Abstract
Here we present the first metagenomic analyses of an uncultured viral community from human feces, using partial shotgun sequencing. Most of the sequences were unrelated to anything previously reported. The recognizable viruses were mostly siphophages, and the community contained an estimated 1,200 viral genotypes.
The human gut is colonized by an abundant, active, and diverse microbiota. This microbiota has been studied extensively using culture-based assays and, more recently, by a variety of molecular methods including fluorescent in situ hybridization, terminal restriction fragment length polymorphism, membrane assays, microarrays, and direct sequencing of 16S libraries (15, 18, 22, 25, 32, 35). These studies have shown that there are 400 to 500 human intestinal microbial species, with 30 to 40 species accounting for 99% of the total population (9, 15, 23, 24, 35).
Bacteriophages likely exert a strong influence on the diversity and population structure of bacterial communities in the human gut. Phages that infect Escherichia coli, Salmonella spp., and Bacteroides fragilis have been isolated from human fecal samples at concentrations ranging from 0 to 105 phages per g of dry feces (4, 7, 11-14, 16, 17, 19, 26, 28, 33). The presence and concentration of these phages differed among individuals and did not correlate with age or sex (12). Studies have also demonstrated prophage induction from human fecal bacteria upon treatment with DNA-damaging agents (5, 20). However, direct counts of bacteriophages in human feces have not been performed, nor have molecular methods been applied to study the phage populations. Here we present the first metagenomic analyses of the composition and population structure of an uncultured viral community from human feces.
Approximately 500 g of freshly voided fecal matter was collected from a 33-year-old healthy male. The fecal matter was resuspended in 5 liters of phosphate-buffered saline and shaken vigorously to dislodge the viral particles from the feces. The supernatant was poured through a Nitex filter (∼100-μm pore size) and then concentrated using a 100-kDa tangential flow filter. The concentrate was loaded onto a cesium chloride step gradient and ultracentrifuged, and the 1.35- to 1.5-g ml−1 fraction was collected. A portion of the viral concentrate was stained with 1× SYBR Gold (Molecular Probes) for 10 min and visualized by using epifluorescent microscopy (Fig. 1). When stained with SYBR Gold, prokaryotic cells are extremely bright and have visible morphologies. Prokaryotic cells are easily distinguishable from viruses, which are distinct pinpricks of light. As shown in Fig. 1, the purified viral concentrate was not contaminated with any microbial cells.
FIG. 1.
SYBR Gold-stained human fecal viral concentrate under epifluorescent microscopy. No contaminating microbial cells were observed.
DNA was extracted from the viral concentrate by using formamide and cetyltrimethylammonium bromide extractions (31). A linker-amplified shotgun library was then created from the human fecal viral DNA as described previously (3; M. Breitbart, B. Felts, S. Kelley, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer, submitted for publication; www.sci.sdsu.edu/PHAGE/LASL/index.htm). Briefly, the total viral community DNA was randomly sheared (HydroShear; GenMachine, San Carlos, Calif.) and end repaired and double-stranded DNA linkers were ligated to the ends. The fragments were amplified using Vent DNA polymerase, ligated into the pSMART vector (Lucigen, Middleton, Wis.), and electroporated into MC12 cells. This method overcomes limitations of modified nucleotides and bactericidal genes in viral genomes.
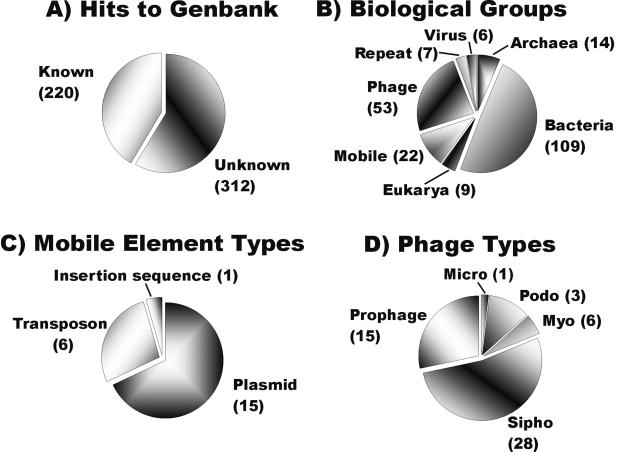
With the use of the AmpL2 forward primer (Lucigen), 532 clones from the library were sequenced (accession no. CC820769 to CC821300). TBLASTX comparison of these sequences against those in GenBank revealed that the majority (59%) of the sequences were not significantly similar (E value of <0.001) to anything previously reported (1, 2) (Fig. 2A). Sequences with significant hits were classified as phages, viruses, mobile elements, repeat elements, Bacteria, Archaea, or Eucarya based on GenBank annotation. Bacterial, archaeal, and eukaryotic hits were examined manually to identify repeat elements and potential prophages.
FIG. 2.
Genomic overview of the uncultured viral community from human feces based on TBLASTX sequence similarities. (A) Numbers of sequences with significant matches (E values of <0.001) in GenBank. (B) Distribution of significant matches among major classes of biological entities. (C) Types of mobile elements recognized in the library. (D) Families of phages identified in the fecal library.
The most common known matches were with bacteria. We do not believe that the presence of these sequences was due to contamination. Cesium chloride purification efficiently separates the viral particles from the prokaryotic cells and free DNA. As shown in Fig. 1, no contaminating microbial cells were present in the viral concentrate. Additionally, the open reading frames in purified cultured phage genomes are often more similar to bacterial open reading frames than to those of other phages (see, e.g., references 27 and 30). For several clones from a previously sequenced marine viral library which were sequenced from both ends, the match with one end of the clone was with a phage while the other end of the same clone had a significant match with a bacterial sequence (3). These bacterial hits may also represent uncharacterized prophages or sequences from transducing phages.
Significant matches with phages were the second most abundant category in the uncultured fecal library (Fig. 2B). Among the phage matches, the majority (81%) were with siphophages and prophages within bacterial genomes. Since many of these phages have the ability to be temperate, this suggests that temperate lifestyles may be important in the human colon. The most common phage matches were with bacteriophage A118 of Listeria monocytogenes, bacteriophage E125 of Burkholderia thailandensis, and bacteriophage bIL285 of Lactococcus lactis. Fifty-three percent of the phage matches were with known proteins, with structural proteins and terminases being the most common (Table 1). Several matches with mobile elements (plasmids, transposons, and insertion sequences) were also observed in the library (Fig. 2C).
TABLE 1.
Categories of phage proteins with significant matches in the uncultured human fecal viral library
| Protein type | No. of matches |
|---|---|
| Unknown | 25 |
| Structural protein | 8 |
| Terminase | 8 |
| Portal protein | 3 |
| Protease | 3 |
| Antirepressor | 1 |
| Endolysin | 1 |
| Endopeptidase | 1 |
| Polyketide synthase | 1 |
| Repressor | 1 |
| Tape measure protein | 1 |
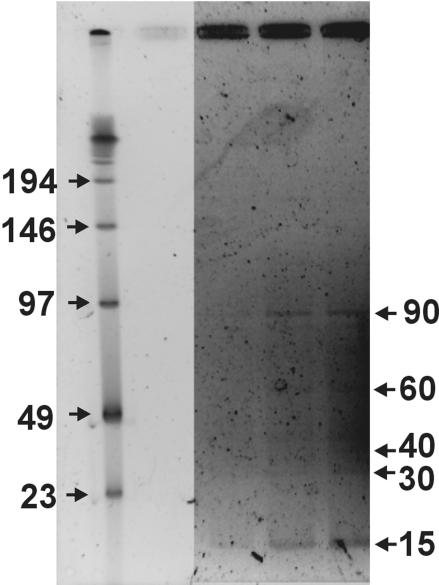
In order to determine the genome size distribution of uncultured bacteriophages in the human fecal sample, a fraction of the viral concentrate was examined using pulsed-field gel electrophoresis as described previously (10). Major bands for the fecal phage population were observed at 15 and 90 kb, with minor bands present at 30, 40, and 60 kb (Fig. 3). This genome size distribution was significantly different than that observed in other environments (e.g., seawater, sediment, and rumen) (10, 21, 36). Especially notable was the presence of a dominant band at a small genome size of approximately 15 kb.
FIG. 3.
Pulsed-field gel displaying the major genome size classes of viruses in the human fecal sample. The three sample lanes (on the right) represent increasing amounts of sample DNA. The DNA ladder is shown in the leftmost lane. The limited amount of DNA made it necessary to enhance the sample bands relative to the ladder by using Corel PHOTO-PAINT. The reported bands were visible by eye on the original gel.
The structure of the uncultured fecal viral population was determined by running Monte Carlo simulations to match the distribution of overlapping sequences (i.e., the contig spectrum) observed in the library (3; Breitbart et al., submitted). Among the 532 sequences from the fecal viral library, there were 18 contiguous sequences made up of two sequences (2-contigs), two 3-contigs, and two 4-contigs, as well as 482 sequences that did not overlap with any other sequences (1-contig). Based on the normalized band intensities of the pulsed-field gel, an average genome size of 30 kb was assumed for the population modeling. The population structure of the uncultured fecal viral community was assumed to follow a power law distribution (ni = ai−b, where ni is the relative frequency of the ith genotype, a is relative abundance of the most abundant genotype, i is the rank index from 1 to the total number of genotypes, and b is the evenness parameter) based on previous results (3). Using these assumptions, and an average fragment size of 699 bp, we calculated that the viral community contained ∼1,200 genotypes. The most abundant virus made up ∼4% of the total (Table 2). The nonparametric estimator Chao1 predicted 162 genotypes for this population (3, 6). Effects of varying the average genome size on the model predictions are shown in Table 2. Based on previous estimates of gut microbial diversity, there are approximately two to five times as many viral genotypes as the number of bacterial species in the human intestinal microbiota. Diversity of the fecal viral community was high, with a Shannon index (Hnats) of 6.4 nats. This is a higher Shannon index than that observed for most microbial communities, but it is lower than the Shannon index found for uncultured viral communities in seawater (Hnats = ∼7) and sediment (Hnats > 9) samples (3; Breitbart et al., submitted).
TABLE 2.
Population structure of the uncultured fecal viral community as determined by mathematical modelinga
| Assumed avg genome size (kb) | Total no. of viral genotypes | % Abundance of most abundant virus (a × 100) | Evenness parameter (b)b | Shannon index (Hnats) |
|---|---|---|---|---|
| 15 | 1,450 ± 700 | 2.4 ± 1.2 | 0.611 ± 0.110 | 6.83 |
| 30 | 1,250 ± 230 | 4.2 ± 0.7 | 0.715 ± 0.041 | 6.45 |
| 50 | 1,930 ± 470 | 6.3 ± 0.8 | 0.831 ± 0.029 | 6.43 |
The total number of viral genotypes is equivalent to species richness. Evenness is a measure of how genotypes are distributed within the population. The Shannon index is a measure of diversity that incorporates both species richness and evenness.
Rank-abundance curve shape, ni = ai−b.
In all the uncultured viral communities examined so far (3; Breitbart et al., submitted), the majority of the sequences have been novel. Similarly, culture-based studies of phage genomes have found that much, if not most, of phage diversity is unsampled (27, 29). Significant matches with each of the major groups of double-stranded DNA phages—siphophages, podophages, and myophages—have been observed in all uncultured viral communities studied. Siphophages and prophages were more abundant in the marine sediment (Breitbart et al., submitted) and fecal libraries than in the seawater libraries (3). The fecal viral community contained very few matches with the T7-like podophage and λ-like siphophage, which were the most abundant groups in the marine viral communities (Breitbart et al., submitted). In the fecal viral community, numerous similarities to phages that are known to infect gram-positive bacteria were seen. This can be explained by the fact that over 62% of the cells in human feces that can be detected with a bacterium-specific probe belong to gram-positive groups (15).
Understanding the population structure and dynamics of the normal human intestinal microbiota has important implications for human health, nutrition, and the development of probiotics for the treatment and prevention of gastrointestinal disorders. Since phages found in human feces are used as indicators of sewage-derived contamination of the environment, it is also important to know the identity of these phages (12). Phages likely influence the composition of bacterial populations in the intestine through specific predation on microbial hosts. As a particular strain becomes dominant, phages can infect and lyse that host, allowing another bacterial strain, either of endogenous or exogenous origin, the opportunity to become abundant (34). Additionally, through lysogenic conversion of resident intestinal bacteria, phages may introduce new phenotypic traits, such as antibiotic resistance and the ability to produce exotoxins (8).
Phages are a diverse and largely unexplored component of the microbial community in human feces. Using culture-independent methods, we have described the population structure and genome size distribution of a human fecal phage community. In addition, we have elucidated the identity of some of these uncultured phages based on sequence similarities. Future studies need to focus on determining the ecological roles of phages in the human intestine and the degree of similarity of the phage communities populating different individuals.
Acknowledgments
We thank Anca Segall, Beltran Rodriguez Brito, David Bangor, and Linda Wegley for helpful comments on the mathematical modeling and manuscript, as well as the San Diego State University Microchemical Core Facility for sequencing.
This work was supported by NSF DEB 03-16518 and DEB-BE0221763.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calci, K., W. Burkhardt, W. Watkins, and S. Rippey. 1998. Occurrence of male-specific bacteriophage in feral and domestical animal wastes, human feces, and human-associated wastewaters. Appl. Environ. Microbiol. 64:5027-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderaro, A., G. Dettori, L. Collini, P. Ragni, R. Grillo, P. Cattani, G. Fadda, and C. Chezzi. 1998. Bacteriophages induced from weakly beta-haemolytic human intestinal spirochaetes by mitomycin C. J. Basic Microbiol. 38:323-325. [PubMed] [Google Scholar]
- 6.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:783-791. [Google Scholar]
- 7.Cornax, R., M. Morinigo, F. Gonzalez-Jaen, M. Alonso, and J. Borrego. 1994. Bacteriophages presence in human faeces of healthy subjects and patients with gastrointestinal disturbances. Zentbl. Bakteriol. 281:214-224. [DOI] [PubMed] [Google Scholar]
- 8.Davis, B. M., and M. K. Waldor. 2002. Mobile genetic elements and bacteria pathogenesis, p. 1040-1055. In N. L. Craig, R. Gragie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 9.Drasar, B., and P. Barrow. 1985. Intestinal microbiology. American Society for Microbiology, Washington, D.C.
- 10.Fuhrman, J. A., J. F. Griffith, and M. S. Schwalbach. 2002. Prokaryotic and viral diversity patterns in marine plankton. Ecol. Res. 17:183-194. [Google Scholar]
- 11.Furuse, K., S. Osawa, J. Kawashiro, R. Tanaka, Z. Osawa, S. Sawamura, Y. Yanagawa, T. Nagao, and I. Watanabe. 1983. Bacteriophage distribution in human faeces: continuous survey of healthy subjects and patients with internal and leukemic diseases. J. Gen. Virol. 64:2039-2043. [DOI] [PubMed] [Google Scholar]
- 12.Gantzer, C., J. Henny, and L. Schwartzbrod. 2002. Bacteroides fragilis and Escherichia coli bacteriophages in human faeces. Int. J. Hyg. Environ. Health 205:325-328. [DOI] [PubMed] [Google Scholar]
- 13.Grabow, W., C. Holtzhausen, and C. De Villiers. 1993. Research on bacteriophages as indicators of water quality. Water Research Commission report N 321/1/93, p. 147. Water Research Commission, Pretoria, South Africa.
- 14.Grabow, W., T. Neubrech, C. Holtzhausen, and R. Jofre. 1995. Bacteroides fragilis and Escherichia coli bacteriophages: excretion by humans and animals. Water Sci. Technol. 31:223-230. [Google Scholar]
- 15.Harmsen, H., G. Raangs, T. He, J. Degener, and G. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havelaar, A. 1993. A bacteriophage standard for bathing waters. Final report, Commission of the European Communities, B4:3040/92/012609. European Commission, Brussels, Belgium.
- 17.Havelaar, A., W. Pot-Hogeboom, K. Furuse, R. Pot, and M. Hormann. 1990. F-specific RNA bacteriophages and sensitive host strains in faeces and wastewater of human and animal origin. J. Appl. Bacteriol. 69:30-37. [DOI] [PubMed] [Google Scholar]
- 18.Hold, G., S. Pryde, V. Russell, E. Furrie, and H. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 19.Kai, S., S. Watanabe, K. Furuse, and A. Osawa. 1985. Bacteroides bacteriophages isolated from human faeces. Microbiol. Immunol. 29:895-899. [DOI] [PubMed] [Google Scholar]
- 20.Kameyama, L., L. Fernandez, J. Calderon, A. Ortiz-Rojas, and T. Patterson. 1999. Characterization of wild lambdoid bacteriophages: detection of a wide distribution of phage immunity groups and identification of a Nus-dependent, nonlambdoid phage group. Virology 263:100-111. [DOI] [PubMed] [Google Scholar]
- 21.Klieve, A. V., and R. A. Swain. 1993. Estimation of ruminal bacteriophage numbers by pulsed-field gel electrophoresis and laser densitometry. Appl. Environ. Microbiol. 59:2299-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Miyamoto, T. Takada, K. Matsumoto, H. Oyaizu, and R. Tanaka. 2002. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore, W., and L. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, W., and L. Moore. 1995. Intestinal floras of populations that have a high risk of colon cancer. Appl. Environ. Microbiol. 61:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagashima, K., T. Hisada, S. Maremi, and J. Mochizuki. 2003. Application of new primer-enzyme combinations to terminal restriction fragment length polymorphism profiling of bacterial populations in human feces. Appl. Environ. Microbiol. 69:1251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osawa, A., K. Furuse, and I. Watanabe. 1981. Distribution of ribonucleic acid coliphages in animals. Appl. Environ. Microbiol. 41:164-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, and N. R. Pannunzio. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 28.Puig, A., N. Queralt, J. Jofre, and R. Araujo. 1999. Diversity of Bacteroides fragilis strains in their capacity to recover phages from human and animal wastes and from fecally polluted wastewater. Appl. Environ. Microbiol. 65:1772-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohwer, F. 2003. Global phage diversity. Cell 113:141. [DOI] [PubMed] [Google Scholar]
- 30.Rohwer, F., A. M. Segall, G. Steward, V. Seguritan, M. Breitbart, F. Wolven, and F. Azam. 2000. The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with non-marine phages. Limnol. Oceanogr. 42:408-418. [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tartera, C., and R. Jofre. 1987. Bacteriophages active against Bacteroides fragilis in sewage-polluted waters. Appl. Environ. Microbiol. 53:1632-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thingstad, T. F., and R. Lignell. 1997. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 13:19-27. [Google Scholar]
- 35.Wang, R., S.-J. Kim, L. Robertson, and C. Cerniglia. 2002. Development of a membrane-array method for the detection of human intestinal bacterial in fecal samples. Mol. Cell. Probes 16:341-350. [DOI] [PubMed] [Google Scholar]
- 36.Wommack, K. E., J. Ravel, R. T. Hill, J. Chun, and R. R. Colwell. 1999. Population dynamics of Chesapeake Bay virioplankton: total-community analysis by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]