophilus RQ-1 in E. coli showed that these genes code for the enzymes responsible for the two-step pathway leading to the synthesis of trehalose. However, we were unable to demonstrate TreS activity in crude extracts of T. thermophilus RQ-1 or RQ-1M6. We cannot explain these negative results, although the treS gene product of strain RQ-1 shares 98 and 99% identity with its homologue in T. thermophilus AT-62 and GK-24, respectively, where maltose-converting activity was detected by using recombinant proteins (16, 39). Our inability to detect TreS activity in the crude extracts of the wild type or the mutant may be due to lack of expression of the gene under the conditions examined.
Sequence analysis showed that otsB was located immediately downstream of otsA, and treS is located immediately downstream of otsB. The three genes are structurally linked and may or may not be organized in an operon-like arrangement, since obvious promoter sequences could not be recognized. However, these promoter sequences are not easily identified in T. thermophilus (26). The presence of a partial otsB sequence and a complete treS sequence in T. thermophilus strain AT-62 (39) as well as a complete otsB and treS sequence in strain T. thermophilus GK-24 (AF135796) had already been reported, but this is the first report of the presence of otsA in T. thermophilus. It should also be noted that several wild-type T. thermophilus strains lack otsA while others lack otsA, otsB, and treS. None of the organisms grow in defined medium containing more than 2% NaCl, confirming the results obtained with RQ-1M6. Moreover, none of the strains examined possess treY or treZ (S. Alarico, Z. Silva, A. Henne, H. Santos, and M. S. da Costa, unpublished results).
The role of trehalose in osmotic adjustment of T. thermophilus RQ-1 was demonstrated by deletion of otsA and otsB, because the mutant lacking these genes was unable to accumulate this compatible solute in a defined medium deprived of the disaccharide or to grow with NaCl above 3%. The accumulation of trehalose was never detected in the mutant when maltose or glucose was supplied to this strain, indicating that maltose could not be converted to trehalose via treS or that glucose, derived from the hydrolysis of maltose, could not be converted to trehalose via otsA and otsB. On the other hand, the accumulation of trehalose by the mutant in trehalose-containing medium showed that it was taken up from the medium. This uptake system has been identified as an ATP binding cassette-type transporter that also takes up maltose (Z. Silva, M. Sampaio, M. S. da Costa, and H. Santos, unpublished results). We are therefore led to conclude that T. thermophilus RQ-1 can synthesize trehalose in a medium lacking trehalose or can take up trehalose from a medium containing yeast extract. Compatible solute uptake is well documented and is preferred to de novo synthesis in many organisms (6, 34).
Exogenous glycine betaine did not serve as a compatible solute in T. thermophilus RQ-1, despite the earlier observation that trace levels of this solute accumulate in a few other strains of T. thermophilus (28). Glycine betaine did not improve growth of mutant RQ-1M6 in media containing salt, nor did it accumulate during salt stress in glycine betaine-containing medium. This observation indicates that one of the most common compatible solutes of mesophilic prokaryotes does not serve as a compatible solute in T. thermophilus or, for that matter, in any of the (hyper)thermophiles examined thus far. We speculate that glycine betaine does not meet the requirements of thermophiles or hyperthermophiles for osmotic adjustment, perhaps because it is not taken up from the environment due to the lack of the appropriate transporters or is unstable at high growth temperatures (34). The accumulation of glucose and maltose did not, as expected, occur when these sugars were added to the medium, since these sugars are only rarely used as compatible solutes in prokaryotes (6).
The inability of the mutant to grow in TD medium with more than 3% NaCl leads us to view the accumulation of trehalose as evidence that this disaccharide serves as a compatible solute. The mutant accumulates progressively higher levels of mannosylglycerate as the salt concentration increases up to 3%, but osmotic adjustment at higher salt concentrations is not possible unless trehalose is taken up by the organism. Growth is restored in defined trehalose-containing medium with 4 or 5% NaCl, although growth is slower than in the parental strain. Moreover, trehalose accumulates to levels as high as those in the wild-type strain. Under these conditions the levels of intracellular mannosylglycerate in the wild type are lower than in the mutant in defined medium without the addition of trehalose.
Mannosylglycerate serves as the sole or major compatible solute under salt stress in other thermophiles and hyperthermophiles in yeast extract media (34), but T. thermophilus RQ-1 appears to use mannosylglycerate only under low-level osmotic adjustment in media containing less than about 3% NaCl. Above these salt levels, trehalose is required for growth and the accumulation of mannosylglycerate becomes less important for osmotic adjustment. The accumulation of mannosylglycerate alone, even during low-level osmotic adjustment, does not fulfill the requirement for trehalose accumulation, since the growth rates and the final yields are lower, at the corresponding salinity, than after uptake of trehalose.
The results presented here show that trehalose plays, under the conditions examined, an important role in osmotic adjustment of T. thermophilus RQ-1. The role of trehalose in the osmotic adjustment of this organism was demonstrated by the inability of the mutant to grow in a defined medium without yeast extract containing more than 3% NaCl. This is the first study using genetic deletion mutants showing that an organic solute is a de facto compatible solute in a thermophilic organism.
Acknowledgments
This research was funded by the European Commission 5th Framework Programme, project QLK3-CT-2000-00640, and FCT/FEDER projects PRAXIS/P/BIO/12082/1998 and POCTI/35715/BIO/2000. Z. Silva acknowledges a Ph.D. scholarship from PRAXIS XXI (BD/21669/99).
We thank Helena Santos and Maria Manuel Sampaio (ITQB, Oeiras, Portugal) for performing some experiments, critical reading of the manuscript, and helpful discussions.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates, New York, N.Y.
- 3.Besra, G. S., and D. Chatterjee. 1994. Lipids and carbohydrates of Mycobacterium tuberculosis, p. 285-306. In B. R. Bloom (ed.), Tuberculosis: pathogenesis, protection and control. American Society for Microbiology, Washington, D.C.
- 4.Bouveng, H., B. Lindberg, and B. Wickberg. 1955. Low-molecular carbohydrates in algae. Structure of the glyceric acid mannoside from red algae. Acta Chem. Scand. 9:807-809. [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.da Costa, M. S., H. Santos, and E. A. Galinski. 1998. An overview of the role and diversity of compatible solutes in Bacteria and Archaea. Adv. Biochem. Eng. Biotechnol. 61:118-153. [DOI] [PubMed] [Google Scholar]
- 7.da Costa, M. S., M. F. Nobre, and F. A. Rainey. 2001. The genus Thermus, p. 404-414. In D. R. Boone and R. W. Castenholtz (ed.), Bergey's manual of systematic bacteriology, vol. 1, 2nd ed. Springer, New York, N.Y.
- 8.De Smet, K. A. L., A. Weston, I. N. Brown, D. B. Young, and B. D. Robertson. 2000. Three pathways for trehalose biosynthesis in mycobacteria. Microbiology 146:199-208. [DOI] [PubMed] [Google Scholar]
- 9.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:287-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Virgilio, C., T. Hottinger, J. Domínguez, T. Boller, and A. Wiemken. 1994. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. I. Genetic evidence that trehalose is a thermoprotectant. Eur. J. Biochem. 219:179-186. [DOI] [PubMed] [Google Scholar]
- 11.Fernandéz-Herrero, L. A., G. Olabarría, J. R. Cáston, I. Lasa, and J. Berenguer. 1995. Horizontal transfer of S-layer genes within Thermus thermophilus. J. Bacteriol. 177:5460-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giæver, H. M., O. B. Styrvold, I. Kaasen, and A. R. Strøm. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 170:2841-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horlacher, R., R. Peist, and W. Boos. 1996. Improved method for the preparative synthesis of labeled trehalose of high specific activity by Escherichia coli. Appl. Environ. Microbiol. 62:3861-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacin, H., and A. R. Mishkin. 1965. Separation of carbohydrates on borate-impregnated silica gel G plates. J. Chromatogr. 18:170-173. [DOI] [PubMed] [Google Scholar]
- 15.Karsten, U., K. D. Barrow, A. S. Mostaert, R. J. King, and J. A. West. 1994. 13C- and 1H-NMR studies on digeneaside in the red alga Caloglossa leprieurii. A reevaluation of its osmotic significance. Plant Physiol. Biochem. 32:669-676. [Google Scholar]
- 16.Koh, S., J. Kim, H.-J. Shin, D. Lee, J. Bae, D. Kim, and D.-S. Lee. 2003. Mechanistic study of intramolecular conversion of maltose to trehalose by Thermus caldophilus GK24 trehalose synthase. Carbohydr. Res. 338:1339-1343. [DOI] [PubMed] [Google Scholar]
- 17.Koyama, Y., T. Oshino, N. Tomizuka, and K. Furukawa. 1986. Genetic transformation of the extreme thermophile Thermus thermophilus and other Thermus spp. J. Bacteriol. 166:338-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamosa, P., L. O. Martins, M. S. da Costa, and H. Santos. 1998. Effects of temperature, salinity, and medium composition on compatible solute accumulation by Thermococcus spp. Appl. Environ. Microbiol. 64:3591-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasa, I., J. R. Cáston, L. A. Fernández-Herrero, M. A. de Pedro, and J. Berenguer. 1992. Insertional mutagenesis in extreme thermophilic eubacteria Thermus thermophilus HB8. Mol. Microbiol. 6:1555-1564. [DOI] [PubMed] [Google Scholar]
- 20.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 21.Martins, L. O., R. Huber, H. Huber, K. O. Stetter, M. S. da Costa, and H. Santos. 1997. Organic solutes in hyperthermophilic Archaea. Appl. Environ. Microbiol. 63:896-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maruta, K., T. Nakada, M. Kubota, H. Chaen, T. Sugimoto, M. Kurimoto, and Y. Tsujisaka. 1995. Formation of trehalose from maltooligosaccharides by a novel enzymatic system. Biosci. Biotechnol. Biochem. 59:1829-1834. [DOI] [PubMed] [Google Scholar]
- 23.Maruta, K., K. Hattori, T. Nakada, M. Kubota, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of trehalose biosynthesis genes from Arthrobacter sp. Q36. Biochim. Biophys. Acta 1289:10-13. [DOI] [PubMed] [Google Scholar]
- 24.Maruta, K., K. Hattori, T. Nakada, M. Kubota, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of trehalose biosynthesis genes from Rhizobium sp. M-11. Biosci. Biotechnol. Biochim. 60:717-720. [DOI] [PubMed] [Google Scholar]
- 25.Maruta, K., K. Hattori, T. Nakada, M. Kubota, H. Chaen, S. Fukuda, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of cluster of genes encoding novel enzymes of trehalose biosynthesis from thermophilic archaebacterium Sulfolobus acidocaldarius. Biochim. Biophys. Acta 1291:177-181. [DOI] [PubMed] [Google Scholar]
- 26.Maseda, H., and T. Hoshino. 1995. Screening and analysis of DNA fragments that show promoter activities in Thermus thermophilus. FEMS Microbiol. Lett. 128:127-134. [DOI] [PubMed] [Google Scholar]
- 27.Mikkat, S., U. Effmert, and M. Hagemann. 1997. Uptake and use of the osmoprotective compounds trehalose, glucosylglycerol, and sucrose by the cyanobacterium Synechocystis sp. PCC6803. Arch. Microbiol. 167:112-118. [PubMed] [Google Scholar]
- 28.Nunes, O. C., C. M. Manaia, M. S. da Costa, and H. Santos. 1995. Compatible solutes in the thermophilic bacteria Rhodothermus marinus and “Thermus thermophilus.” Appl. Environ. Microbiol. 61:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 31.Reed, R. H., D. L. Richardson, S. R. C. Warr, and W. D. P. Stewart. 1984. Carbohydrate accumulation and osmotic stress in cyanobacteria. J. Gen. Microbiol. 130:1-4. [Google Scholar]
- 32.Reinders, A., N. Bürckert, S. Hohmamm, J. M. Thevelein, T. Boller, A. Wiemken, and C. De Virgilio. 1997. Structural analysis of the subunits of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae and their function during heat shock. Mol. Microbiol. 24:687-695. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Santos, H., and M. S. da Costa. 2002. Compatible solutes of organisms that live in hot saline environments. Environ. Microbiol. 4:501-509. [DOI] [PubMed] [Google Scholar]
- 35.Silva, Z., N. Borges, L. O. Martins, R. Wait, M. S. da Costa, and H. Santos. 1999. Combined effect of the growth temperature and the salinity of the medium on the accumulation of compatible solutes by Rhodothermus marinus and Rhodothermus obamensis. Extremophiles 3:163-172. [DOI] [PubMed] [Google Scholar]
- 36.Simola, M., A. L. Hänninen, S. M. Stranius, and M. Makarow. 2000. Trehalose is required for conformational repair of heat-denatured proteins in yeast endoplasmic reticulum but not for maintenance of membrane traffic functions after severe heat stress. Mol. Microbiol. 37:42-53. [DOI] [PubMed] [Google Scholar]
- 37.Singer, M. A., and S. Lindquist. 1998. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol. Cell 1:639-648. [DOI] [PubMed] [Google Scholar]
- 38.Tsusaki, K., T. Nishimoto, T. Nakada, M. Kubota, H. Chaen, T. Sugimoto, and M. Kurimoto. 1996. Cloning and sequencing of trehalose synthase gene from Pimelobacter sp. R48. Biochim. Biophys. Acta 1290:1-3. [DOI] [PubMed] [Google Scholar]
- 39.Tsusaki, K., T. Nishimoto, T. Nakada, M. Kubota, H. Chaen, S. Fukuda, T. Sugimoto, and M. Kurimoto. 1997. Cloning and sequencing of trehalose synthase gene from Thermus aquaticus ATCC33923. Biochim. Biophys. Acta 1334:28-32. [DOI] [PubMed] [Google Scholar]
- 40.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S., Makarova, L. Aravind, M. J. Daly, and C. M. Fraser, et al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams, R. A. D., and M. S. da Costa. 1992. The genus Thermus and related microorganisms, p. 3745-3753. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The procaryotes, 2nd ed. Springer, New York, N.Y.
- 42.Xavier, K. B., L. O. Martins, R. Peist, M. Kossmann, W. Boos, and H. Santos. 1996. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 178:4773-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]