Abstract
We report the isolation and partial characterization of three new mutants of Streptomyces coelicolor that are defective in morphogenesis and antibiotic production. The genes identified by the mutations were located and cloned by using a combination of Tn5 in vitro mutagenesis, cotransformation, and genetic complementation. Mutant SE69 produces lower amounts of antibiotics than the wild type produces, produces spores only after prolonged incubation on rich media, and identifies a gene whose predicted protein product is similar to the GntR family of transcriptional regulators; also, production of aerial mycelia on both rich and poor media is significantly delayed in this mutant. Mutant SE293 is defective in morphogenesis, overproduces antibiotics on rich media, fails to grow on minimal media, and identifies a gene whose predicted protein product is similar to the TetR family of transcriptional regulators. Preliminary evidence suggests that the SE293 gene product may control a molybdopterin binding protein located immediately adjacent to it. Mutant SJ175 sporulates sooner and more abundantly than the wild type and overproduces antibiotics on rich media, and it identifies a gene whose predicted protein product contains regions of predominantly hydrophobic residues similar to those of integral membrane proteins.
Understanding the signals and pathways that allow cells to sense changes in their environment and transmit this information at the molecular level to make changes in gene expression is fundamental to understanding development in both prokaryotes and eukaryotes. Streptomyces spp. are gram-positive soil bacteria that, upon sensing starvation, release small molecules from the growing substrate mycelia (37) that signal initiation of a program of gene expression evidenced by the erection of developmental structures called aerial hyphae. These aerial hyphae coil and ultimately septate to form uninucleoid mature spores. Coincident with these complex morphological changes is the production of a number of medically important natural product antibiotics. Most of what is known about the pathways that contribute to morphogenesis and antibiotic production has come from studies of mutants that are defective in one or both of these processes. Mutants blocked in the production of aerial hyphae, the first visible sign of morphogenesis, are called bald (bld) mutants. Many of these mutants are also defective in antibiotic production. Mutants that are able to initiate morphogenesis but are defective in the ability to form mature spores are called white (whi) mutants. The genes identified by these bld and whi mutants have diverse functions, and while a clear picture of how they participate in morphogenesis and antibiotic production is still emerging, it is clear from the study of these mutants that they hold the key to understanding both morphogenesis and antibiotic production in these complex bacteria.
The bld gene products include a tRNA (encoded by bldA) that recognizes a rare leucine codon (19), a DNA binding protein (encoded by bldD) that acts as a transcriptional regulator of its own transcription, as well as that of whiG and bldN, a sigma factor (encoded by bldN) (3), an anti-anti-sigma factor (encoded by bldG) (4), an oligopeptide importer (encoded by bldK) (23), a response regulator required for morphogenesis (encoded by bldM) (3, 21), and a putative DNA binding protein (encoded by bldB) required for catabolite repression, morphogenesis, and antibiotic production (25). While no clear pathway emerges from a description of these genes, the ability to form aerial hyphae can be restored by extracellular complementation between some bld mutants, suggesting that at least one pathway controlling the initiation of morphogenesis involves a cascade of extracellular signals (36). Since bldN and bldB do not fit into this cascade, it is possible that these genes are involved in additional pathways that signal development. Similarly, the ram gene cluster has been shown to activate development independent of the bld gene cascade (22), clearly indicating that our understanding of how these genes function and interact to facilitate the initiation of morphogenesis and antibiotic production is at an early stage.
Six whi loci have been identified that are needed early in development to form normal sporulation septa, and two of these genes have been extensively characterized at the molecular level. whiG encodes a sporulation-specific sigma factor that resembles the fliA-encoded sigma factor of enteric bacteria and σD of Bacillus subtilis (6), and, of special interest to this study, whiH encodes a member of the GntR family of transcriptional regulators, many of which are repressors that respond to carboxylate-containing intermediates in carbon metabolism (29).
The complexity of these pathways and the fact that several of the bld and whi genes were identified on the basis of single alleles suggest that many more genes have yet to be identified. In fact, a recent genome-wide screen for morphological mutants led to isolation of several previously uncharacterized genes (9). Recent advances by Gehring et al. (9) have led to adaptation of an effective method of in vitro transposon mutagenesis in Streptomyces in which a Tn5 derivative was used (10). We have used this method to perform a genome-wide screen for mutants using two new transposons, each containing a reporter gene (either EGFP or xylE), to generate transcriptional fusions upon insertion of the transposon. The xylE gene encodes a catechol dioxygenase, which converts colorless catechol to a yellow oxidation product. The EGFP gene encodes a protein that fluoresces green when it is exposed to UV light (7) and has been used successfully in Streptomyces as a reporter gene (34).
Here we describe identification of three previously uncharacterized genes that affect, directly or indirectly, both morphogenesis and antibiotic production. Linkage between the drug resistance marker in a transposon and the mutant phenotype was established by cotransformation. Primers homologous to the ends of Tn5 were used to determine the chromosomal DNA sequence adjacent to the insertion and the address of the insertion in the genome. The wild-type allele of each mutant was cloned and used to test for genetic complementation of the mutant phenotype.
MATERIALS AND METHODS
Strains and growth conditions.
Streptomyces coelicolor M145 (SCP1− SCP2−) genomic DNA was used for in vitro transposon mutagenesis with derivatives of Tn5. S. coelicolor M145 was used as a recipient of the M145 genomic library with transposon insertions. Growth and manipulation of these strains were performed as described previously (16). R2YE, MS, and MYM were used for growth on solid medium, and YEME was used for growth in liquid medium (16). Morphogenic and antibiotic production phenotypes were examined on MYM, minimal medium with glucose as the sole carbon source (MM-glucose), minimal medium with mannitol as the sole carbon source (MM-mannitol), minimal medium with maltose as the sole carbon source (MM-maltose), and R2YE agar. When appropriate, the medium was supplemented with 50 μg of apramycin per ml, 200 μg of spectinomycin per ml, or 50 μg of hygromycin per ml. Escherichia coli strain XL-10 gold was used for preparation of plasmid DNA as described previously (20). E. coli ET12567/pUB307 (dam dcm) was used for conjugation with S. coelicolor. E. coli DH5α was used for construction of the Tn5EGFP insertion library, E. coli XL-1 blue was used for construction of the Tn5xylE library, and E. coli strain XL-10 gold was used for construction of the Tn5apr library.
Construction of modified Tn5 transposons for use in vitro.
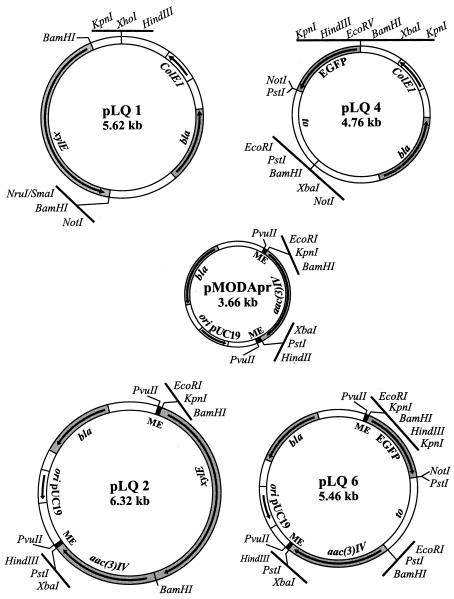
Three versions of Tn5 were used in this study, Tn5apr (9), Tn5xylE, and Tn5EGFP. To construct Tn5xylE, the 2.66-kb HindIII-NruI fragment containing the promoterless copy of the xylE gene from pXE4 (14) was ligated into pBluescript II KS(+) that had been digested with HindIII and SmaI, resulting in plasmid pLQ1 (Fig. 1). The 2.4-kb BamHI fragment of pLQ1 was cloned into the BamHI site of pMODApr (9) to insert the promoterless copy of the xylE gene immediately adjacent to the 19-bp end of Tn5, resulting in plasmid pLQ2 (Fig. 1). To construct Tn5EGFP, a 1.8-kb EcoRI-EcoRV fragment containing a promoterless copy of the EGFP gene from pIJ8668 (34) was ligated into pBluescript II KS(+) that had been digested with EcoRI and EcoRV, resulting in plasmid pLQ4 (Fig. 1). The 1.8-kb BamHI fragment from pLQ4 was ligated into pMODApr to insert the EGFP gene immediately downstream of the 19-bp end of Tn5 to generate pLQ6 (Fig. 1). All plasmid constructions were verified by restriction digestion and analysis on agarose gels.
FIG. 1.
Construction of transposon Tn5xylE, which contains a promoterless copy of the xylE reporter gene immediately downstream of the 19-bp repeat at the end of Tn5, and transposon Tn5EGFP, which contains a promoterless copy of the EGFP reporter gene in the same position with respect to the end of the transposon.
In vitro transposon mutagenesis.
To create plasmid libraries, M145 genomic DNA was digested with Sau3A1 and cloned into the BamHI site of pSpecoriT. One of the genomic libraries used in this study was kindly provided by Amy Gehring and Rich Losick, and its construction is described elsewhere (9). These genomic libraries were used in in vitro transposition reactions with Tn5xylE and Tn5EGFP contained on the PvuII fragments from pLQ2 and pLQ6, respectively. The reaction conditions used were those described by the manufacturer (Epicenter Technologies, Madison, Wis.). After incubation, each transposon containing a plasmid library was transformed into E. coli, and transformants were selected on Luria-Bertani (LB) agar (20) containing 50 μg of apramycin sulfate (Sigma) per ml and 200 μg of spectinomycin (Sigma) per ml. The resulting transformants were pooled, and plasmid DNA was transformed either into E. coli ET12567/pUB307 for mating with S. coelicolor or into E. coli ET12567 for preparation of plasmid DNA for transformation of S. coelicolor. Selection of plasmid DNA in E. coli ET12567/pUB307 was performed as described above except that 30 μg of kanamycin per ml was added. Mating and transformation were carried out as described previously (16). Colonies were picked after 5 to 7 days of incubation at 30°C, retested on MYM containing apramycin (50 μg/ml), and screened for spectinomycin sensitivity (150 μg/ml), as well as for defects in antibiotic production and/or morphogenesis. Spectinomycin-resistant strains with obvious morphological defects were serially passaged on MYM containing 50 μg of apramycin per ml and screened for spectinomycin sensitivity.
Cotransformation.
Genomic DNA from the mutants that were generated were prepared by the salting-out procedure for isolation of genomic DNA (16) and alkali denatured (24), and 1 μg of each DNA was used to transform M145 protoplasts. Transformants were selected on R2YE overlaid with 50 μg of apramycin per ml.
Identification of the site of transposon insertion.
Genomic DNA was isolated from the verified transposon mutants, digested with ApaI (Tn5EGFP and Tn5xylE mutants) or SacII (Tn5apr mutants), and ligated into pBluescript II KS(+) digested with the same enzymes. The ligated DNA was then used to transform E. coli XL-10 gold with selection on LB agar containing 50 μg of apramycin per ml. Plasmid DNA was recovered from the Aprr transformants and was sequenced with primers complementary to the ends of the transposon. Primers 5′ TCGACCTGCAGGCATGCAAGCTT3′ and 5′GGGTACCGAGCTCGAATTCATCG 3′ generated sequences with SJ175 and SE293, but these primers did not generate a sequence with SE69. For SE69 primers 5′ GATGCCGCTCGCCAGTCGATTG 3′ and 5′ GAACAGCTCCTCGCCCTTGCTC 3′ complementary to regions of the EGFP and apramycin resistance genes were used. Sequencing reactions were carried out with a Big Dye terminator Ready Reaction cycle sequencing kit (version 3.0), and samples were analyzed with an ABI Prism 310 sequencer (Applied Biosystems) used according to the manufacturer's instructions.
Complementation of insertional mutants.
Wild-type alleles of each mutant were obtained by PCR amplification of S. coelicolor A3 (2) chromosomal DNA. For SJ175 primers 5′ ACAAGCGCCTGCGAGGCAAGGA 3′ and 5′ TCACCGCGTAGTACCACAAGCTG 3′ were used; for SE293 primers 5′ TCGACGTCATCCCACGGCTGCG 3′ and 5′ GACGGCGAGGTTCAGGCAGGAG 3′ were used; and for SE69 primers 5′ GCCGCGAACTCCTCGATGATGAC 3′ and 5′ CTTCCCCGCATGTCCTGCACCTTG 3′ were used. Each PCR was performed with a DyNAzyme EXT PCR kit (Finnzymes) used according to the manufacturer's instructions. The PCR program consisted of 94°C for 5 min and then 35 cycles of 94°C for 45 s, 68°C for 30 s, and 72°C for 2 min, followed by incubation at 72°C for 10 min and then holding at 4°C. The reaction mixtures were supplemented with 5% dimethyl sulfoxide and 5 mM MgCl2. The PCR products were subjected to electrophoresis, extracted from the agarose gels, and purified with a Qiagen QIAquick gel extraction kit. The fragments containing wild-type alleles of SJ175 and SE293 were treated with the Klenow polymerase and then digested with either SacII (SJ175) or BclI (SE293). The fragment containing the wild-type allele of SE69 was digested with PvuII and BglII. Each fragment was then ligated into pHygoriT that had been digested with SacII and EcoRV (SJ175) or BamHI and EcoRV (SE293 and SE69). To construct pHygoriT, a fragment containing the hygromycin resistance gene (hygB) was generated by PCR amplification of Streptomyces hygroscopicus chromosomal DNA. A DNA linker containing the recognition sequence for NcoI was ligated to the ends of the PCR product, and the resulting fragment was digested with NcoI and ligated to pUC19 that had been digested with AflIII to generate pUH19. A 1.98-kb SapI-AluNI fragment from pUH19 containing the hygromycin resistance gene was treated with the Klenow polymerase (20) and cloned into the 4.96-kb SacI fragment of pSET152, which was also treated with the Klenow polymerase. The ligation reaction mixtures were transformed into E. coli XL-10 gold, and transformants were selected on LB agar containing 50 μg of hygromycin (Sigma) per ml. All plasmid constructions were verified by restriction digestion and analysis on agarose gels. Plasmid DNA was recovered and used to transform E. coli strain ET12567/pUB307. Conjugation between E. coli and Streptomyces was performed as described previously (16). The conjugation mixtures were plated on MS agar containing 10 mM MgCl2, and after 18 h of incubation at 30°C, nalidixic acid (0.5 mg) and hygromycin (2 mg) were applied to the plates in an agar overlay.
RESULTS AND DISCUSSION
Construction of Tn5 insertions in the S. coelicolor chromosome by using in vitro transposition and marker replacement.
Three transposons based on Tn5 were used in our experiments. Tn5apr was constructed by Gehring et al. (9) and contains an apramycin resistance gene flanked by the 19-bp repeats of Tn5. Tn5xylE is a derivative of Tn5apr that contains a promoterless copy of the xylE reporter immediately downstream of the 19-bp repeat at the end of the transposon so that insertion of Tn5xylE downstream of a regulatory region generates a transcriptional fusion between the regulatory region of the gene identified and the xylE gene. Tn5EGFP is a derivative of Tn5apr that generates similar fusions to the EGFP reporter gene. A library of S. coelicolor M145 genomic DNA was constructed in pSpecoriT, which has no origin of replication for Streptomyces but which does contain an E. coli origin of replication, as well as oriT, which allows plasmid transfer by mating between E. coli and S. coelicolor. This genomic library was exposed to one of the three transposons under conditions that allowed the transposons to insert into the genomic DNA in vitro (9, 10). S. coelicolor M145 was then either mated with E. coli containing the mutagenized genomic libraries or transformed with plasmid DNA. The four mutants described here were, in fact, generated by transformation. Exconjugants or transformants were selected by using the apramycin resistance (Aprr) gene located within the transposon. Since pSpecoriT does not contain an origin of replication for S. coelicolor, recombinants arose either by integration of the entire vector via a single-crossover event or by marker replacement of the chromosomal DNA with the DNA contained on the vector via a double-crossover event. These two events were distinguished from each other by the fact that a single crossover resulting in insertion of the entire plasmid would produce recombinants that were Aprr (contained within the transposon) and spectinomycin resistant (Spcr) (contained on the vector), while marker replacement between the vector and the chromosome via a double-crossover event would result in recombinants that were Aprr and Spcs. These cells would contain a replacement of the region of the chromosome contained on the vector with a fragment that contained a transposon insertion generating a transposon library. The recombinants were screened for morphological or antibiotic production phenotypes that potentially arose from insertions of the transposon into genes required for these functions.
When the library mutagenized with Tn5apr was used, 276 Aprr exconjugants were selected, all of which were also Spcr. Sixty-nine of these exconjugants had phenotypes of interest, and they were serially passaged in the presence of apramycin and tested for spectinomycin sensitivity. Ultimately, 11 Aprr Spcs strains were generated. When the library mutagenized with Tn5xylE was used, 5,216 Aprr exconjugants were selected; 151 of these exconjugants were Spcs, and 14 of them had phenotypes of interest. When the library mutagenized with Tn5EGFP was used, 8,096 Aprr exconjugants were selected; 158 of these exconjugants were Spcs, and 35 of them had phenotypes of interest (Table 1).
TABLE 1.
Identification of Tn mutants
| Library | No. of exconjugants or transformants | No. with phenotypes of interest | No. linked to transposon |
|---|---|---|---|
| Tn5apr | 276 | 11 | 1 |
| Tn5xylE | 5,216 | 14 | 0 |
| Tn5EGFP | 8,096 | 35 | 3 |
Establishment of linkage between mutant phenotypes and the apramycin resistance marker contained in the transposon.
The process of generating and regenerating protoplasts, especially protoplasts of S. coelicolor, results in generation of a significant number of colonies with morphological defects. To distinguish this background from mutations caused by transposon insertion, linkage between the apramycin resistance marker and the phenotype of interest was examined by cotransformation (24). For cotransformation experiments, genomic DNA was isolated from each Aprr Spcs strain that had either an antibiotic production or a morphological phenotype different from that of the wild type, and this DNA was used for transformation into S. coelicolor M145 protoplasts. Apramycin-resistant transformants were then screened for the mutant phenotype of the donor. Only the transformants that showed 100% cotransformation between the apramycin resistance marker and the phenotype of the donor were characterized further.
Of the 11 potential mutants generated by Tn5apr, 7 yielded transformants, and 1 of these (SJ175) was linked to the transposon (four of four transformants obtained showed the mutant phenotype). Of the 14 potential mutants generated by Tn5xylE and the 35 potential mutants generated by Tn5EGFP, 37 yielded transformants; none of these were linked to Tn5xylE, but 3 of them, SE69 (15 of 15 transformants), SE220 (8 of 8), and SE293 (2 of 2), were linked to Tn5EGFP (Table 1).
Characterization and complementation of the mutant phenotypes.
The phenotypes of each mutant were examined on MYM and R2YE, which are relatively rich media, as well as MM-glucose, MM-mannitol, and MM-maltose, which are minimal media. Wild-type S. coelicolor grows well on R2YE, sporulates abundantly on MYM, and grows relatively well on minimal media with glucose, mannitol, or maltose as the sole carbon source. S. coelicolor M145, the parental strain used in these experiments, sporulates relatively poorly on minimal media. Mannitol was used in these experiments because growth on mannitol actually rescues the phenotype of some morphological mutants of S. coelicolor, such as bldA, bldG, and bldH mutants (5).
Figure 2 shows a comparison of the wild-type parental strain, strain M145, with three of the four mutants identified in this screening analysis, SE69, SE293, and SJ175. As described below, subsequent investigation revealed that the fourth mutant, SE220, had been identified previously. To confirm that the phenotypes of these mutants were caused by the transposon insertions (linkage was established by cotransformation) and to test the limits of the genes affected by the insertions, wild-type copies of the regions of the chromosome identified by the insertions were cloned by PCR amplification into pHygoriT and mated into the corresponding mutant strain to test for genetic complementation. The regions used for complementation are shown in Fig. 3. Mating of pHygoriT without an insert was used as a negative control.
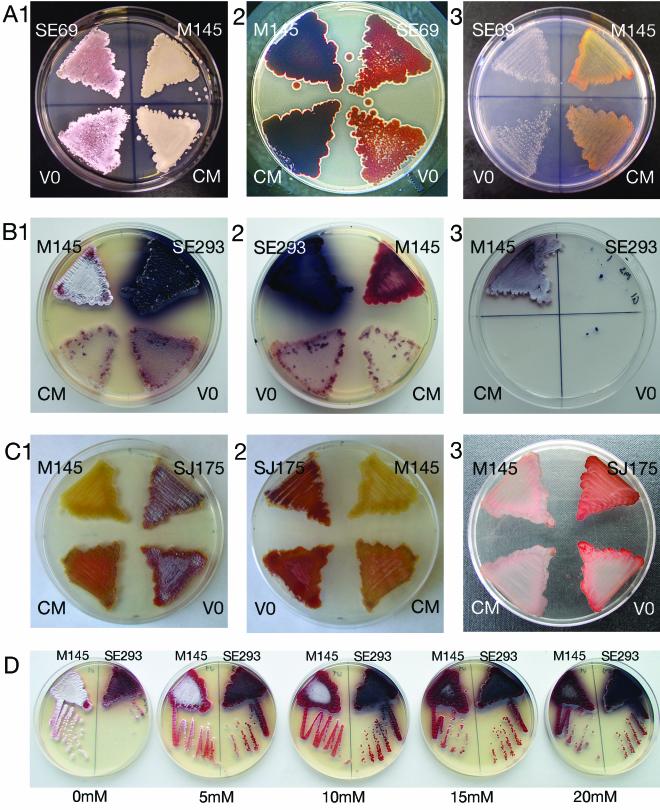
FIG. 2.
S. coelicolor insertional mutants. M145 is the wild-type parental strain. SE69, SE293, and SJ175 are insertional mutants. CM indicates the relevant mutant strain containing a vector with a DNA fragment containing the region of the insertion from the wild-type strain for genetic complementation. VO indicates the mutant containing the same vector used for complementation but without insert DNA. The strains were compared after growth on MYM (A1, B1, and C1, photographed from the top of the plate, and A2, B2, and C2, photographed from the back of the plate) or MM-glucose (A3, B3, and C3). (D) SE293 and M145 grown on MYM agar with different concentrations of tungstate.
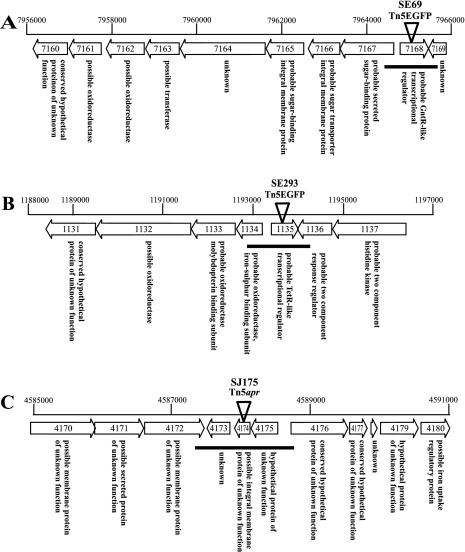
FIG. 3.
Organization of the S. coelicolor chromosome near the sites of transposon insertion. For gene designations we used the nomenclature of the Sanger Centre genome sequencing project. The numbers above each diagram indicate the base pair positions of the genes in the cosmid DNA sequence, and the Sanger Centre annotation is given in the arrow delineating the position of each gene. The chromosomal location of each transposon insertion is indicated by a triangle. The DNA regions that were amplified by PCR and cloned into vectors for genetic complementation are indicated by solid lines below the open reading frames. The presumed function of each open reading frame is shown below the arrow indicating its position.
SE69 was morphologically defective on both MYM and R2YE and failed to make the blue pigment associated with actinohorodin production in the parental strain. As shown in Fig. 2A1, on MYM at times when and under conditions in which the wild type produced abundant spores (pigmented grey), SE69 produced no spores and very few aerial mycelia. As shown in Fig. 2A2 (a view of the back of the plate), SE69 was also defective in actinohorodin production. On relatively poor media, such as, MM-glucose (Fig. 2A3), MM-mannitol, and MM-maltose, the mutant grew more slowly than wild type and failed to produce pigment. As also shown in Fig. 2A, both the morphological and antibiotic production defects of this mutant were restored by complementation with the wild-type allele of the gene disrupted in the SE69 mutant. No complementation was observed with the same vector without an insert.
Mutant SE293 was defective in morphogenesis on MYM and R2YE (Fig. 2B1), producing virtually no aerial mycelia, but it dramatically overproduced the blue pigment associated with actinohorodin production on both these media (Fig. 2B2). Interesting, SE293 failed to grow at all on MM-glucose (Fig. 2B3), MM-mannitol, or MM-maltose. Standard tests for auxotrophy (16) revealed that SE293 required arginine for growth on minimal medium. Somewhat surprisingly, neither the morphological nor the antibiotic production defects of this mutant were complemented by a fragment that included the gene identified by the transposon insertion. There are several possible explanations for this. It is possible that the defect created by this insertion was not complemented in trans or that it was a dominant mutation. While cotransformation established linkage between the apramycin resistance marker within the transposon and the mutant phenotype, it is also possible that this strain contained two closely spaced transposons. Experiments to generate a null allele of SCO1135 by marker replacement with a constructed deletion are in progress.
SJ175 sporulated more abundantly than the wild type and produced aerial mycelia and the pigments associated with antibiotic production at least 24 h sooner than the wild type produced aerial mycelia and these pigments on MYM (Fig. 2C1 and 2C2). SJ175 also produced the pigments associated with antibiotic production at least 24 h sooner than the wild type produced them on MM-glucose (Fig. 2C3). Complementation of SJ175 with the wild-type allele of the gene disrupted in the SJ175 mutant on MYM partially restored the timing of aerial mycelium production and antibiotic production to that of the wild type (Fig. 2C1 and 2C2). Complementation with the wild-type allele on MM-glucose restored the timing of antibiotic production (Fig. 2C3). No complementation was observed with the same vector without an insert.
Chromosomal locations of the transposon insertions and preliminary characterization of the genes likely to be affected.
Primers complementary to the 19-bp repeats at the end of Tn5 were used to determine sequences from the transposon insertions in both directions into the surrounding chromosomal DNA. Attempts to obtain sequences directly from chromosomal DNA were unsuccessful. To eliminate this problem, the transposon and chromosomal DNA adjacent to it were cloned into pBluescript II KS(+), with selection for the drug resistance marker on the transposon, prior to sequencing. At least 100 bp of DNA sequence generated with each primer was used as a query to BLAST for the S. coelicolor genome.
Mutant SE220 resulted from an insertion located within SCE59.12c, a gene recently described by Gehring et al. (9) as rsuA. Our insertion was located approximately 300 bp upstream of the insertion isolated by Gehring et al., and like rsuA, SE220 was bld and failed to produce either the red or blue pigments associated with undecylprodigiosin and actinorhodin production on R2YE (data not shown). On MYM, SE220 was unpigmented and bald.
Mutant SE69 resulted from an insertion located within SC9A4.30 (SCO7168), and the annotated region of the chromosome at this address is shown in Fig. 3A. The predicted 224-amino-acid protein encoded by this open reading frame exhibited sequence similarity to the GntR family of transcriptional regulators, which also includes WhiH. These proteins contain conserved helix-turn-helix (DNA binding) motifs which are slightly different for each subfamily, as well as effector binding and/or dimerization domains that also vary with the subfamily. As shown in Fig. 4, a comparison of the predicted protein encoded by SCO7168 with several GntR family proteins revealed homology and similarity throughout the protein with other members of this family, particularly members of the FadR subfamily (27). The GntR family of regulators includes almost 300 members that are distributed among a diverse group of bacteria and are involved in the regulation of many different biological processes. Of special interest in this context is the whiH gene of S. coelicolor, which is required for sporulation in this organism. While the genome sequence of S. coelicolor suggests that there are potentially 10 GntR-like proteins in this organism, only the whiH mutants and SE69 have been identified by mutation.
FIG. 4.
End-to-end alignment of conserved (black background) and similar (gray background) amino acids in inferred sequences of proteins with similarity to the predicted protein encoded by the SCO7168 (SE69) open reading frame. Abbreviations: SE69-Sc, putative GntR family transcriptional regulator identified by transposon insertion in this study from S. coelicolor; GntR-Bs, gluconate operon transcriptional repressor from B. subtilis; WhiH-Sc, sporulation-specific transcription factor from S. coelicolor; AphS-Ct, transcriptional regulator of a phenol utilization operon from Comamonas testosteroni; BphS-Ps, transcriptional regulator of genes involved in polychlorinated biphenyl-biphenyl degradation in Pseudomonas sp. strain KKS102.
The morphological phenotype of SE69 is similar to that of the whiH mutants in that it fails to produce spores. Unlike the whiH mutants, however, which make wild-type levels of aerial mycelia with normal timing, aerial mycelium formation in SE69 is delayed. The whiH locus is one of several known genes required for septation of the aerial hyphae of S. coelicolor. Like SE69, whiH mutants initiate morphogenesis, as shown by the emergence of aerial hyphae, but they fail to develop the grey spore pigment associated with mature spores. Transcription of whiH is dependent on whiG, and whiH mutants are defective in expression of some late sporulation genes, including whiE and sigF. The whiH gene product has also been implicated in regulation of its own synthesis (29). While many GntR family proteins have been shown to act as repressor proteins that are responsive to carboxylate-containing intermediates in carbon metabolism, some have been shown to bind sites other than typical operator sequences, and at least one, FadR, can act as a transcriptional activator (8).
The SE69 (SCO7168) gene is located directly adjacent to a cluster of genes that may be translationally coupled (Fig. 3A). The products of these genes show amino acid sequence similarity to ATP binding cassette (ABC)-type membrane transporters. SCO7167 encodes a putative 425-amino-acid protein similar to a class of solute binding proteins. The mostly closely related protein with a known function is MsmE of Streptococcus mutans (35). MsmE is a lipoprotein involved in transport of multiple sugars. A highly conserved cysteine residue in this class of proteins, including the SCO7167 protein, allows attachment of the glyceride fatty acid lipid moiety of the protein responsible for membrane attachment (12). SCO7166 (encoding 310 amino acids) and SCO7165 (encoding 303 amino acids) encode putative integral transmembrane components of the ABC transporter. Like other members of this class of protein permeases, the SCO7166 and SCO7165 products contain a highly conserved signature region located near the C terminus, as well as membrane-spanning helixes typical of integral membrane proteins and cytosolic loops that extend outside the membrane (31). SCO7164 encodes a hypothetical protein of unknown function. SCO7163 encodes a putative protein with similarity to acyl/methyl transferases and aldolases. SCO7162 encodes a putative protein with similarity to aldo/keto reductases and sugar dehydrogenases. These proteins may be involved in the utilization of transported solutes. SCO7161 encodes a probable NAD- or NADP-dependent oxidoreductase similar to members of the short-chain dehydrogenase/reductase family. SCO7160 encodes a hypothetical protein of unknown function. Although there is no gene in this cluster that would be predicted to bind ATP, it would not be uncommon for the ATP binding protein component that participates in the transporter to be located elsewhere. While the ATP binding protein, MsiK, is not part of a cluster that encodes transport proteins, it has been shown to participate in both cellobiose and maltose transport in Streptomyces reticuli (32). Interestingly, the S. coelicolor genome has 81 predicted proteins that resemble typical ABC permeases and 141 predicted proteins that resemble ATP binding proteins. ABC transporters, regulated by GntR-like transcription factors, have been found in B. subtilis (38) and Streptomyces griseus (33).
ABC transporters have a wide variety of functions in solute transport, including transport of sugars, peptides, and amino acids, as well as roles in several possible drug efflux pathways. Perhaps most relevant in this context are the relatively well characterized bldK (23) and dasABC (33) ABC transport clusters that have been shown to be required for morphogenesis in S. coelicolor and S. griseus, respectively. Mutations in these clusters result in a Bld phenotype. The bldK cluster of S. coelicolor is responsible for the import of a small molecule, morphogen, which is required for sporulation (23). The organization of the dasABC cluster is very similar to that of the genes adjacent to SCO7168 (identified as the SE69 insertion), and dasR, like SCO7168, is a GntR-like transcriptional regulator. A detailed analysis of mutations in dasR and dasABC strongly suggested that dasR regulates transcription of the dasABC cluster in S. griseus (33).
We emphasize that the suggested functions of these proteins are tentative and are based entirely on predicted protein sequence similarities. One interesting possibility, however, is that the GntR-like protein identified by the SE69 mutation regulates genes required for the transport of glycosylated peptides and/or small molecules involved in morphogen-mediated signaling or the transport and utilization of sugars as carbon sources. In fact, many of the previously described bld genes in S. coelicolor have been shown to be defective in regulation of carbon utilization (26). The genes immediately adjacent to SCO7168 might also encode proteins involved in sugar transport. The fact that the SE69 mutant grows poorly on glucose, mannitol, or maltose as a carbon source may be the result of an effect of the mutation on the transport of these sugars.
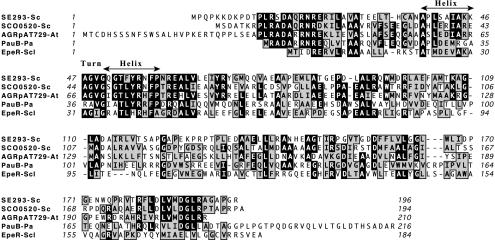
The SE293 mutant resulted from an insertion located within 2SCG38.28 (SCO1135), and the annotated region of the chromosome at this address is shown in Fig. 3B. The predicted 196-amino-acid protein encoded by the open reading frame identified by this insertion exhibited sequence similarity to the TetR family of transcriptional regulators (1, 2). TetR, AraC, MarR, and MerR represent families of transcriptional regulators involved in bacterial drug transport. They are present in distantly related species and show little correlation with the family of drug pumps whose expression they control. Assignment of these regulatory proteins to their families is based solely on similarities in the helix-turn-helix DNA binding domains (11). As shown in Fig. 5, a comparison of several TetR family proteins revealed particularly strong similarity between SE293 and the conserved TetR helix-turn-helix region, which is responsible for binding of these proteins to target DNA sequences. The best-characterized member of this family is the TetR protein encoded by Tn10. In the absence of tetracycline, TetR protein binds the tet operator and represses transcription of the genes for a tetracycline efflux pump. When tetracycline is present, it binds TetR, causing a conformational change in the protein, a loss of TetR binding to the Tet operator, and induction of genes required for tetracycline efflux (13, 30). While most TetR-like proteins function as repressors, HapR of Vibrio cholerae, which exhibits the highest levels of homology with both TetR and SE293 in the helix-turn-helix DNA binding region of the protein, has been shown to act as a transcriptional activator (15), leaving open the possibility that the protein encoded by the gene identified by SE293 may act as either an activator or a repressor. The genome sequence of S. coelicolor suggests that there are potentially 18 proteins in this strain with similarity to TetR that may bind DNA to affect transcriptional regulation.
FIG. 5.
End-to-end alignment of conserved (black background) and similar (gray background) amino acids in inferred sequences of proteins with similarity to the predicted protein encoded by the SCO1135 (SE293) open reading frame. Abbreviations: SE293-Sc, putative TetR family transcriptional regulator identified by transposon insertion in this study from S. coelicolor; SCO0520-Sc, putative TetR family transcriptional regulator from S. coelicolor; AGRpAT729-At, TetR family transcriptional regulator from Agrobacterium tumefaciens; PauB-Pa, TetR-like transcriptional regulator from Pseudonocardia autotrophica; EpeR-Scl, repressor protein of a gene predicted to be involved in multidrug import from Streptomyces clavuligerus.
SE293 (SCO1135) is located directly adjacent to a cluster of genes that are apparently translationally coupled and whose protein products exhibit amino acid sequence similarity to domains of proteins that function as oxidoreductases, ferredoxins, dehydrogenases, and carbon monoxide dehydrogenases (Fig. 3B). This cluster specifically contains genes which encode proteins that are similar to a class of enzymes that are members of the xanthine oxidase family. SCO1134 encodes a putative protein with an iron-sulfur binding signature region typical of 2Fe-2S ferredoxins. SCO1133 encodes a putative protein similar to a flavin adenine dinucleotide binding domain similar to those found in molybdopterin-containing proteins. SCO1132 encodes a putative protein similar to a molybdopterin binding domain. These proteins represent a special class of molybdenum cofactor binding proteins that require molybdenum in a complex with pterin. SCO1131 encodes a putative highly conserved protein of unknown function.
The xanthine oxidase family of proteins is conserved in eukaryotes, prokaryotes, and archaea and is one of the largest and most diverse families of proteins that contain molybdenum as a cofactor. These enzymes have broad and partially overlapping substrate specificities and typically contain two iron-sulfur binding clusters, one flavin adenine dinucleotide binding domain, and a molybedenum binding domain. The enzyme may consist of a single polypeptide or several polypeptides that function together (17). One possibility is that the proteins encoded by the genes adjacent to the mutation identified by SE293 function together as a single enzyme and that their transcription is regulated by a TetR-like transcription factor encoded by SCO1135. The fact that molybdenum binding is essential for the function of such a complex suggested a test for this hypothesis. Molybdopterin is one of two forms of biologically active molybdenum, and the biosynthesis and/or biological activity of molybdopterin is specifically inhibited by tungsten because of the chemical similarity between the two elements (18). If the mutation in SCO1135 eliminates a TetR-like transcriptional activator required for expression of the genes adjacent to it and if these genes encode a complex required for morphogenesis, then the addition of tungsten to wild-type cells might mimic the effect of the SE293 mutation. As shown in Fig. 2D, addition of tungstate to wild-type cells (at levels similar to those used in other studies to inactivate dimethyl sulfoxide reductase in E. coli [28] without affecting aerobic growth and viability) resulted in a phenotype similar to that of SE293.
Mutant SJ175 resulted from an insertion located within SCD66.12c (SCO4174) on the S. coelicolor chromosome, and the annotated region of the chromosome at this address is shown in Fig. 3C. The open reading frame identified by this insertion encodes a hypothetical 83-amino-acid protein with no significant similarity to any known protein in the database. This protein is postulated to be an integral membrane protein based on the fact that it contains hydrophobic residues similar to those found in the membrane-spanning regions of integral membrane proteins. The open reading frame immediately downstream of the SJ175 mutation encodes a hypothetical 115-amino-acid protein of unknown function without similarity to any known protein. The open reading frame immediately upstream of the SJ175 mutation encodes a hypothetical protein of unknown function but with similarity to another S. coelicolor protein of unknown function, TR:CAB697132. In addition to BLAST analysis, for SCO4174 and the genes surrounding it neighbor-joining trees were constructed (data not shown) for the 150 proteins most similar to those identified by the BLAST search. No obvious relationships between any of these predicted proteins and proteins having known functions were revealed.
Summary and conclusions.
Using an in vitro method of mutagenesis based on Tn5 developed by Goryshin and Reznikoff (10) and adapted for Streptomyces by Gehring et al. (9), we preformed a genome-wide screening analysis for mutants involved in sporulation and/or antibiotic production in S. coelicolor. Three new mutants were detected that identify previously uncharacterized genes: SE69 contains a mutation in a putative GntR-like transcriptional regulator, SE293 contains a mutation in a putative TetR-like transcriptional regulator, and SJ175 contains a mutation in a probable membrane protein of unknown function. Cotransformation was used to establish linkage between the transposon insertion and the phenotype of each mutant, and genetic complementation of two of the mutants confirmed that the transposon, in fact, identified a gene required for wild-type morphogenesis and antibiotic production. One of the mutants, the SE69 mutant, was complemented by a clone that contained a single intact open reading frame, suggesting that the phenotype resulting from the insertion was due to inactivation of a single gene.
One of the most interesting aspects of this analysis is that we did not identify alleles of bldA or bldB, which are by far the most common genes identified in mutant screens when chemical mutagenesis and UV mutagenesis are used (5), and although in part of this study we used the same genomic library that was used by Gehring et al., we recovered only one of the mutants that Gehring et al. recovered.
While the phenotypes of each of the mutants identified in this screen may be rationalized by their predicted functions, we emphasize that a complete study of these genes and their effects is needed to determine a definite mode of action or mechanism. As with other searches for genes required for morphogenesis and antibiotic production in these complex bacteria, the genes identified lead to more questions than answers, and the findings emphasize the fact that the network of pathways that these bacteria use to interact with their environment is indeed complex.
Acknowledgments
We thank Amy Gehring and Rich Losick for providing strains and materials prior to publication and for their help and advice throughout the course of this work, Mike Adams for suggesting the tungsten experiment and his help in executing it, Karen Stirrett for a critical review of the manuscript, and David Brown for help with preparation of the figures.
REFERENCES
- 1.Aramaki, H., N. Yagi, and M. Suzuki. 1995. Residues important for the function of a multihelical DNA binding domain in the new transcription factor family of Cam and Tet repressors. Protein Eng. 8:1259-1266. [DOI] [PubMed] [Google Scholar]
- 2.Beck, C. F., R. Mutzel, J. Barbe, and W. Muller. 1982. A multifunctional gene (tetR) controls Tn10-encoded tetracycline resistance. J Bacteriol. 150:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibb, M. J., V. Molle, and M. J. Buttner. 2000. σBldN, an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J. Bacteriol. 182:4606-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bignell, D. R., J. L. Warawa, J. L. Strap, K. F. Chater, nad B. K. Leskiw. 2000. Study of the bldG locus suggests that an anti-anti-sigma factor and an anti-sigma factor may be involved in Streptomyces coelicolor antibiotic production and sporulation. Microbiology 146:2161-2173. [DOI] [PubMed] [Google Scholar]
- 5.Champness, W. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J. Bacteriol. 170:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chater, K. F., C. J. Bruton, K. A. Plaskitt, M. J. Buttner, C. Mendez, and J. Helmann. 1989. The developmental fate of S. coelicolor hyphae depends crucially on a gene product homologous with the motility sigma factor of B. subtilis. Cell 59:133-143. [DOI] [PubMed] [Google Scholar]
- 7.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 8.DiRusso, C. C., A. K. Metzger, and T. L. Heimert. 1993. Regulation of transcription of genes required for fatty acid transport and unsaturated fatty acid biosynthesis in Escherichia coli by FadR. Mol. Microbiol. 7:311-322. [DOI] [PubMed] [Google Scholar]
- 9.Gehring, A. M., J. R. Nodwell, S. M. Beverley, and R. Losick. 2000. Genomewide insertional mutagenesis in Streptomyces coelicolor reveals additional genes involved in morphological differentiation. Proc. Natl. Acad. Sci. USA 97:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goryshin, I. Y., and W. S. Reznikoff. 1998. Tn5 in vitro transposition. J. Biol. Chem. 273:7367-7374. [DOI] [PubMed] [Google Scholar]
- 11.Grkovic, S., M. H. Brown, and R. A. Skurray. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi, S., and H. C. Wu. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 22:451-471. [DOI] [PubMed] [Google Scholar]
- 13.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 14.Ingram, C., M. Brawner, P. Youngman, and J. Westpheling. 1989. xylE functions as an efficient reporter gene in Streptomyces spp.: use for the study of galP1, a catabolite-controlled promoter. J. Bacteriol. 171:6617-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 16.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomuces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 17.Kisker, C., H. Schindelin, D. Baas, J. Retey, R. U. Meckenstock, and P. M. Kroneck. 1998. A structural comparison of molybdenum cofactor-containing enzymes. FEMS Microbiol. Rev. 2:503-521. [DOI] [PubMed]
- 18.Kletzin, A., and M. W. Adams. 1996. Tungsten in biological systems. FEMS Microbiol. Rev. 18:5-63. [DOI] [PubMed] [Google Scholar]
- 19.Lawlor, E. J., H. A. Baylis, and K. F. Chater. 1987. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1:1305-1310. [DOI] [PubMed] [Google Scholar]
- 20.Maniatis, T., J. Sambrook, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Molle, V., and M. J. Buttner. 2000. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol. Microbiol. 36:1265-1278. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen, K. T., J. Willey, L. D. Nguyen, L. T. Nguyen, P. H. Viollier, and C. J. Thompson. 2002. A central regulator of morphological differentiation in the multicellular bacterium Streptomyces coelicolor. Mol. Microbiol. 46:1223-1238. [DOI] [PubMed] [Google Scholar]
- 23.Nodwell, J. R., K. McGovern, and R. Losick. 1996. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22:881-893. [DOI] [PubMed] [Google Scholar]
- 24.Oh, S. H., and K. Chater. 1997. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J. Bacteriol. 179:122-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pope, M. K., B. Green, and J. Westpheling. 1998. The bldB gene encodes a small protein required for morphogenesis, antibiotic production, and catabolite control in Streptomyces coelicolor. J. Bacteriol. 180:1556-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pope, M. K., B. D. Green, and J. Westpheling. 1996. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell-cell signaling. Mol. Microbiol. 19:747-756. [DOI] [PubMed] [Google Scholar]
- 27.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 28.Rothery, R., J. L. S. Grant, J. Johnson, K. V. Rajagopalan, and J. H. Weiner. 1995. Association of molybdopterin guanine dinucleotide with Escherichia coli dimethyl sulfoxide reductase: effect of tungstate and a mob mutation. J. Bacteriol. 177:2057-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryding, N. J., G. H. Kelemen, C. A. Whatling, K. Flardh, M. J. Buttner, and K. F. Chater. 1998. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 29:343-357. [DOI] [PubMed] [Google Scholar]
- 30.Saenger, W., P. Orth, C. Kisker, W. Hillen, and W. Hinrichs. 2000. The tetracycline repressor—a paradigm for a biological switch. Angew. Chem. Int. Ed. Engl. 39:2042-2052. [DOI] [PubMed] [Google Scholar]
- 31.Saurin, W., W. Koster, and E. Dassa. 1985. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol. Microbiol. 12:993-1004. [DOI] [PubMed] [Google Scholar]
- 32.Schlosser, A., T. Kampers, and H. Schrempf. 1997. The Streptomyces ATP-binding component MsiK assists in cellobiose and maltose transport. J. Bacteriol. 179:2092-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo, J. W., Y. Ohnishi, A. Hirata, and S. Horinouchi. 2002. ATP-binding cassette transport system involved in regulation of morphological differentiation in response to glucose in Streptomyces griseus. J Bacteriol. 184:91-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun, J., G. H. Kelemen, J. M. Fernandez-Abalos, and M. J. Bibb. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145:2221-2227. [DOI] [PubMed] [Google Scholar]
- 35.Sutcliffe, I. C., L. Tao, J. J. Ferretti, and R. R. Russell. 1993. MsmE, a lipoprotein involved in sugar transport in Streptococcus mutans. J. Bacteriol. 175:1853-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willey, J., J. Schwedock, and R. Losick. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7:895-903. [DOI] [PubMed] [Google Scholar]
- 37.Willey, J., R. Santamaria, J. Guijarro, M. Geistlich, and R. Losick. 1991. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell 65:641-650. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida, K. I., Y. Fujita, and S. D. Ehrlich. 2000. An operon for a putative ATP binding cassette transport system involved in acetoin utilization in Bacillus subtilis. J. Bacteriol. 182:5454-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]