Abstract
In order to obtain particulate methane monooxygenase (pMMO)-enriched membranes from Methylococcus capsulatus (Bath) with high activity and in high yields, we devised a method to process cell growth in a fermentor adapted with a hollow-fiber bioreactor that allows easy control and quantitative adjustment of the copper ion concentration in NMS medium over the time course of cell culture. This technical improvement in the method for culturing bacterial cells allowed us to study the effects of copper ion concentration in the growth medium on the copper content in the membranes, as well as the specific activity of the enzyme. The optimal copper concentration in the growth medium was found to be 30 to 35 μM. Under these conditions, the pMMO is highly expressed, accounting for 80% of the total cytoplasmic membrane proteins and having a specific activity as high as 88.9 nmol of propylene oxide/min/mg of protein with NADH as the reductant. The copper stoichiometry is ∼13 atoms per pMMO molecule. Analysis of other metal contents provided no evidence of zinc, and only traces of iron were present in the pMMO-enriched membranes. Further purification by membrane solubilization in dodecyl β-d-maltoside followed by fractionation of the protein-detergent complexes according to molecular size by gel filtration chromatography resulted in a good yield of the pMMO-detergent complex and a high level of homogeneity. The pMMO-detergent complex isolated in this way had a molecular mass of 220 kDa and consisted of an αβγ protein monomer encapsulated in a micelle consisting of ca. 240 detergent molecules. The enzyme is a copper protein containing 13.6 mol of copper/mol of pMMO and essentially no iron (ratio of copper to iron, 80:1). Both the detergent-solubilized membranes and the purified pMMO-detergent complex exhibited reasonable, if not excellent, specific activity. Finally, our ability to control the level of expression of the pMMO allowed us to clarify the sensitivity of the enzyme to NADH and duroquinol, the two common reductants used to assay the enzyme.
In the methanotroph Methylococcus capsulatus (Bath), the oxidation of methane to methanol is catalyzed by methane monooxygenase. The process is driven by either NADH (1, 2) or duroquinol (2, 3, 22, 25). In addition, there are two distinct forms of the enzyme (14) that are associated with different gene products. One is the soluble methane monooxygenase (sMMO) expressed in the cytosolic portion of cells grown under copper-limiting conditions (10, 12, 13, 14). The other enzyme is the particulate methane monooxygenase (pMMO), a membrane-associated protein that is expressed in the presence of high copper-to-biomass ratios.
Membrane proteins are difficult to purify, and the pMMO has resisted numerous attempts to purify it for detailed characterization. It is well known that M. capsulatus has difficulty surviving at copper concentrations in the range from 10 to 20 μM because of the instability of the system under these conditions (15, 17). On the other hand, copper ions become toxic to the cells at concentrations higher than 50 μM (15). Expression of the pMMO is accompanied by the formation of an extensive network of intracytoplasmic membranes, where the membrane-bound pMMO resides. Three polypeptides with apparent molecular masses of 45, 27, and 23 kDa have been observed in the membrane fractions when M. capsulatus (Bath) switches from expressing the sMMO to expressing the pMMO. Addition of increasing amounts of copper ions to the growth medium leads to synthesis of additional intracytoplasmic membranes, increased expression of the pMMO, increased growth yields, and a concomitant loss of sMMO activity (15, 19, 24).
The regulation of gene expression by copper ions in methanotrophs for the production of the two methane monooxygenases is a complex process. A large decrease in the copper ion concentration in the growth medium accompanies culturing of methanotrophs in a fermentor, particularly at high cell densities or a high biomass level, during cell growth. In addition, metabolites and toxins that are toxic to the cells accumulate in the growth medium. Processing the cells in a fermentor adapted with a hollow-fiber bioreactor that allows easy control and quantitative adjustment of the copper ion concentration in NMS medium over the time course of culturing of the cells could alleviate these problems. In addition, the same hollow-fiber membrane bioreactor could be used to remove deleterious substances from the growth medium to minimize the interference of these metabolites with cellular growth. In the present study, we devised a reproducible procedure for preparation of high-quality pMMO from M. capsulatus (Bath) in high yields in a Bioflo 3000 fermentor that was adapted with a hollow-fiber membrane bioreactor.
Production of high-quality pMMO-enriched membranes would greatly facilitate purification of the pMMO. In addition, such pMMO-enriched membranes would allow characterization of the pMMO with greater precision and with less interference from other proteins. Almost a decade ago, it was proposed on the basis of electron paramagnetic resonance (EPR) and X-ray absorption spectroscopy experiments that pMMO is a multicopper protein (16). In addition, it was hypothesized that the copper ions are arranged in five trinuclear copper clusters. Two of these clusters were subsequently found to participate in dioxygen chemistry, and it was noted that the remaining copper ions were in the reduced d10 state. Accordingly, the two types of copper ions were referred to as C-clusters and E-clusters, respectively. The E-clusters were thought to be responsible for channeling electrons to the C-clusters from NADH, where the hydroxylation chemistry takes place. These copper ions, when reduced, could also act as an electron reservoir to store reducing equivalents and maintain a high internal electron pressure within the enzyme. As expected, the copper ions in the two C-clusters had to be fully reduced before the enzyme was functional (15). Since the suicide substrate acetylene was known to modify the 27-kDa subunit (4, 18), it was generally assumed that the C-clusters were associated with this subunit (16). The three E-clusters were found to be associated with the water-exposed domains of the 45-kDa subunit (6).
MATERIALS AND METHODS
Culturing of bacteria.
M. capsulatus (Bath) strain ATCC 33009 was maintained on petri plates containing NMS medium (1306 nitrate mineral salts medium [American Type culture Collection]) solidified with 1.7% agar. Cultures were maintained under an atmosphere consisting of 20% methane in air and were streaked onto fresh plates every 4 to 6 weeks. The organisms were transferred first from petri plates to 250-ml flasks containing 30 ml of NMS medium and subsequently to 2-liter Erlenmeyer flasks containing 300 ml of NMS medium; in each case the atmosphere consisted of 20% methane in air. After 48 h of incubation with continual shaking, each resulting cell suspension was used to seed a Bioflo 3000 fermentor with a 5-liter vessel containing 3 liters of NMS medium. The iron concentration in the growth medium was typically 18 μM throughout the experiments. No extra iron was added. When the optical density at 595 nm of the culture medium reached 1.2 to 1.6, typically after 24 h of incubation, additional NMS medium was added to increase the total culture volume to 5 liters for further semicontinuous growth.
It was usually necessary to replenish the cell-enriched medium with fresh NMS medium by draining the used medium in the bioreactor two or three times every 12 to 24 h. This medium replenishment kept the cell culture in the mid-log to late-log phase during the period used. With M. capsulatus (Bath), it was possible to keep the cells growing at pH 6.8 to 7.4 for 12 to 24 h by agitating the cellular suspension at 200 to 800 rpm and controlling the methane and air feed rates (0.7 to 1.3 liters/min) in order to maintain the dissolved oxygen content at 2 to 5% of saturation. During the culturing process, cell growth was always accompanied by an increase in the pH of the culture medium. Increasing the agitation speed increased the oxygen content in the solution and resulted in a temporary decrease in the pH. However, the pH always increased following more bacterial growth and after more methane and oxygen were consumed. We harvested the cells when the agitation speed reached 800 rpm and the pH was 7.4 (optical density at 595 nm after 25-fold dilution, 0.35). After this point, we would have risked coprecipitation of the buffering phosphate salts in the presence of the dissolved copper and calcium ions at the high pH values, a chemical instability that could culminate in a precipitous decrease in the pH of 3 to 4 U and could eliminate the buffering effect of the culture medium instantaneously.
After the cells had been cultured at a specific copper concentration (see below) for 12 to 24 h and were ready for harvest, about 4 liters of the medium was removed with the assistance of the hollow-fiber filter. Three-quarters of the resulting concentrated cell suspension was removed and centrifuged, and the harvested pellet was retained for further characterization. The remainder of the concentrated cell suspension was then filtered almost to dryness. The cells were resuspended in the fermentor with 5 liters of fresh NMS medium injected through the inlet tube into the hollow-fiber filter for the next round of culturing. Typically, about 35 to 40 g of cells was harvested during the medium renewal process described above. During the next round of culturing, the cell concentration usually recovered to 10 to 12 g/liter within the next 12 h.
Regulation of the copper concentration.
At the beginning of the culturing experiments, the M. capsulatus (Bath) cells were cultured in the copper-limiting NMS medium. After 2 days of culturing in this medium, the copper concentration was increased gradually by adding either copper sulfate (less than 10 μM) or both copper sulfate and copper EDTA, each in 5 μM increments, to refresh the NMS medium during the medium renewal process. It was possible to maintain the total copper concentration at a specific level for 2 to 3 days. By controlling the copper ion concentration during cell growth in this manner, we found that the cells could survive even in the presence of 60 μM copper ions in NMS medium.
Preparation of pMMO-enriched membranes.
The harvested cell paste was suspended in 25 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer at pH 7.0 (1.2 ml of buffer/g of cell paste) containing 0.01 mg of DNase I per ml. Cell suspensions were passed three times through a French pressure cell at 20,000 lb/in2 to separate the cytosolic and membrane fractions. Unlysed cells and cell debris were removed by centrifugation at 27,000 × g for 20 min. The supernatant was then ultracentrifuged at 220,000 × g for 40 min to pellet the membrane fraction. The clear supernatant obtained after ultracentrifugation was used as the cytosolic fraction. The pelleted membranes often had distinct layers. The minor bottom layer containing bluish and black materials and the thin white top layer were discarded. Only the middle layer, the translucent intracytoplasmic membrane fraction, which contained the bulk of the membrane proteins, was collected. The translucent membranes were washed by suspending them in washing buffer containing 25 mM PIPES, 5 mM ascorbate, and 25 μg of catalase/ml (pH 7.0) by using a Dounce homogenizer, repelleted by ultracentrifugation, and resuspended in washing buffer whose volume was two to three times the volume of the original cell suspension. This process was repeated two or three times until the supernatant was virtually free of soluble proteins.
Metal contents of the pMMO-enriched membranes derived from cells cultured in the presence of various copper concentrations in the growth media.
Analysis of metal ions (copper, iron, and zinc) was performed by inductively coupled plasma mass spectroscopy (ICP-MS). Samples of membrane fractions were digested and dissolved in distilled Q-water containing 1 N HNO3 (Supra-pure grade; Merck) at 45°C in acid-washed crucibles. Each digested sample was diluted by using 0.1 N HNO3 solutions prior to copper analysis. The copper concentrations of the samples were determined by comparison with standard solutions of Cu(NO3)2 in 0.1 N HNO3. A solution of 0.1 N HNO3 in distilled water was used as the copper-free control. The values reported below are the averages of three separate determinations. The same samples were used for iron and zinc analyses. The standards were prepared by diluting an iron and zinc atomic absorption standard (Sigma) with 0.1 N HNO3.
The protein concentration in the membranes was determined by the method of Lowry et al. (11). The pMMO content, expressed as a percentage of the 40-μg total protein sample applied to the gel, was quantified by imaging the preformed sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gel (sum of the bands at 45, 27, and 23 kDa) with Scion Image for MS Windows. An uncalibrated optical density was determined for each of the bands by converting the average gray value measured by the image scanner for each band, and this optical density was then normalized to the optical density for the total protein. The average optical density for the total membrane protein (40.0 μg) for the various copper concentrations was determined to be 0.40 (average of lanes containing 0.3 to 60 μM), and the standard deviation was 0.08. For the optical density of 0.11 measured for the six protein markers (11.6 μg of protein) (Amersham Pharmacia Inc.), a 40-μg sample of membrane protein gave an optical density of 0.38, which is in reasonable agreement with the observed uncalibrated value. This result was used to renormalize the optical densities of the bands to obtain the protein contents in the various membrane protein gels. The metal contents of the pMMO were derived from the results of the metal analysis together with the pMMO contents determined for the total proteins in the membranes as described above.
pMMO activity assays.
The methane monooxygenase activities of the samples were determined by propylene epoxidation assays. In the case of the membranes, 0.4 ml of 25 mM PIPES buffer (pH 7) was typically added to 0.1 ml of a stock solution of a pMMO-enriched membrane suspension. Typically, the reaction mixtures contained 100 μM CuSO4 and 5 mM NADH. It was shown previously that extra CuSO4 added to the assay medium enhanced the stability of the pMMO when the cells were grown at copper ion concentrations below 10 μM (15, 21). No effects on the stability or the activity of the protein were noted when the cells were grown at higher copper ion concentrations (20 to 60 μM). Typically, the effective copper ion concentration for the pMMO used in the activity assays was about 5 mM, so the added CuSO4 was only incremental.
When the duroquinol-dependent pMMO activity was examined, NADH was replaced by duroquinol, and the reductant concentrations studied were 1, 2, and 5 mM. Since duroquinol is not soluble in water, enough of the reductant was added as a solid to the assay medium to give the desired overall concentration (22). The duroquinol was preferentially partitioned into the membrane, and the actual concentration of the duroquinol in the membrane was estimated to be ∼10-fold higher. The effect of duroquinone on the NADH-dependent activity was also determined. Again, the duroquinone should have been associated with the membrane only due to its limited solubility in the aqueous phase.
The assays were performed at 45°C with a gas consisting of air and propylene at a ratio of 50/50 (vol/vol). After 0.1, 0.2, 0.5, 1, 3, 5, 10, 20, 30, and 60 min of incubation, 0.10-ml aliquots of the reaction mixtures were withdrawn and placed in 1.5-ml Eppendorf tubes. To the reaction mixture in each of the Eppendorf tubes, 0.10 ml of methylene chloride was added and then centrifuged with a Sorval microcentrifuge at 8,000 rpm for 5 min. The organic layer was dried over anhydrous MgSO4 and filtered through a pad of silica gel. Oxidation products (propylene oxide; retention time (tR), 5.98 min) were identified and quantified by calibration with an internal standard (cyclohexane; tR, 7.42 min) by gas chromatography. The activity of the pMMO was determined from the limiting initial slope of the product-versus-time plot. The specific activity was then obtained by dividing the observed activity by the total amount of pMMO as determined by the method of Lowry et al. by using the detergent-compatible reagent (Bio-Rad Inc.) and the gel electrophoresis-image quantitation procedure described above.
Duroquinone and preparation of duroquinol.
Duroquinone (tetramethyl-p-benzoquinone) was obtained from Sigma and was used without purification. The duroquinone was reduced to the corresponding quinol by using a 1.5-fold molar excess of solid NaBH4 in an acidified ethanol solution. Duroquinol was isolated as a white powder by using the procedure of Shiemke et al. (22).
In-gel protein digestion and analysis of the peptide fragments of the pMMO subunits.
The identities of the pMMO subunits were verified by peptide mass fingerprinting by using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). The subunits isolated from SDS-PAGE gels were each subjected to protease digestion by using trypsin. Each stained band of interest was sliced into 1-mm cubes and washed three times with 50% acetonitrile in 25 mM ammonium bicarbonate buffer (pH 8.0) for 15 min at room temperature. The gels were soaked in 100% acetonitrile for 5 min and then dried over a lyophilizer for 20 to 30 min. Before proteolytic digestion, the gels were rehydrated in 25 mM ammonium bicarbonate buffer (pH 8.0) containing 10 μg of trypsin per ml (35 μl; ratio of protein to trypsin, ∼100/1) until the gel pieces were fully immersed. After incubation for 18 to 20 h at 37°C, the trypsin solution was transferred into a new Eppendorf tube. The gel pieces remaining were reextracted with 50% acetonitrile in 5.0% trifluoroacetic acid (TFA) for 60 min. The two extracts were combined and then dried over a lyophilizer. Occasionally, the sample was also rehydrated and treated with C18 ZipTip for desalting and removal of residual acrylamide and other condensates.
Prior to MALDI-TOF MS analysis, all the samples treated by in-gel digestion were dissolved in a 0.1% TFA solution and then treated with a matrix solution consisting of saturated (10 mg/ml) α-cyano-4-hydroxycinnamic acid premixed in a 30% acetonitrile-0.1% TFA solution at a ratio of 1:10 (vol/vol). The sample was then spotted onto the MALDI target plate, dried, and subjected to analysis by MALDI-TOF MS.
Database searching of MALDI-TOF mass spectra was accomplished as follows. Monoisotopic mass values were used to search the possible amino acid compositions of the peptides based on the National Center for Biotechnology Information database by using the MS-Fit search engine (http://prospector.ucsf.edu). The tolerance for the input mass was set at 100 ppm, and the number of missed cleavages was set at 1 to 3.
Membrane solubilization and protein purification by size exclusion chromatography.
Since almost 80% of the membrane protein was the desired pMMO, it was not necessary to resort to complex chromatography to purify the protein, as was done previously (17). In principle, it should be possible to solubilize the membranes in detergent and fractionate the proteins according to molecular size to recover the pMMO-detergent complex.
The membrane suspension in PIPES buffer was first degassed with several vacuum-argon cycles to remove dioxygen. Sufficient dodecyl β-d-maltoside was then added to obtain a final concentration of 5% (wt/vol) or ∼1 mg of detergent/mg of protein. The mixture was mixed vigorously, incubated on ice for 1 h, and then centrifuged at 37,000 × g for 45 min to remove unsolubilized material. The clear supernatant was considered the solubilized membranes and was used for subsequent steps. The detergent concentration was critical in these experiments as the pMMO exhibited instability at both low and high detergent concentrations. At detergent/protein ratios much less than 1 mg of detergent/mg of protein, there was insufficient detergent to form the solubilized protein-detergent complex. At significantly higher concentrations of the detergent (for example, concentrations greater than 3 mg of detergent/mg of protein), the enzyme was inactivated. Under these conditions, the protein dissociated due to differential solubility of the protein subunits in the detergent, and the 45-kDa subunit precipitated out of solution (17).
To obtain the purified pMMO, the solubilized membranes were then subjected to Äkta FPLC by using an S-300HR gel filtration column (Amersham Pharmacia Inc.) that was previously equilibrated with buffer containing 50 mM PIPES, 150 mM NaCl, 50 mM imidazole, 5 mM ascorbate, and 0.05% (wt/vol) dodecyl β-d-maltoside (pH ∼7.2). The pMMO-detergent complex eluted according to its molecular size. As the major protein in the membrane suspension, it eluted at the maximum of the eluting fraction, as expected.
Electrophoresis.
Electrophoresis was performed on a 13.3% acrylamide-SDS gel covered with a 4.5% acrylamide stacking gel. Both the 4.5% acrylamide stacking gel and the 13.3% acrylamide-SDS gel were prepared by using a stock solution containing 38.67% (wt/wt) acrylamide and 1.33% (wt/wt) N,N′-bis(methylene acrylamide). Electrophoresis was carried out at 100 V until the bromophenol blue marker reached the bottom of the gel (about 2.5 h). The gel was stained in a 0.25% (wt/vol) Coomassie brilliant blue G-250 solution containing 25% methanol for 20 to 30 min and then destained in a destaining solution containing 25% methanol and 7% acetic acid.
Molecular mass identification of the pMMO subunits.
In order to determine the composition of the purified pMMO-detergent complex, the molecular masses of the subunits were determined by MALDI-TOF MS. A sample of the complex was dissolved in a 0.1% TFA solution and then treated with a matrix solution consisting of saturated (10 mg/ml) α-cyano-4-hydroxycinnamic acid premixed in a 30% acetonitrile-0.1% TFA solution at a ratio of 1:10 (vol/vol). The sample was then spotted onto the MALDI target plate, dried, and subjected to analysis by MALDI-TOF MS.
Instrumentation.
A Bioflo 3000 fermentor (New Brunswick Inc.) equipped with a 5-liter vessel and a hollow-fiber filter (pore size, 0.2 μm; 25 by 1.25 in.; capacity, 2,790 cm3; A/G Technology Co.) was used throughout the culture experiments. The methane-air feeding rate was adjusted by keeping the oxygen content of the culture medium at around 2 to 5% of the dissolved oxygen at saturation by using a gas dispersion system.
Metal analysis was performed by atomic absorption spectroscopy (Z-8200; Hitachi), ICP-MS (SCIEX-ELAN 5000; Perkin-Elmer), and inductively coupled plasma optical emission spectroscopy (OPTIMA 2000; Perkin-Elmer).
X-ray absorption spectroscopy data were collected at a sample temperature of 10 K at the National Synchrotron Radiation Research Center in Hsinchu, Taiwan, by using an Si(III) double-crystal monochromator on beamline Wiggler 17C. Fluorescence data were collected by using an Ar-filled ionization chamber. The edge energies were calibrated by assigning the first inflection points of Fe, Cu, and Zn foils to 7112.0, 8980.3, and 9659.0 eV, respectively. For metal analysis, the edge jumps were calibrated by using CuSO4, FeSO4, and ZnSO4 solution standards to relate the observed fluorescence intensity to the metal content for each metal.
EPR spectra were obtained at the X-band by using a Bruker E500 spectrometer equipped with a Bruker TE102 cavity. The sample temperature was maintained at 77 K by immersing the EPR tube in a liquid nitrogen-containing finger dewar.
Gas chromatography analyses were performed by using a Hewlett-Packard 6890plus gas chromatograph equipped with a flame ionization detector. The gas chromatography conditions were as follows: fused-silica capillary column that was 30 m long and had an inside diameter of 0.25 mm; stationary phase, HP-5; carrier gas, nitrogen at a flow rate of 0.5 ml/min; oven temperature, isothermal at 35°C; injection temperature, 250°C; and detector temperature, 250°C.
All MALDI-TOF spectra were acquired with a Voyager-DE PRO (PerSeptive Biosystems) operating in reflector, delayed extraction mode. The instrument was equipped with a 337-nm nitrogen laser source that delivered pulses of light at 3 Hz to the matrix spot. The spectra were recorded in reflector mode at an accelerating voltage of 20 kV with 70% grid voltage, 0% guide wire voltage, a 100-ns delay, and a low mass gate (500 Da). Fifty laser shots were averaged per spectrum. External mass calibration was usually used, based on a mixture of three reference peptides (human angiotensin I, adrenocorticotropin clip 1-17, and adrenocorticotropin clip 18-39) covering the m/z range from 500 to 3500. All MALDI-TOF mass spectra were analyzed by using Data Explorer (PE Biosystems).
RESULTS
Copper regulation and hollow-fiber bioreactor.
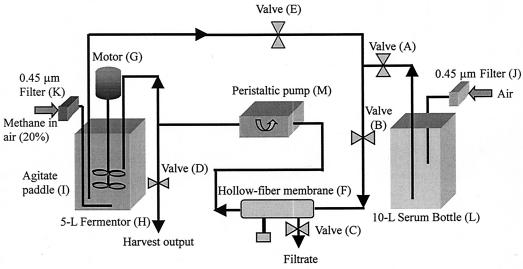
We attempted to solve two technical problems as part of this study. One problem was to grow M. capsulatus cells in the presence of well-defined copper concentrations so that the cells were not subjected to gross variations in the copper ion levels during the time of culturing. We needed to grow the cells in the presence of a wide range of specific copper concentrations but under otherwise identical conditions. Since such cells do not tolerate large changes in the copper concentration (17), particularly at copper ion concentrations in the range from 50 to 60 μM, it was possible to reach these high copper concentrations only by first incubating the cells at lower copper concentrations and then gradually increasing the copper concentration. The second technical problem was associated with the quality of the cell culture, mostly notably its heterogeneity, as it was gradually exposed to a continually changing chemical environment during the prolonged culture period. We brought these two problems together and solved them by adapting a hollow-fiber filter to the Bioflo 3000 fermentor (Fig. 1). The hollow-fiber membrane bioreactor enabled us to filter off used NMS medium in order to discard cellular waste and toxic metabolites from the cellular milieu, to replenish the cellular medium in order to maintain an essentially constant environment, and to adjust the copper concentration in the growth medium in a controlled manner in increments that were not harmful to the cells.
FIG. 1.
Schematic diagram of a fermentor adapted with a hollow-fiber membrane bioreactor to culture M. capsulatus (Bath) for the production of high-quality pMMO in high yields.
In our setup, the cell medium was continuously drained through a hollow-fiber membrane bioreactor consisting of a large number of tubular fiber membranes encased in a cylindrical shell with a pore size that was much smaller than the size of bacteria but large enough to allow passage of CuSO4 and small metabolites. In this dual reactor, the bulk of the bacterial cells were continuously flowing through the high-surface-area lumen of the fiber and the used NMS medium containing the waste and small molecules was filtered away from the cell medium across the hollow-fiber membranes for removal. At the same time, the discarded medium was replenished with fresh growth medium with a controlled CuSO4 concentration to maintain the copper ions at a constant level.
The combined fermentor-hollow-fiber bioreactor proved to be a high-capacity and highly efficient system. With this system we could harvest more than 50 g of cells per day. In addition, the system was very robust, lasting for months. Taking advantage of these features, we performed a detailed study of the effects of copper concentration in the growth medium on the pMMO-enriched membranes and the pMMO protein. Cells could be incubated in the presence of various copper concentrations up to 60 μM. When the copper concentration exceeded 60 μM, the cells could no longer be activated, even after replenishment of the growth medium at a lower copper concentration. Otherwise, high-quality pMMO-enriched membranes were obtained in high yields.
Copper uptake analysis.
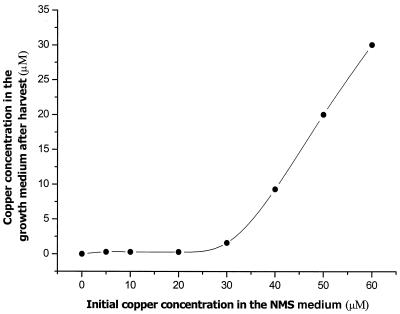
In addition to being a transcriptional switch, copper ions serve as a metabolic activator to produce large quantities of pMMO, as well as cell membranes to support the pMMO. In addition, since pMMO is a multicopper protein that requires many copper ions for biological function (4, 15, 16, 17, 23, 24), there must be a threshold copper concentration in the growth medium during cell culturing before the expressed pMMO can incorporate the full complement of copper ions to assemble the fully functional enzyme. To illustrate this point, the copper uptake by the cells was examined by measuring the difference between the copper concentration of an applied fresh medium and the copper concentration remaining in the used medium after growth of the cell suspension to the mid-log phase. The concentration of the copper ions in the growth medium was determined by ICP-MS after the cells were harvested, and it was compared with the copper ion concentration in the original replenishing medium. The results are shown in Fig. 2. The cells took up almost all of the copper ions when the initial copper concentration was below 30 μM. On the other hand, no additional copper ions were taken up from the replenishment medium when the cells were grown at copper concentrations approaching 40 μM; that is, the cells were apparently saturated with copper. These important results underscore not only the role of copper ions as a metabolic activator but also the finding that pMMO is a multicopper protein that requires many copper ions for full activity.
FIG. 2.
Comparison of the copper contents of NMS medium before medium replenishment and after harvesting of the cells.
Quantitation of the pMMO in the membranes.
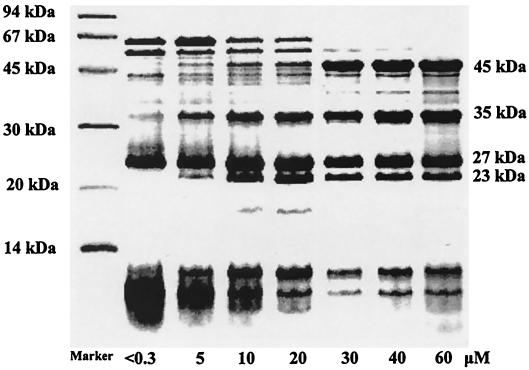
The pMMO in the purified membrane fractions was examined by SDS-PAGE (Fig. 3). The results (Table 1) indicated that the pMMO accounted for no more than 20% of the total protein in the membrane fraction when copper was limiting in the growth medium (concentration, <0.3 μM). When the copper concentration was increased to 20 μM in the growth medium, a substantially larger amount (68%) of pMMO was expressed, as shown by the appearance of the peptide subunit bands at 45, 27, and 23 kDa on the SDS-PAGE gel. Moreover, a proteolytic subunit with an apparent molecular mass of 35 kDa (17) was also found to be one of the major constituents in the membrane. At even higher copper concentrations (30 μM), the pMMO accounted for 80% of the total membrane proteins.
FIG. 3.
pMMO expression under various conditions: SDS-PAGE of pMMO-enriched membranes obtained by culturing M. capsulatus (Bath) in the presence of various copper concentrations in the growth medium. The positions of molecular weight markers are indicated on the left.
TABLE 1.
Effect of copper concentration in the culture medium on the pMMO protein contents of pMMO-enriched membranes, the specific activity of the pMMO-enriched membranes when NADH is the reductant and the average copper content of the pMMO protein
| Cu concn in growth medium (μM) | pMMO/ (%)a | Sp Act (nmol/min/mg of protein)b | Copper/ pMMOc |
|---|---|---|---|
| <0.3 | 19 | 19.6 | 9.4 |
| 5 | 27 | 30.5 | 10.5 |
| 10 | 44 | 42.6 | 10.3 |
| 20 | 68 | 59.6 | 10.8 |
| 30 | 80 | 65.9 | 13.4 |
| 40 | 73 | 88.9 | 13.8 |
| 60 | 65 | 63.5 | 15.3 |
Amount of pMMO expressed as a percentage of the total protein.
The values are averages of three determinations obtained with the same batch of pMMO-enriched membranes.
Moles of copper per mole of protein, determined by assuming that the pMMO hydroxylase is a monomer with an αβγ subunit composition and a molecular mass of 99 kDa.
Specific activity of the pMMO in the pMMO-enriched membranes.
The quality of the pMMO was assessed by examining the methane monooxygenase activity of the membranes. The pMMO activity was examined by performing propylene epoxidation assays with NADH as the reductant. The specific activities of the pMMO-enriched membranes obtained with various copper ion concentrations in the growth medium are summarized in Table 1. From these results, we concluded that the pMMO derived from an M. capsulatus (Bath) culture incubated in the presence of 40 μM copper exhibited the highest specific activity (88.9 nmol of propylene oxide/min/mg of protein). As shown in Table 1, the number of copper ions taken up per pMMO molecule was 13.3 under these conditions. This value must be close to the value for the full complement of copper ions associated with the fully functional pMMO for maximal NADH-driven pMMO activity. Previously, Nguyen et al. (17) reported a stoichiometry of 12 to 15 copper ions per pMMO molecule on the basis of protein isolation, solubilization, and reconstitution studies.
Other metals: Fe and Zn.
The Fe and Zn contents in the cytoplasmic membranes were also determined by ICP-MS, inductively coupled plasma optical emission spectroscopy, and atomic absorption spectroscopy. There was no evidence of zinc in the pMMO-enriched membranes grown in the presence of 40 μM copper. However, a small amount of iron was found in the cytoplasmic membranes, and the copper-to-iron ratio was 11 to 13. These data were corroborated by Kα X-ray absorption edge data recorded in the region of the copper, iron, and zinc absorption edges (data not shown). From the edge jumps or a comparison of the relative absorbance values obtained for the copper, iron, and zinc edges, the copper/iron/zinc ratio was determined to be 11.8:1.0:0.1.
NADH versus duroquinol activity.
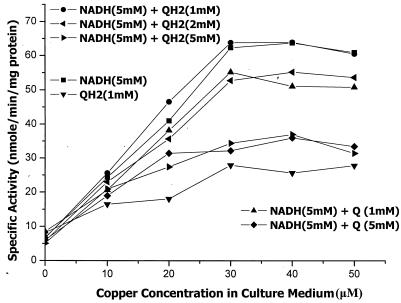
Duroquinol is often used as the reductant to assay pMMO activity (2, 9, 22, 25), although it is not a natural substrate. Nevertheless, workers have asked whether a membrane pool of plastoquinol might be the source of the reducing equivalents for pMMO (22) in addition to or in lieu of NADH. We compared the specific activities of pMMO determined by using NADH and duroquinol as the reductants, and the results are summarized in Fig. 4. Note that the data are the averages of three assays per data point and were obtained by using a somewhat less active batch of pMMO-enriched membranes than the batch used to obtain the data summarized in Table 1. From these data, it is clear that our pMMO-enriched membranes showed sensitivity toward both NADH and duroquinol. In fact, the specific activity increased with the level of pMMO in the pMMO-enriched membranes and leveled off only after all the pMMO molecules had obtained their full complement of copper ions when the cells were cultured and harvested in the presence of copper ion concentrations greater than 30 μM. However, the specific activity determined with 5 mM NADH was more than twofold higher than the specific activity determined with 1 mM duroquinol. While 1 mM duroquinol supported reasonable activity by itself, this amount of quinol had no effect on the pMMO activity in the presence of 5 mM NADH. More significantly, increasing levels of duroquinol (2 to 5 mM) were increasingly inhibitory. Duroquinone had a similar but greater inhibitory effect. These results suggest that duroquinol or duroquinone might be a noncompetitive substrate or inhibitor of NADH in pMMO (see below).
FIG. 4.
Specific activities of pMMO-enriched membranes grown in the presence of various copper concentrations in the culture medium. Each data point represents the average of three determinations made with the same batch of pMMO-enriched membranes. The curves were obtained by using either NADH or duroquinol or both as the reductant. The concentration of the reductant(s) for each curve is indicated. Other details are given in the text. The effects of duroquinone on the NADH-dependent activity are also shown. QH2, duroquinol; Q, duroquinone.
Peptide mass fingerprinting of the pMMO subunits.
In order to ascertain whether the protein that was highly expressed in the pMMO-enriched membranes corresponded to the pMMO that was isolated and purified previously by Nguyen et al. (17), we performed peptide mass fingerprinting of the pMMO subunits derived from the pMMO-enriched membranes and compared the peptides with the amino acid sequences predicted by the gene sequences of pmoA, pmoB, and pmoC deduced by Semrau et. al (20). SDS-PAGE bands of the 45-, 27-, and 23-kDa subunits were subjected to in-gel trypsin protease digestion, and the peptides were analyzed by peptide mass fingerprinting by MALDI-TOF MS. Some of the results are summarized in Table 2. The data support the conclusion that we studied the same pMMO that was studied in the previous work.
TABLE 2.
Identification of the pMMO subunits by in-gel trypsin digestion and peptide mass fingerprinting by MALDI-TOF MS
| pMMO subunit | Peptide sequence | Molecular mass (atomic mass units)
|
||
|---|---|---|---|---|
| Measured | Calculated | Coverage (%) | ||
| PmoB (45 kDa) | 121TYDFR125 | 701.3038 | 701.3259 | 51 |
| 37SQAAFMR43 | 810.3948 | 810.3932 | ||
| 286VEDATYR292 | 853.3803 | 853.4056 | ||
| 216LLMVDAGR223 | 874.4755 | 874.4820 | ||
| 209RPIFIPR215 | 898.5452 | 898.5627 | ||
| 314LGEFYTASVR323 | 1,142.5853 | 1,142.5846 | ||
| 102ESYIGGQLVPR112 | 1,218.6485 | 1,218.6483 | ||
| 376LSDIIYDPDSR386 | 1,293.6451 | 1,293.6327 | ||
| 302LTITNHGNSPIR313 | 1,322.7264 | 1,322.7181 | ||
| 286VEDATYRVPGR296 | 1,262.6533 | 1,262.6493 | ||
| 101KESYIGGQLVPR112 | 1,346.7341 | 1,346.7432 | ||
| 46TIHWTDLSWSK56 | 1,435.6887 | 1,435.7010 | ||
| 156WITVEGSMSEFR167 | 1,441.6822 | 1,441.6786 | ||
| 156WITVEGSMSEFR167 (1Met-ox) | 1,457.7163 | 1,457.6735 | ||
| 258YPITIPLQAGTMR270 | 1,460.7933 | 1,460.7935 | ||
| 258YPITIPLQAGTMR270 (1Met-ox) | 1,476.7681 | 1,476.7885 | ||
| 271GMKPLELPAPTVSVK285 | 1,566.8771 | 1,566.8929 | ||
| 387FAGLLFFFDATGNR400 | 1,575.7949 | 1,575.7960 | ||
| 361TVDVTASDAAWEVYR375 | 1,682.7987 | 1,682.8026 | ||
| 132RPGDWHVHTMMNVQGGGPIIGPGK155 | 2,541.2614 | 2,541.2607 | ||
| 132RPGDWHVHTMMNVQGGGPIIGPGK155 | 2,557.2309 | 2,557.2556 | ||
| 332DTTGYPEDLLAEDGLSVSDNSPLAPGETR360 | 3,019.3530 | 3,019.4020 | ||
| PmoA (27 kDa) | 243FLQST247 | 595.3074 | 595.3092 | 15 |
| 1SAAQSAVR9 (Acet N) | 831.4285 | 831.4325 | ||
| 237WFSNER242 | 838.3761 | 838.3848 | ||
| 1MSAAQSAVR9 (1Met-ox) | 936.4794 | 936.4573 | ||
| 191TGTPEYIR198 | 936.4794 | 936.4791 | ||
| 10SHAEAVQVSR19 | 1,083.5303 | 1,083.5547 | ||
| PmoC (23 kDa) | 126EELLRR130 | 702.3650 | 702.3899 | 7 |
| 199LPFFAK204 | 722.3874 | 722.4241 | ||
| 118NLAALTPR125 | 855.4657 | 855.5052 | ||
| 197TRLPFFAK204 | 979.5252 | 979.5729 | ||
| 116DRNLAALTPR125 | 1,126.5821 | 1,126.6333 | ||
Purification of the pMMO from pMMO-enriched membranes.
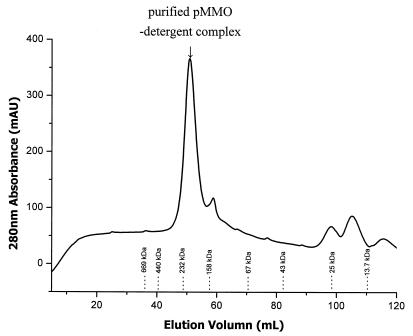
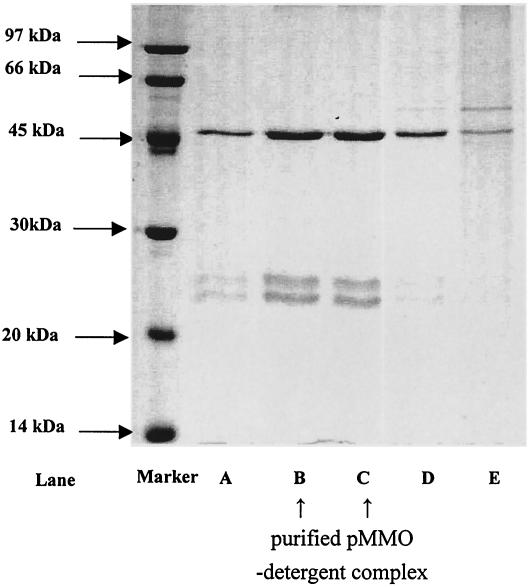
When high-quality pMMO accounted for 80 to 90% of the total proteins in the membranes, protein isolation and purification were greatly simplified. Figure 5 shows the elution profile of the proteins after a preparation of the membranes from cells cultured and harvested at a copper concentration of 30 μM was solubilized in 5% (wt/vol) dodecyl β-d-maltoside and subjected to size exclusion column chromatography. SDS-PAGE gels of different cuts near the peak of the elusion profile are shown in Fig. 6. In particular, the cut derived from the peak of the elusion profile showed no contamination with other proteins. It is clear that a homogeneous pMMO preparation was derived by this procedure.
FIG. 5.
Elution of the purified pMMO-detergent complex on an S-300HR size exclusion column. The elution buffer contained 50 mM PIPES, 150 mM NaCl, 50 mM imidazole, 5 mM ascorbate, and 0.05% (wt/vol) dodecyl β-d-maltoside (pH ∼7.2). The positions of molecular mass markers are indicated at the bottom. The standard proteins used were proteins from molecular mass calibration kits (Amersham Pharmacia Inc.) and included thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and RNase A (13.7 kDa).
FIG. 6.
SDS-PAGE of several cuts of the elution profile of the detergent-solubilized pMMO-enriched membranes from an S-300HR gel filtration column around the peak of the elution profile. A 5-μl aliquot of eluant was taken from each cut. The protein concentration was 0.5 mg/ml in lanes B and C. Lane A, elution volume, 46 to 48 ml; lane B, elution volume, 49 to 51 ml; lane C, elution volume, 52 to 54 ml; lane D, elution volume, 55 to 57 ml; lane E, elution volume, 58 to 60 ml.
Molecular masses of the three subunits of pMMO.
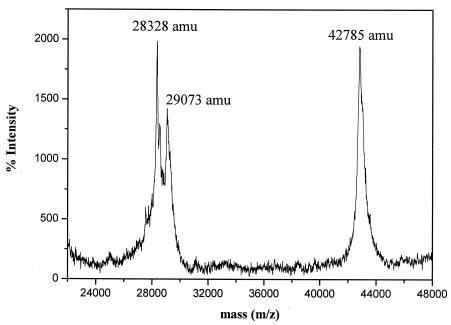
The MALDI-TOF MS spectra for the purified pMMO-detergent complex are shown in Fig. 7. The data indicate that only three polypeptides were associated with the purified pMMO-detergent complex, and their actual molecular masses were 42.785 kDa (subunit α or PmoΒ), 29.073 kDa (subunit β or PmoA), and 28.328 kDa (subunit γ or PmoC), in excellent agreement with the peptide sequences predicted by the gene sequences in the National Center for Biotechnology Information database (Table 3). The relative intensities of the peptide mass peaks are consistent with equal stoichiometry of the three subunits in the pMMO-detergent complex.
FIG. 7.
MALDI-TOF MS map of the purified pMMO-detergent complex.
TABLE 3.
Molecular masses of the three subunits associated with the purified pMMO-detergent complex as identified by MALDI-TOF MS
| Subunit | Molecular mass
|
|
|---|---|---|
| Measured | Predicted | |
| PmoB | 42,785 | 42,786 |
| PmoA | 29,073 | 29,063 |
| PmoC | 28,328 | 28,376 |
Properties of the solubilized and purified pMMO.
The purified pMMO-complex eluted on an S-300HR size exclusion column at a molecular mass of 220 kDa according to the molecular mass calibration kits (Amersham Pharmacia Inc.) (Fig. 5). Thus, the pMMO-detergent complex was isolated either as a micelle of an α2β2γ2 dimer with ∼40 detergent molecules, as suggested by Lieberman et al. (9), or as an αβγ monomer with ∼240 detergent molecules. Since there are ∼100 molecules per dodecyl β-d-maltoside micelle, the latter possibility is more reasonable and was, in fact, confirmed by the composition of the protein-detergent complex eluted by gel filtration. Metal analysis of three samples yielded average copper and iron contents of 0.143 and ∼0.002 μmol/mg of protein, respectively, or a copper content of 13.6 g-atom/mol of protein and a copper-to-iron ratio of 80:1. Thus, the pMMO hydroxylase is a copper protein only.
We compared the copper Kα X-ray absorption edges, as well as the 77 K EPR spectra, of the purified pMMO-detergent complex, detergent-solubilized pMMO-enriched membranes, and isolated pMMO-enriched membranes (data not shown). The copper Kα spectra were almost identical; in particular, the intensities of the Cu(I) feature at 8,984 eV (due mostly to the E-cluster copper ions) (16) were similar for all three preparations. Similarly, the three preparations gave almost identical 77 K EPR spectra for the partially oxidized C-cluster copper ions (16). Thus, the catalytic site was not modified by membrane solubilization and purification of the pMMO-detergent complex.
Finally, we measured the specific activities of the pMMO in the detergent-solubilized membranes and in the purified pMMO-detergent complex using both NADH and duroquinol as reductants. The results for three samples from the same batch of pMMO-enriched membranes were averaged. For the pMMO in the detergent-solubilized membranes, the specific activities were 10.2 nmol of propylene oxide/min/mg of protein with 5 mM NADH and 93.5 nmol of propylene oxide/min/mg of protein with 1 mM duroquinol. The corresponding specific activities of the purified pMMO-detergent complex were 21.5 nmol of propylene oxide/min/mg of protein with NADH and 15.6 nmol of propylene oxide/min/mg of protein with duroquinol. It is well known that specific activities of membrane proteins are strongly dependent on the physiochemical environment in which they are embedded, particularly the microscopic fluidity, and hence the specific activity of a given membrane protein is expected to be dependent on the substrates used to assay the activity and whether the protein is vectorially oriented in a phospholipid bilayer, solubilized in a phospholipid-detergent mixture, or encapsulated in a protein-detergent complex. In light of this, it is clear that we obtained both an active detergent-solubilized pMMO and a purified pMMO-detergent complex. The pMMOs in both preparations were functional, irrespective of whether NADH or duroquinol was used to assay the activity.
DISCUSSION
In this study, we improved the conventional method of culturing bacteria in a batch fermentor by coupling a hollow-fiber membrane bioreactor to the fermentor. The hollow-fiber bioreactor provided a rather precise way to control the chemical composition of the growth medium. In particular, the hollow-fiber bioreactor provided a convenient mechanism to maintain defined copper concentrations in NMS medium and to adjust the copper concentration (or the concentrations of other nutrients) incrementally when desired (for example, in order to optimize the copper concentration for enhanced production of the pMMO protein and to obtain pMMO with the full complement of copper ions to become functional). We found that this procedure allowed the cells to grow well near the mid-log phase or even to the late-log phase, increasing the cell density and the overall yield. Thus, with this improved technology, it was possible to obtain cell cultures in higher yields and membrane proteins with greater reproducibility. High-quality and highly enriched pMMO membranes facilitate pMMO protein isolation and purification, as we demonstrated here.
Quality of pMMO.
The quality of pMMO is perhaps best assessed by its specific activity in the membranes. The highest specific activity was obtained when the cells were grown in the presence of 30 to 40 μM copper ions in the growth medium. This specific activity (as high as 88.9 nmol of propylene oxide/min/mg of protein), as determined from the rate of propylene epoxidation by using NADH as the reducing agent, was almost 30% higher than the highest specific activity (60 nmol of propylene oxide/min/mg of protein) previously obtained by growing the cells in the presence of 50 μM copper ions (16). Cultures of M. capsulatus are seldom incubated in the presence of initial copper concentrations higher than 20 μM, despite the fact that the organism has unusual tolerance for higher copper concentrations. When cultures are incubated in the presence of a copper concentration of 20 μM, the medium runs out of copper ions so that many of the pMMO molecules synthesized are assembled without the full complement of copper ions. Replenishing the copper ions at the same concentration should merely stimulate the replication of additional cells that synthesize more pMMO to take up the copper ions added. More membranes are produced, but the average copper content of the pMMO membranes stays essentially the same. On the other hand, the fact that the specific activity could be improved by increasing the initial copper concentration in the growth medium was evident in previous studies (15, 18, 21). Thus, during culturing of the cells, the methanotroph attempts to grow or express a level of protein that is commensurate with the final level of copper ions in the growth medium, and a compromise is reached between the level of cell growth and/or protein expression and the uptake of the copper ions by the pMMO to assemble the functional enzyme in the membranes.
The pMMO-enriched membranes obtained from harvesting of methanotroph cultures grown in the presence of 30 to 40 μM copper ions in the growth media exhibited high specific activities when NADH was used as the reductant. However, the same membranes also responded with reasonable activity when duroquinol was the reducing substrate. Thus, it is clear that pMMO could derive its reducing equivalents from both NADH and duroquinol. From the study of the specific activities in the presence of different levels of NADH and duroquinol, as well as duroquinone, it is apparent that NADH and duroquinol are noncompetitive substrates and that duroquinone is a noncompetitive inhibitor; that is, there must be two electron input ports in the pMMO, and the reducing equivalents for the monooxygenase reaction could be derived from either NADH or duroquinol. Recently, we obtained evidence that the NADH activity requires the E-cluster copper ions to shuttle the reducing equivalents to the active site. Since the reduction potential of NADH is −390 mV (more negative than that of duroquinol) (22), it is very likely that the reducing equivalents from duroquinol enter the protein at a different point, perhaps bypassing the E-clusters, to rereduce the C-cluster copper ions following turnover. This scenario could well account for the sensitivity of the enzyme to both NADH and duroquinol. In fact, the partial inhibition of the NADH-driven pMMO activity by duroquinone could simply result from bleeding of a portion of the reducing equivalents from NADH to bound duroquinone to produce duroquinol, which is subsequently released to the membrane when the concentrations of duroquinol and duroquinone are low. Such branching of the electron flow competes with the dioxygen chemistry and the hydroxylation reaction mediated by the C-clusters.
Similar observations were made for the pMMO in the detergent-solubilized membranes and the purified pMMO-detergent complex, where the specific activity observed for the enzyme with either NADH or duroquinol as the source of reducing equivalents again attested to its functional integrity. For the enzyme in the detergent-solubilized membranes, significantly higher activity was observed with duroquinol than with NADH, in contrast to the enzyme in the pMMO-enriched membranes. For the purified pMMO-detergent complex, the corresponding specific activities with NADH and duroquinol were more comparable. Differences in the specific activities among the various preparations aside, observation of simultaneous NADH-dependent and duroquinol-dependent activities in each of the preparations was possible only when the fully functional pMMO hydroxylase was isolated. As noted above, the specific activity of a given membrane protein is dependent on the microscopic fluidity of the environment in which the protein resides, reflecting the requirement for breathing motions within the enzyme to facilitate its turnover. The different specific activities observed with NADH and duroquinol in the various preparations could also have arisen from the differential accessibility of the two reductants to the electron input sites in the protein or from different solubilities of NADH and duroquinol in the detergent micelle or the lipid bilayer supporting the protein.
Copper content of pMMO.
One of the most important findings of the present study is that the pMMO protein expressed in the membrane could acquire its full complement of copper ions only when the cells were grown in a culture containing copper ions at a concentration of 30 μM or more. The maximum NADH-dependent methane monooxygenase activity was obtained when the cells were harvested after growth in NMS medium supplemented with 35 μM copper. The pMMO obtained under these conditions contained the full complement of ∼13 copper ions per protein molecule, the same copper stoichiometry as that of the solubilized, purified, and reconstituted pMMO previously reported by Nguyen et al. (17).
We repeated the protein solubilization procedure (detergent solubilization of the pMMO-enriched membranes) and purified the pMMO-detergent complex by size exclusion chromatography using the high-quality pMMO-enriched membranes obtained with the improved technology developed in this study. Although there was on average one iron atom per 12 to 15 copper ions associated with the pMMO in the membranes, metal analysis of the purified pMMO-detergent complex revealed negligible amounts of iron and a copper-to-iron ratio of 80:1. Thus, the pMMO hydroxylase is clearly a copper-only protein, and the iron observed in the pMMO-enriched membranes was associated with other proteins.
Since the original work of Nguyen et al. (17), a number of other attempts to purify the pMMO have also been reported. In almost all of these studies (2, 4, 9, 15, 22, 23, 24, 25), NADH activity was observed only in membranes, and the enzyme following detergent solubilization and purification by column chromatography exhibited activity only with duroquinol, usually with a significant loss of copper ions during the purification process. We believe that the fully functional protein was not obtained in these studies. During solubilization of membrane proteins, the soluble activity of a protein typically increases and then decreases as the detergent concentration is increased (5). Unless the detergent-to-protein ratio that gives the maximal activity is used for solubilization, there is a loss of activity. As noted above, the pMMO solubilized at a high detergent-to-protein ratio is inactivated due to dissociation of the subunits and loss of the bulk of the copper ions.
Even in our hands, when the cells were grown in NMS medium containing copper at a concentration of 20 μM or less, the pMMO was heterogeneous, and the copper ions were distributed among the protein molecules. Although the pMMO was highly expressed, not all the pMMO molecules contained the full complement of copper ions necessary to function as an NADH-driven methane monooxygenase. From the average specific activity and the average copper content determined for the pMMO (namely, 9 to 10 copper ions per pMMO molecule), it is evident that only 30 to 40% of the pMMO molecules contained the complete or nearly complete complement of copper ions necessary to function as an NADH-driven methane monooxygenase. In this analysis, we assumed that a fully functional pMMO molecule contains at least 12 copper ions and that even a protein molecule without the full complement of copper ions contains the six copper ions that form the two catalytic C-clusters (16). However, the latter form of the pMMO, with only the catalytic sites, cannot support NADH activity. On the other hand, there is no reason why this form of the pMMO could not support a quinol-driven methane monooxygenase activity by employing the quinol pool that is part of the electron transport chain in the membrane. The C-clusters of pMMO are buried in the plasma membrane, and their redox potentials are properly poised relative to the quinols; hence, the copper ions are accessible to the reducing equivalents of the quinol pool. This scenario could account for the recently reported isolation of a duroquinol-sensitive pMMO from cells grown in the presence of 10 μM copper ions (2).
It is well known that at sufficiently high levels copper is toxic to organisms (7, 8). While copper ions in the growth medium are critical to expression of the pMMO in the membranes and for the methane metabolism process, excess copper inactivates the cells. When M. capsulatus (Bath) cells are cultured in a growth medium containing copper at an initial concentration higher than 40 μM, extra copper ions are adsorbed by the membranes. This copper adsorption results in thickening of the cytoplasmic membrane (24) and eventually inactivates the cells. Thus, the optimal initial copper concentration for culture of this methanotroph should be around 30 to 35 μM.
Acknowledgments
This work was supported by grants NSC 90-2113-M-001-006, NSC 90-2113-M-001-080, and NSC 91-2113-M-006-006 from the National Science Council.
We are grateful to Jyh-Fu Lee of the Research Division of the National Synchrotron Radiation Research Center, Hsinchu, Taiwan, for his kind assistance with the X-ray absorption measurements.
REFERENCES
- 1.Anthony, C. 1982. The biochemistry of methylotrophs, p. 296-379. Academic Press, London, United Kingdom.
- 2.Basu, P., B. Katterle, K. K. Andersson, and H. Dalton. 2003. The membrane-associated form of methane monooxygenase from Methylococcus capsulatus (Bath) is a copper/iron protein. Biochem. J. 369:417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bédard, C., and R. Knowles. 1989. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol. Rev. 53:68-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook S. A., and A. K. Shiemke. 1996. Evidence that copper is a required cofactor for the membrane-bound form of methane monooxygenase. J. Inorg. Chem. 63:273-284. [Google Scholar]
- 5.Daniel R. M., J. T. Kadonaga, R. R. Burgess, M. W. Knuth, W. A. Brennan, and S. H. Lin. 1996. Strategies for protein purification and characterization, p. 275-308. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 6.Elliott, S. J. 2000. The copper clusters of particulate methane monooxygenase: differentiation of C- and E-clusters. Ph. D. thesis. California Institute of Technology, Pasadena.
- 7.Finney, L. A., and T. V. O'Halloran. 2003. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300:931-936. [DOI] [PubMed] [Google Scholar]
- 8.Gadd, G. M. 1990. Metal tolerance, p. 178-210. In E. Edwards (ed.), Microbiology of extreme environments. Open University Press, Milton Keynes, United Kingdom.
- 9.Lieberman, R. L., D. B. Sherstha, P. E. Doan, B. M. Hoffman, T. L. Stemmler, and A. C. Rosenzweig. 2003. Purified particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a dimer with both mononuclear copper and a copper-containing cluster. Proc. Natl. Acad. Sci. USA 100:3820-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd, J. S., P. D. Marco, H. Dalton, and J. C. Murrell. 1999. Heterologous expression of soluble methane monooxygenase genes in methanotrophs containing only particulate methane monooxygenase. Arch. Microbiol. 171:364-370. [DOI] [PubMed] [Google Scholar]
- 11.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 12.Malashenko, Y., I. Sokolov, and V. Romanovskaya. 2000. Role of monooxygenase reaction during assimilation of non-growth substrates by methanotrophs. J. Mol. Catal. B Enzymes 10:305-312. [Google Scholar]
- 13.Morton, J. D., K. F. Hayes, and J. D. Semrau. 2000. Effect of copper speciation on whole-cell soluble methane monooxygenase activity in Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 66:1730-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murrell, J. C., I. R. McDonald, and B. Gilbert. 2000. Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol. 8:221-225. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen, H.-H. T., A. K. Shiemke, S. J. Jacobs, B. J. Hales, M. E. Lidstrom, and S. I. Chan. 1994. The nature of the copper ions in the membranes containing the particulate methane monooxygenase from Methylococcus capsulatus (Bath). J. Biol. Chem. 269:14995-15005. [PubMed] [Google Scholar]
- 16.Nguyen, H.-H. T., K. H. Nakagawa, B. Hedman, S. J. Elliott, M. E. Lidstrom, K. O. Hodgson, and S. I. Chan. 1996. X-ray absorption and EPR studies on the copper ions associated with the particulate methane monooxygenase from Methylococcus capsulatus (Bath). Cu(I) ions and their implications. J. Am. Chem. Soc. 118:12766-12776. [Google Scholar]
- 17.Nguyen, H.-H. T., S. J. Elliott, J. H. Yip, and S. I. Chan. 1998. The particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a novel copper-containing three-subunit enzyme. Isolation and characterization. J. Biol. Chem. 273:7957-7966. [DOI] [PubMed] [Google Scholar]
- 18.Prior, S. D., and H. Dalton. 1985. Acetylene as a suicide substrate and active site probe for methane monooxygenase from Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 29:105-109. [Google Scholar]
- 19.Prior, S. D., and H. Dalton. 1985. The effect of copper ion on membrane content and methane monooxygenase activity in methanol-grown cells of Methylococcus capsulatus (Bath). J. Gen. Microbiol. 131:155-163. [Google Scholar]
- 20.Semrau, J. D., A. Chistoserdov, J. Lebron, A. Costello, J. Davagnino, E. Kenna, A. J. Holmes, R. Finch, J. C. Murrel, and M. E. Lidstrom. 1995. Particulate methane monooxygenase genes in methanotrophs. J. Bacteriol. 177:3071-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semrau, J. D., D. Zolandz, M. E. Lidstrom, and S. I. Chan. 1995. The role of copper in the pMMO of Methylococcus capsulatus (Bath): a structural vs. catalytic function. J. Inorg. Chem. 58:235-244. [DOI] [PubMed] [Google Scholar]
- 22.Shiemke, A. K., S. A. Cook, T. Miley, and P. Singleton. 1995. Detergent solubilization of membrane-bound methane monooxygenase requires plastoquinol analogs as electron donors. Arch. Biochem. Biophys. 321:421-428. [DOI] [PubMed] [Google Scholar]
- 23.Takeguchi, M. K. M., and I. Okura. 1998. Purification and properties of particulate methane monooxygenase from Methylosinus trichosporium OB3b. J. Mol. Catal. A Chem. 132:145-153. [DOI] [PubMed] [Google Scholar]
- 24.Xin J. Y., J. R. Cui, X. X. Hu, S. B. Li, C. G. Xia, L. M. Zhu, and Y. Q. Wang. 2002. Particulate methane monooxygenase from Methylosinus trichosporium is a copper-containing enzyme. Biochem. Biophys. Res. Commun. 295:182-186. [DOI] [PubMed] [Google Scholar]
- 25.Zahn, J. A., and A. A. DiSpirito. 1996. The membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J. Bacteriol. 178:1018-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]