Abstract
FNR is an Escherichia coli transcription factor that regulates the transcription of many genes in response to anaerobiosis. We have constructed a series of artificial FNR-dependent promoters, based on the melR promoter, in which a consensus FNR binding site was centered at position −41.5 relative to the transcription start site. A second consensus FNR binding site was introduced at different upstream locations, and promoter activity was assayed in vivo. FNR can activate transcription from these promoters when the upstream FNR binding site is located at many different positions. However, sharp repression is observed when the upstream-bound FNR is located near positions −85 or −95. This repression is relieved by the FNR G74C substitution mutant, previously identified as being defective in transcription repression at the yfiD promoter. A parallel series of artificial FNR-dependent promoters, carrying a consensus FNR binding site at position −61.5 and a second upstream DNA site for FNR, was also constructed. Again, promoter activity was repressed by FNR when the upstream-bound FNR was located at particular positions.
The regulator of fumarate and nitrate reduction (FNR) and the cyclic AMP receptor protein (CRP) are related transcription activators which control the expression of networks of Escherichia coli genes in response to oxygen starvation and glucose starvation, respectively. Both FNR and CRP bind as dimers to specific 22-bp sequences located at target promoters, and the specificity of DNA recognition and the mechanisms by which FNR and CRP activate transcription have been extensively studied (reviewed in references 8 and 13). Both FNR and CRP can activate transcription by recruiting holo RNA polymerase (RNAP) to target promoters via direct interactions with the C-terminal domain of the RNAP α subunit (αCTD) (reviewed in reference 7). Many target promoters contain just one DNA site for FNR or CRP, and they can be grouped into two classes according to the location of this site. At Class I promoters the activator binds to a site located upstream from the promoter −35 element and makes direct contact with one αCTD, which binds immediately downstream of the bound FNR or CRP dimer. At Class II promoters the activator binds to a site that overlaps the −35 element and makes direct contact with the αCTD that binds immediately upstream of the bound FNR or CRP. Interestingly, although RNAP contains two α subunits and hence two αCTDs, activation at these promoters requires contact with only one αCTD (18).
Many FNR- and CRP-regulated promoters contain two DNA sites for FNR or CRP (8, 13). At some of the CRP-regulated promoters, optimal expression depends on the binding of CRP to both target sites. To account for this, Busby and Ebright (7) proposed that each bound CRP contacts one of the two αCTDs. Systematic studies of promoters carrying tandem DNA sites for CRP were performed by Belyaeva et al. (3) and Tebbutt et al. (24). Belyaeva et al. (3) started with a Class II CRP-dependent promoter carrying a single DNA site for CRP (centered at position −41.5) and introduced a second site at different upstream locations. Activation by the tandem-bound CRP was increased when the upstream-bound CRP was located at certain positions. Similarly, Tebbutt et al. (24) studied a Class I promoter carrying a single DNA site for CRP (centered at position −61.5) and observed increased activation when a second DNA site for CRP was located at certain upstream positions.
The many similarities between FNR and CRP suggest that tandem-bound FNR molecules should also be able to cooperate at a target promoter to activate transcription synergistically. However, present evidence suggests that this is not the case. For example, the E. coli yfiD promoter is activated by FNR binding to a target site centered at position −40.5, but this activation is suppressed rather than enhanced by FNR binding to an additional upstream site, located at position −93.5 (12). Green and collaborators have presented evidence that this down-regulation results from specific interactions between the two tandem-bound FNR molecules and that these interactions are dependent on the spacing between the bound FNR molecules (11, 20). The main purpose of this study was to make a systematic investigation of promoters with tandem DNA sites for FNR. One aim was to search for promoter architectures where tandem-bound FNR molecules would cooperate to activate transcription. Thus, starting with the Class II CRP-dependent promoters described by Belyaeva et al. (3) that carry a second DNA site for CRP at different upstream locations, we generated a related set of promoters with two DNA sites for FNR. Studies with these promoters show that upstream-bound FNR suppresses rather than enhances transcription. Similarly, starting with the Class I CRP-dependent promoters described by Tebbutt et al. (24), which carry a second upstream DNA site for CRP, we generated a second set of promoters with two DNA sites for FNR. Studies with these promoters show that upstream-bound FNR can either suppress or enhance transcription according to its location.
MATERIALS AND METHODS
Strains, plasmids, and materials.
The bacterial strains and plasmids used in this study are listed in Table 1. Cultures of E. coli were grown in Luria Bertani medium (LB) (20 g of tryptone, 10 g of NaCl, 10 g of yeast extract per liter) supplemented with appropriate antibiotics (ampicillin, 100 μg ml−1; tetracycline, 35 μg ml−1). For agar plates, 16 g of agar per liter was added or MacConkey Agar (Difco) was used. DNA was isolated and manipulated by using standard methods. All of the new promoter constructs were checked by using the automated DNA sequencing facility at the Birmingham Functional Genomics Laboratory. Synthetic oligonucleotides for PCR and DNA sequencing were purchased from Alta Bioscience, University of Birmingham, or from Sigma.
TABLE 1.
Bacterial strains and plasmids
| Strains and plasmids | Description | Source or reference |
|---|---|---|
| Strains | ||
| M182 | E. coli K-12 Δlac fnr+ | 9 |
| M182 fnrA1 | E. coli K12 Δlac, fnr::Tn10 cured using fusaric acid, tetracycline sensitive | A. Bell |
| RLG221 | E. coli recA56 araD139 (ara-leu)7697 lacX74 galU galK hsdR strA | R. Gourse |
| XL1-Blue | E. coli recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | 6 |
| Plasmids | ||
| pAA121 | pBR322-based cloning vector for EcoRI-HindIII promoter fragments; confers resistance to ampicillin | 14 |
| pRW50 | Broad-host-range low-copy-number lac expression vector for cloning EcoRI-HindIII promoter fragments; confers resistance to tetracycline | 19 |
| pFNR | fnr gene cloned in pBR322 derivative; confers resistance to ampicillin | 1 |
| pFNR G74C | fnr derivative encoding FNR G74C substitution mutant cloned in pBR322 derivative; confers resistance to ampicillin | 11 |
| pSR | pBR322 derivative for cloning EcoRI-HindIII promoter fragments, with an oop terminator sequence downstream of the HindIII site; also encodes a control RNA and RNA1 and confers resistance to ampicillin | 15 |
| pQE60 FNR D154A | Overexpression plasmid encoding C-terminally His-tagged FNR D154A; confers resistance to ampicillin | 26 |
Promoter constructions.
All the promoters used in this study were cloned on EcoRI-HindIII fragments and were shuttled between pAA121 (for construction), pRW50 (for assay), and pSR (for in vitro studies). The starting point was the CC(−41.5) promoter and the series of derivatives described by Belyaeva et al. (3) carrying a second upstream DNA site for CRP. CC(−41.5) is a derivative of the E. coli melR promoter carrying a consensus DNA site for CRP centered at position −41.5; expression is completely dependent on CRP. In the first set of experiments, the DNA sites for CRP in these promoters were changed to DNA sites for FNR to give FF(−41.5) and a series of promoter derivatives carrying tandem DNA sites for FNR (Fig. 1a). To do this, oligo-directed mutagenesis was used to change the two TGTGA motifs in each 22-bp DNA site for CRP to TTTGA to create 22-bp DNA sites for FNR. These base changes are sufficient to switch binding specificity from CRP to FNR (2).
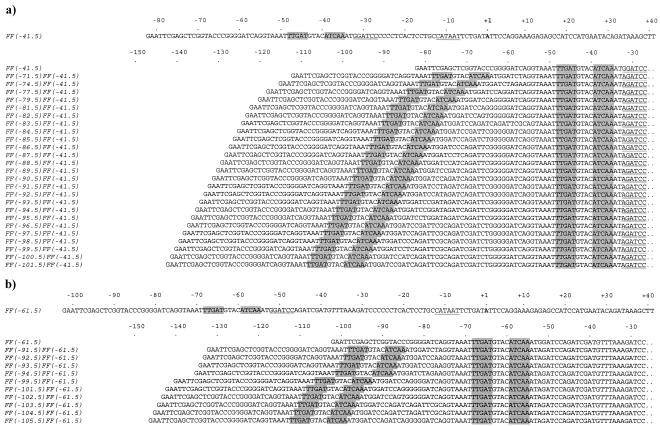
FIG. 1.
Nucleotide sequences of the FF(−n)FF(−41.5) and FF(−n)FF(−61.5) promoter series. (a) The FF(−n)FF(−41.5) promoter series. The upper part of the figure shows the complete sequence of the FF(−41.5) promoter from the EcoRI site (GAATTC) to the HindIII site (AAGCTT) located at position +36 relative to the transcription start point (+1, in boldface). The FNR consensus binding site (FF; TTGAT-n4-ATCAA) is shaded. The −10 sequence (CATAAT) is underlined, and the BamHI site (GGATCC) located immediately downstream of the FNR binding site is underlined twice. The lower part of the panel shows the sequences of the FF(−n)FF(−41.5) promoter series, aligned at the FF site centered around position −41.5. The name of each promoter reflects the location of the upstream FF site. The full sequences between the EcoRI site (GAATTC) and position −27, showing the BglII/BamHI hybrid site (AGATCC), are shown. The promoter sequences are identical to the FF(−41.5) sequence between position −27 and the HindIII site. (b) The FF(−n)FF(−61.5) promoter series. The upper part of the panel shows the complete sequence of the FF(−61.5) promoter from the EcoRI site to the HindIII site located at position +36 relative to the transcription start point (+1, in boldface). The FNR consensus binding site is shaded, the −10 sequence is underlined, and the BamHI site is underlined twice. The lower part of the panel shows the sequences of the FF(−n)FF(−61.5) promoter series, aligned at the FF site centered around position −61.5. The name of each promoter reflects the location of the upstream FF site. The full sequences between the EcoRI site and position −27, showing the BglII/BamHI hybrid site, are shown. The promoter sequences are identical to the FF(−61.5) sequence between position −27 and the HindIII site.
Oligos used in the constructions are listed in Table 2, and the PCRs used to make the different promoters are described in Table 3. The FF(−41.5) promoter (Fig. 1a) was created from CC(−41.5) by PCR with primer AB1 (which covers the upstream EcoRI site and converts the DNA site for CRP into a site for FNR) and primer D2591 (which covers the downstream HindIII site). The PCR product was digested with EcoRI and HindIII, and the FF(−41.5) EcoRI-HindIII fragment was cloned into pAA121.
TABLE 2.
Oligonucleotide primers
| Primers | Description | Source or reference |
|---|---|---|
| Mutagenic primers used to convert CC sites to FF or EE | ||
| AB1 | GGA ATT CGA GCT CGG TAC CCG GGG ATC AGG TAA ATT TGA TGT ACA TCA AAT GGA TC; converts upstream CC site to FF, in bold; the EcoRI site is underlined | This work |
| AB2 | GGA ATT CGA GCT CGG TAC CCG GGG ATC AGG TAA ATT TCA TGT ACA TGA AAT GGA TC; converts upstream CC site to EE, in bold; the EcoRI site is underlined | This work |
| AB5 | GAA TTA TGG CAG GAG TGA GGG GAG ATC TAT TTG ATG TAC ATC AAA TTT CCC T; converts CC at position −41.5 to FF, in bold; the BglII site is underlined | This work |
| AB6 | GGG GGG ATC TTT AAA CAT CGA TCT AGA TCT ATT TGA TGT ACA TCA AAT TTA CCT; converts CC at position −61.5 to FF, in bold; the BglII site is underlined | This work |
| General primers annealing to pAA121 or pRW50 | ||
| AB4 | GGC GTA TCA CGA GGC CCT TTC G; used to amplify promoter fragments from pAA121, anneals 5′ of EcoRI site | This work |
| D2591 | AGC AAG CTT TAT CTG TAT TCA TGG; used to amplify promoter fragments from pAA121 or pRW50; the HindIII site is underlined | 24 |
| Primers used to decrease or increase spacing between FF sites | ||
| FFΔ6 | ATG GAT CCG ATT CGG GGG ATC AGG TA; deletes 6 bases after the highlighted C in the spacer of FF(−90.5)FF(−41.5); used to make FF(−84.5)FF(−41.5) | This work |
| FFΔ4 | ATG GAT CCC AGA TTC GGG GGA TCA GG; deletes 4 bases after the highlighted C in the spacer of FF(−90.5)FF(−41.5); used to make FF(−86.5)FF(−41.5) | This work |
| FFΔ3 | ATG GAT CCT CAG ATT CGG GGG ATC AG; deletes 3 bases after the highlighted C in the spacer of FF(−90.5)FF(−41.5); used to make FF(−87.5)FF(−41.5) | This work |
| FFΔ2 | ATG GAT CCA TCA GAT TCG GGG GAT CA; deletes 2 bases after the highlighted C in the spacer of FF(−90.5)FF(−41.5); used to make FF(−88.5)FF(−41.5) | This work |
| FFΔ1 | ATG GAT CCG ATC AGA TTC GGG GGA TC; deletes 1 bases after the highlighted C in the spacer of FF(−90.5)FF(−41.5); used to make FF(−89.5)FF(−41.5) | This work |
| FF+1 | ATG GAT CCT AGA TCA GAT TCG GGG GAT; inserts 1 base shown in bold into the spacer of FF(−90.5)FF(−41.5); used to make FF(−91.5)FF(−41.5) | This work |
| FF+2 | ATG GAT CCA TAG ATC AGA TTC GGG GGA T; inserts 2 bases shown in bold into the spacer of FF(−90.5)FF(−41.5); used to make FF(−92.5)FF(−41.5) | This work |
| FF+3 | ATG GAT CCG ATA GAT CAG ATT CGG GGG AT; inserts 3 bases shown in bold into the spacer of FF(−90.5)FF(−41.5); used to make FF(−93.5)FF(−41.5) | This work |
| FF+4 | ATG GAT CCG GAT AGA TCA GAT TCG GGG GAT; inserts 4 bases shown in bold into the spacer of FF(−90.5)FF(−41.5); used to make FF(−94.5)FF(−41.5) | This work |
| FF+5 | ATG GAT CCT GGA TAG ATC AGA TTC GGG GGA T; inserts 5 bases shown in bold into the spacer of FF(−90.5)FF(−41.5); used to make FF(−95.5)FF(−41.5) | This work |
| FF(−96) | ATG GAT CCG ATT CGC AGA TCG ATC TG; deletes 6 bases after the highlighted C in the spacer of FF(−102.5)FF(−41.5); used to make FF(−96.5)FF(−41.5) | This work |
| FF(−97) | ATG GAT CCA GAT TCG CAG ATC GAT CT; deletes 5 bases after the highlighted C in the spacer of FF(−102.5)FF(−41.5); used to make FF(−97.5)FF(−41.5) | This work |
| FF(−98) | ATG GAT CCC AGA TTC GCA GAT CGA TC; deletes 4 bases after the highlighted C in the spacer of FF(−102.5)FF(−41.5); used to make FF(−98.5)FF(−41.5) | This work |
| FF(−99) | ATG GAT CCT CAG ATT CGC AGA TCG ATC; deletes 3 bases after the highlighted C in the spacer of FF(−102.5)FF(−41.5); used to make FF(−99.5)FF(−41.5) | This work |
| FF(−100) | ATG GAT CCA TCA GAT TCG CAG ATC GAT C; deletes 2 bases after the highlighted C in the spacer of FF(−102.5)FF(−41.5); used to make FF(−100.5)FF(−41.5) | This work |
| FF(−101) | ATG GAT CCG ATC AGA TTC GCA GAT CG; deletes 1 base after the highlighted C in the spacer of FF(−102.5)FF(−41.5); used to make FF(−101.5)FF(−41.5) | This work |
TABLE 3.
Promoters used in this work
| Promoter | Promoter construction
|
||
|---|---|---|---|
| Starting promoter | Primers used in first PCR | Primers used in second PCR | |
| FF(−41.5) | CC(−41.5)a | AB1, D2591 | |
| FF(−71.5)FF(−41.5) | CC(−71.5)CC(−41.5)a | AB1, D2591 | AB4, AB5 |
| FF(−74.5)FF(−41.5) | CC(−74.5)CC(−41.5)a | AB1, D2591 | AB4, AB5 |
| FF(−77.5)FF(−41.5) | CC(−77.5)CC(−41.5)a | AB1, D2591 | AB4, AB5 |
| FF(−79.5)FF(−41.5) | CC(−79.5)CC(−41.5)a | AB1, D2591 | AB4, AB5 |
| FF(−81.5)FF(−41.5) | CC(−81.5)CC(−41.5)a | AB1, D2591 | AB4, AB5 |
| FF(−82.5)FF(−41.5) | CC(−82.5)CC(−41.5)a | AB1, D2591 | AB4, AB5 |
| FF(−83.5)FF(−41.5) | CC(−83.5)CC(−41.5)a | AB1, D2591 | AB4, AB5 |
| FF(−84.5)FF(−41.5) | FF(−90.5)FF(−41.5)c | FFΔ6, D2591 | |
| FF(−85.5)FF(−41.5) | CC(−85.5)CC(−41.5)a | AB1, D2591 | AB4, AB5 |
| FF(−86.5)FF(−41.5) | FF(−90.5)FF(−41.5)c | FFΔ4, D2591 | |
| FF(−87.5)FF(−41.5) | FF(−90.5)FF(−41.5)c | FFΔ3, D2591 | |
| FF(−88.5)FF(−41.5) | FF(−90.5)FF(−41.5)c | FFΔ2, D2591 | |
| FF(−89.5)FF(−41.5) | FF(−90.5)FF(−41.5)c | FFΔ1, D2591 | |
| FF(−90.5)FF(−41.5) | CC(−90.5)CC(−41.5)a | AB1, D2591 | AB4, AB5 |
| FF(−91.5)FF(−41.5) | FF(−90.5)FF(−41.5)c | FF+1, D2591 | |
| FF(−92.5)FF(−41.5) | FF(−90.5)FF(−41.5)c | FF+2, D2591 | |
| FF(−93.5)FF(−41.5) | FF(−90.5)FF(−41.5)c | FF+3, D2591 | |
| FF(−94.5)FF(−41.5) | FF(−90.5)FF(−41.5)c | FF+4, D2591 | |
| FF(−95.5)FF(−41.5) | FF(−90.5)FF(−41.5)c | FF+5, D2591 | |
| FF(−96.5)FF(−41.5) | FF(−102.5)FF(−41.5)c | FF(−96), D2591 | |
| FF(−97.5)FF(−41.5) | FF(−102.5)FF(−41.5)c | FF(−97), D2591 | |
| FF(−98.5)FF(−41.5) | FF(−102.5)FF(−41.5)c | FF(−98), D2591 | |
| FF(−99.5)FF(−41.5) | FF(−102.5)FF(−41.5)c | FF(−99), D2591 | |
| FF(−100.5)FF(−41.5) | FF(−102.5)FF(−41.5)c | FF(−100), D2591 | |
| FF(−101.5)FF(−41.5) | FF(−102.5)FF(−41.5)c | FF(−101), D2591 | |
| FF(−102.5)FF(−41.5) | CC(−102.5)CC(−41.5)a | AB1, D2591 | AB4, AB5 |
| EE(−85.5)FF(−41.5) | FF(−85.5)FF(−41.5)c | AB2, D2591 | |
| EE(−90.5)FF(−41.5) | FF(−90.5)FF(−41.5)c | AB2, D2591 | |
| FF(−61.5) | CC(−61.5)b | AB1, D2591 | |
| FF(−91.5)FF(−61.5) | CC(−91.5)CC(−61.5)b | AB1, D2591 | AB4, AB6 |
| FF(−92.5)FF(−61.5) | CC(−92.5)CC(−61.5)b | AB1, D2591 | AB4, AB6 |
| FF(−93.5)FF(−61.5) | CC(−93.5)CC(−61.5)b | AB1, D2591 | AB4, AB6 |
| FF(−94.5)FF(−61.5) | CC(−94.5)CC(−61.5)b | AB1, D2591 | AB4, AB6 |
| FF(−99.5)FF(−61.5) | CC(−99.5)CC(−61.5)b | AB1, D2591 | AB4, AB6 |
| FF(−101.5)FF(−61.5) | CC(−101.5)CC(−61.5)b | AB1, D2591 | AB4, AB6 |
| FF(−102.5)FF(−61.5) | CC(−102.5)CC(−61.5)b | AB1, D2591 | AB4, AB6 |
| FF(−103.5)FF(−61.5) | CC(−103.5)CC(−61.5)b | AB1, D2591 | AB4, AB6 |
| FF(−104.5)FF(−61.5) | CC(−104.5)CC(−61.5)b | AB1, D2591 | AB4, AB6 |
| FF(−105.5)FF(−61.5) | CC(−105.5)CC(−61.5)b | AB1, D2591 | AB4, AB6 |
The FF(−71.5)FF(−41.5), FF(−74.5)FF(−41.5), FF(−77.5)FF(−41.5), FF(−79.5)FF(−41.5), FF(−81.5)FF(−41.5), FF(−82.5)FF(−41.5), FF(−83.5)FF(−41.5), FF(−85.5)FF(−41.5), FF(−90.5)FF(−41.5), and FF(−102.5)FF(−41.5) promoters were derived from the corresponding CC(−n)CC(−41.5) promoters described by Belyaeva et al. (3). To do this, first the upstream DNA site for CRP was changed to a site for FNR by PCR, using primers AB1 and D2591 as described above. The PCR products were digested with EcoRI and HindIII, and the resulting fragments were cloned into pAA121. These FF(−n)CC(−41.5) constructs were then used as templates in a second PCR amplification using primers AB4 (which anneals to pAA121 vector sequence just upstream of the EcoRI site) and AB5 (which changes the downstream DNA site for CRP to a site for FNR and introduces a BglII restriction site immediately downstream of the new FNR binding site). The resulting fragments were digested with EcoRI and BglII and were ligated to the EcoRI-BamHI vector fragment from pAA121 carrying the FF(−41.5) promoter. This created a set of pAA121 derivatives carrying different promoters with tandem DNA sites for FNR and a BglII-BamHI hybrid sequence located immediately downstream of the FF site at position −41.5 (Fig. 1a).
Additional promoters were made from pAA121 derivatives carrying the FF(−90.5)FF(−41.5) or FF(−102.5)FF(−41.5) promoters by exploiting the single BamHI site located between the two DNA sites for FNR. PCRs were performed with different upstream primers designed to increase or decrease the spacing between the two FF sites and primer D2591. The resulting DNA fragments were digested with BamHI and HindIII and were ligated to the BamHI-HindIII vector fragment made from the pAA121 derivative carrying the FF(−90.5)FF(−41.5) promoter.
The control EE(−85.5)FF(−41.5) and EE(−90.5)FF(−41.5) promoters were derived from FF(−85.5)FF(−41.5) and FF(−90.5)FF(−41.5), respectively, by PCR using primers AB2 and D2591. The products were digested with EcoRI and HindIII and were cloned into pAA121. The EE sequence is a derivative of the consensus DNA site for FNR, to which FNR is unable to bind (each TTTGA motif is replaced by TTTCA).
In a second series of constructions, the starting point was the CC(−61.5) promoter and the derivatives described by Tebbutt et al. (24) carrying a second upstream DNA site for CRP. CC(−61.5) is a derivative of the E. coli melR promoter carrying a consensus DNA site for CRP centered at position −61.5. The DNA sites for CRP in these promoters were changed to DNA sites for FNR to give FF(−61.5) and a series of promoter derivatives carrying tandem DNA sites for FNR (Fig. 1b). Oligos used in the constructions are listed in Table 2, and the PCRs used to make the different promoters are described in Table 3. The FF(−61.5) promoter (Fig. 1b) was created from CC(−61.5) by PCR using primer AB1 and primer D2591. The PCR product was digested with EcoRI and HindIII, and the FF(−61.5) EcoRI-HindIII fragment was cloned into pAA121.
The different FF(−n)FF(−61.5) promoters were derived from the corresponding CC(−n)CC(−61.5) promoters described by Tebbutt et al. (24). To do this, first the upstream DNA site for CRP was changed to a site for FNR by PCR using primers AB1 and D2591 as described above. The PCR products were digested with EcoRI and HindIII, and the resulting fragments were cloned into pAA121. These FF(−n)CC(61.5) constructs were then used as templates in a second PCR amplification using primers AB4 and AB6 (which changes the downstream DNA site for CRP to a site for FNR and introduces a BglII restriction site immediately downstream of the new FNR binding site). The resulting fragments were digested with EcoRI and BglII and were ligated to the EcoRI-BamHI vector fragment from pAA121 carrying the FF(−61.5) promoter. This created a set of pAA121 derivatives carrying different promoters with tandem DNA sites for FNR with the downstream site at position −61.5 (Fig. 1b).
Assay of promoter activity in vivo.
To measure promoter activities in vivo, promoters were cloned into the lac expression vector, pRW50, as previously described (19), and the constructs were used to transform Δlac E. coli strains. β-Galactosidase activities of the transformants were assayed according to Miller (22) as follows: 10-ml cultures were grown overnight in 25-ml conical flasks in a shaking water bath at 37°C. The growth medium was LB supplemented with 0.2% fructose and appropriate antibiotics. The following morning 100 μl of the overnight cultures was used to inoculate 10 ml of fresh medium contained in narrow capped tubes. The cultures were grown anaerobically, without shaking, at 37°C for 4 to 5 h to an optical density at 650 nm of 0.3 to 0.4 and was lysed with a mixture of toluene and 1% sodium deoxycholate prior to the assay. β-Galactosidase activities are reported relative to the activity obtained using the FF(−41.5) or FF(−61.5) promoters (approximately 10,000 and 500 Miller units, respectively). Each activity is the average of at least three independent determinations. Error bars represent one standard deviation from the mean.
Purification of FNR D154A.
The aerobically active FNR* derivative, FNR D154A (16), was purified as a C-terminally His-tagged fusion protein by using a method adapted from that described by Wing et al. (26) for use with the ÄKTAprime protein purification system (Amersham Biosciences). E. coli XL1-Blue cells were transformed with a pQE60 derivative encoding the C-terminally His-tagged FNR D154A fusion protein. Transformants were grown at 37°C in 100 ml of LB supplemented with 100 μg of ampicillin ml−1 to an optical density at 650 nm of 0.4. Overexpression of the His-tagged protein was then induced by the addition of 0.1 M isopropyl-1-thio-d-galactopyranoside (IPTG) for 1 h. Cells were harvested, and pellets were sonicated in 10 ml of lysis buffer at 4°C (1 mg of lysozyme ml−1, 50 mM NaH2PO4-Na2HPO4 [pH 8.0], 0.75 M NaNO3, 10 mM imidazole, 10 mM benzamidine). Sonicates were centrifuged at 10,000 × g and were passed through a 0.2-μm-pore-size filter before being applied to a 1-ml HiTrap Chelating HP column (Amersham Biosciences), which had been equilibrated with 1 M NiSO4 followed by FNR wash buffer (50 mM NaH2PO4-Na2HPO4 [pH 8.0], 0.75 M NaNO3). His-tagged FNR D154A was eluted from the column by applying FNR elution buffer with a gradient of imidazole (50 mM NaH2PO4-Na2HPO4 [pH 8.0], 0.75 M NaNO3, to 250 mM imidazole over 30 min). The purity of the protein fractions was estimated by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Fractions containing His-tagged FNR D154A were pooled and concentrated by using a Vivaspin 50,000-molecular-weight cutoff concentrator (Vivascience) before the addition of glycerol to a final concentration of 50% (vol/vol). Protein concentration was estimated by the Bradford method (4).
In vitro transcription assays.
Derivatives of pSR carrying different FNR-regulated promoters cloned upstream of the lambda oop transcription terminator were used as templates for in vitro transcription. Plasmid DNA (8 nM) was incubated at 37°C for 20 min with various concentrations of FNR D154A (0 to 5 μM). The reaction mixture contained 40 mM Tris-Cl (pH 7.9), 10 mM MgCl2, 50 mM KCl, 0.1 mM dithiothreitol (DTT), 0.2 μg of bovine serum albumin (BSA) μl−1, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.05 mM UTP, and 5 μCi of [α-32P]UTP in a final volume of 20 μl. Following the incubation, E. coli RNAP (Epicentre) was added to a final concentration of 50 nM, and the mixture was incubated at 37°C for a further 20 min. Reactions were stopped by the addition of 25 μl of formamide buffer (95% [vol/vol] deionized formamide, 20 mM EDTA, 0.05% [wt/vol] bromophenol blue, 0.05% [wt/vol] xylene cyanol FF). Samples were run in 1× Tris-borate-EDTA (TBE) on a 5.5% denaturing polyacrylamide gel at 12 V cm−1 and were analyzed by using a phosphorimager and Bio-Rad Quantity One software. FNR-dependent transcripts were quantified with reference to the FNR-independent RNA1 transcript, encoded by the pSR vector.
DNaseI footprint analysis.
DNaseI footprinting was performed essentially as described by Savery et al. (23). The reaction mixtures contained AatII-HindIII promoter fragments that had been labeled at the HindIII site with [γ-32P]ATP and 0 to 3 μM purified FNR D154A. The reaction buffer consisted of 20 mM HEPES (pH 8.0), 5 mM MgCl2, 50 mM potassium glutamate, 1 mM DTT, 0.5 mg of BSA ml−1, and 0.1 mg of herring sperm DNA ml−1. After treatment with DNaseI (approximately 0.5 ng μl−1 for 30 to 60 s), the reactions were stopped by the addition of DNaseI Stop buffer (10 mM EDTA, 0.3 M sodium acetate). Footprinting reactions were resuspended in formamide buffer and were analyzed on 6% polyacrylamide sequencing gels calibrated with Maxam-Gilbert G+A sequencing ladders and visualized by using a phosphorimager and Bio-Rad Quantity One software.
Electromobility shift assays.
Purified EcoRI-HindIII promoter fragments were prepared from cesium chloride preparations of DNA. These fragments were end-labeled with [γ-32P]ATP, and 0.5 to 2.5 ng of each fragment was incubated with 0 to 5.6 μM concentrations of purified FNR D154A. The sample buffer contained 0.1 M potassium glutamate, 1 mM EDTA, 10 mM potassium phosphate buffer (pH 7.5), 50 μM DTT, 5% glycerol, 0.5 μg of BSA ml−1, and 25 ng of herring sperm DNA (Gibco) ml−1 in a 10-μl final reaction volume. Following incubation at 37°C for 20 min, samples were run in 0.25× TBE on a 6% polyacrylamide gel at 12 V cm−1 and were analyzed by using a phosphorimager and Bio-Rad Quantity One software.
Permanganate footprint analysis.
Reaction mixtures contained AatII-HindIII promoter fragments that had been labeled at the HindIII site with [γ-32P]ATP, 0 to 1 μM purified FNR D154A, and 0 to 50 nM RNAP. The reaction buffer consisted of 20 mM HEPES (pH 8.0), 5 mM MgCl2, 50 mM potassium glutamate, 1 mM DTT, and 0.5 mg of BSA ml−1. After treatment with potassium permanganate (10 mM final concentration for 4 min) to modify single stranded T residues, reactions were quenched by the addition of 2.5 volumes of stop solution (3 M ammonium acetate, 0.1 mM EDTA, and 1.5 M β-mercaptoethanol). Following phenol-chloroform extraction, ethanol precipitation, and treatment with 1 M piperidine (90°C for 30 min), samples were resuspended in formamide buffer. Permanganate cleavage patterns were analyzed by using 6% polyacrylamide sequencing gels and were visualized with a phosphorimager and Bio-Rad Quantity One software.
RESULTS
Activity of promoters with tandem sites for FNR: the FF(−n)FF(−41.5) series.
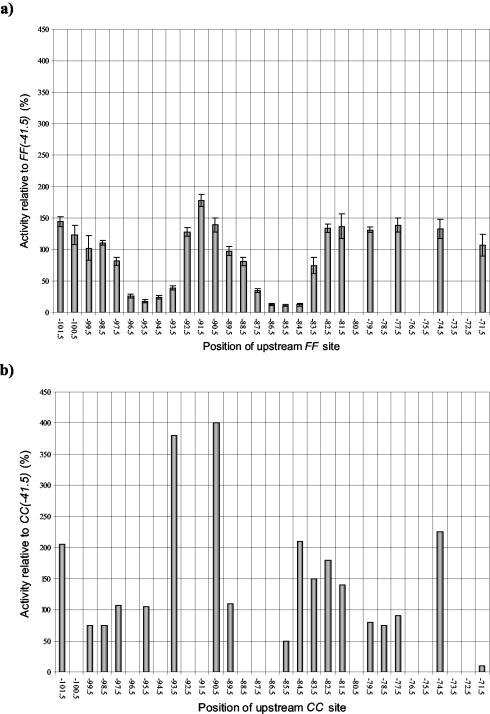
We constructed a series of related promoters carrying one consensus DNA site for FNR centered at position −41.5 and a second consensus DNA site for FNR located further upstream [the FF(−n)FF(−41.5) series] (Fig. 1a). The starting point for these constructions was the CC(−n)CC(−41.5) promoter series described in a previous study (Table 3) (3). Each new FF(−n)FF(−41.5) promoter was transferred into the low-copy lac expression vector pRW50. Measurements of β-galactosidase activities showed that expression from each promoter is anaerobically induced and is totally dependent on FNR (data not shown). Figure 2a illustrates the expression from each promoter in an fnr+ background during anaerobic growth. The results show that expression is dependent upon the spacing between the two DNA sites for FNR.
FIG. 2.
Transcription activation by FF(−n)FF(−41.5) and CC(−n)CC(−41.5) promoters. (a) The anaerobic β-galactosidase activities of M182 (Δlac) cells transformed with each of the FF(−n)FF(−41.5) promoters cloned in pRW50, displayed as a percentage of the activity achieved using the FF(−41.5) promoter. The relative activities are the means of three independent determinations, and error bars depict one standard deviation from the mean. The x axis indicates the location of the center of the upstream 22-bp FF binding site. (b) For comparison, the activities of M182 cells transformed with the CC(−n)CC(−41.5) promoter series reported by Belyaeva et al. (3) are also shown, relative to the activity achieved using the CC(−41.5) promoter. The x axis indicates the location of the center of the upstream 22-bp CC binding site.
For comparison, Fig. 2b illustrates expression from the previously constructed promoters carrying tandem DNA sites for CRP [the CC(−n)CC(−41.5) series] (3). As described before, in many cases the upstream DNA site for CRP hardly affects expression, but when located at certain positions, expression is increased two- to fourfold. Thus, when the upstream DNA site for CRP is located near positions −74.5, −84.5, −90.5, −93.5, and −101.5, the upstream-bound CRP activates transcription synergistically with the downstream-bound CRP at position −41.5. At intermediate locations, the upstream-bound CRP has little or no effect, and the observed expression is similar to that found at the CC(−41.5) promoter, with a single DNA site for CRP at position −41.5.
Comparison of data in Fig. 2a and b shows that the effects of upstream-bound FNR at the FF(−n)FF(−41.5) promoters are different from the effects of upstream-bound CRP at the CC(−n)CC(−41.5) promoters. First, the upstream-bound FNR causes only small increases in expression compared to that with the FF(−41.5) promoter (with a single DNA site for FNR), suggesting little or no synergy between tandem-bound FNR molecules. Second, sharp repression of expression is found when the upstream FNR is located near position −85.5 or −95.5 (Fig. 2a). Promoter activity is reduced to 10 to 20% of the activity of the FF(−41.5) promoter, and this reduction appears to depend on the helical juxtaposition of the tandem-bound FNR molecules.
As controls, we selected the FF(−85.5)FF(−41.5) and FF(−90.5)FF(−41.5) promoters and converted the upstream DNA sites for FNR to EE sequences, which are unable to bind either FNR or CRP. The resulting EE(−85.5)FF(−41.5) and EE(−90.5)FF(−41.5) promoters were transferred into pRW50, and their activities were measured. Results in Table 4 show that expression from these promoters is dependent on FNR. Expression from both the EE(−85.5)FF(−41.5) and EE(−90.5)FF(−41.5) promoters is similar to that observed with FF(−41.5). This argues that both the sharp decrease due to the upstream DNA site for FNR at the FF(−85.5)FF(−41.5) promoter and the small increase due to the upstream DNA site for FNR at the FF(−90.5)FF(−41.5) promoter are due to FNR binding rather than to an artifact of the promoter context.
TABLE 4.
FNR-dependent transcription using FF(−n)FF(−41.5) and EE(−n)FF(−41.5) promoters
| Promotera | Relative activity (%)b of:
|
|||
|---|---|---|---|---|
|
E. coli M182 (fnr+)
|
E. coli M182 fnrA1 (lacking fnr)
|
|||
| Activity | Error | Activity | Error | |
| FF(−85.5)FF(−41.5) | 11 | 1 | 8 | 2 |
| EE(−85.5)FF(−41.5) | 97 | 20 | 7 | 2 |
| FF(−90.5)FF(−41.5) | 139 | 11 | 7 | 0.4 |
| EE(−90.5)FF(−41.5) | 102 | 15 | 6 | 2 |
Promoters were cloned on EcoRI-HindIII fragments in the lac expression vector pRW50.
Cells were grown anaerobically in LB supplemented with 35 μg of tetracycline ml−1 and 0.2% fructose, and β-galactosidase activity was determined as described in Materials and Methods. Activity is shown relative to the β-galactosidase activity obtained using the FF(−41.5) promoter in the M182 background and is the means of three independent determinations. The error is one standard deviation from the mean.
Suppression of FNR-dependent transcription repression by FNR G74C.
Green and colleagues reported that the E. coli yfiD promoter is activated by FNR binding to a target site centered at position −40.5. They found that this activation is suppressed by FNR binding to a second upstream site at position −93.5, but that the suppression can be partially relieved by the G74C substitution in FNR (11, 12). This change appears to interfere with the FNR determinant responsible for down-regulation by upstream-bound FNR. Thus, we tested whether the G74C substitution could also relieve the suppression of the activity of the FF(−n)FF(−41.5) promoters due to upstream-bound FNR. To do this, we introduced pRW50 derivatives carrying selected FF(−n)FF(−41.5) promoter constructs into the M182 fnrA1 strain containing a plasmid encoding either wild-type FNR (pFNR) or FNR G74C (pFNR G74C). Promoter activities were measured and are listed in Table 5. Consistent with the results using the fnr+ M182 strain (Fig. 2a) with M182 fnrA1 containing pFNR, transcription repression was observed with the FF(−84.5)FF(−41.5), FF(−85.5)FF(−41.5), FF(−86.5)FF(−41.5), FF(−87.5)FF(−41.5), and FF(−95.5)FF(−41.5) promoters. However, with M182 fnrA1 containing pFNR G74C, the repression was substantially relieved.
TABLE 5.
Transcription activation at FF(−n)FF(−41.5) promoters by FNR and FNR G74C
| Promotera | Relative activity (%)b of:
|
|||
|---|---|---|---|---|
| pFNR
|
pFNR G74C
|
|||
| Activity | Error | Activity | Error | |
| FF(−41.5) | 100 | 13 | 99 | 17 |
| FF(−82.5)FF(−41.5) | 131 | 4 | 168 | 12 |
| FF(−83.5)FF(−41.5) | 115 | 13 | 196 | 20 |
| FF(−84.5)FF(−41.5) | 24 | 0.2 | 79 | 7 |
| FF(−85.5)FF(−41.5) | 19 | 1 | 67 | 10 |
| FF(−86.5)FF(−41.5) | 26 | 3 | 84 | 8 |
| FF(−87.5)FF(−41.5) | 62 | 5 | 102 | 7 |
| FF(−88.5)FF(−41.5) | 103 | 3 | 102 | 4 |
| FF(−89.5)FF(−41.5) | 110 | 8 | 112 | 5 |
| FF(−90.5)FF(−41.5) | 116 | 9 | 164 | 13 |
| FF(−95.5)FF(−41.5) | 34 | 1 | 101 | 10 |
| EE(−85.5)FF(−41.5) | 97 | 20 | 139 | 9 |
| EE(−90.5)FF(−41.5) | 102 | 15 | 109 | 17 |
Promoters were cloned on EcoRI-HindIII fragments in the lac expression vector pRW50.
β-Galactosidase activities were measured in M182 fnrA1 cells containing plasmids encoding either wild-type FNR (pFNR) or FNR G74C (pFNR G74C) and grown anaerobically in LB supplemented with 35 μg of tetracycline ml−1, 100 μg of ampicillin ml−1, and 0.2% fructose, as described in Materials and Methods. Activities are expressed relative to the activity obtained using the FF(−41.5) promoter in the M182 fnrA1 (pFNR) background (7,059 Miller units). The errors shown are one standard deviation from the means of three independent determinations.
In vitro studies. To investigate tandem-bound FNR in vitro we focused on three promoters: the starting promoter, FF(−41.5), an FNR-repressed promoter, FF(−85.5)FF(−41.5), and FF(−90.5)FF(−41.5), which is not repressed by upstream-bound FNR (Fig. 1a and 2a). DNA fragments carrying the promoters were transferred to the vector pSR for these experiments. To facilitate in vitro studies we used the FNR* mutant, FNR D154A, which can dimerize, bind to DNA sites for FNR, and activate transcription by RNAP at FNR-dependent promoters in aerobic conditions (16, 17).
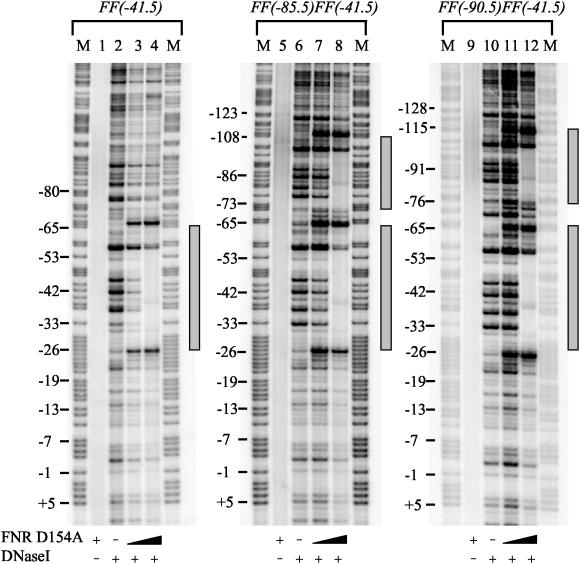
First, we used DNaseI footprinting to monitor FNR binding to the different sites at the promoters. Clear footprints due to FNR D154A binding are observed (Fig. 3). At the FF(−41.5) promoter, FNR D154A protects a single zone from positions −26 to −65 relative to the transcription start point, while at the FF(−85.5)FF(−41.5) and FF(−90.5)FF(−41.5) promoters FNR D154A protects two zones that correspond to the tandem binding sites. Parallel electromobility shift assays confirmed that FNR D154A can bind to two sites at the FF(−85.5)FF(−41.5) and FF(−90.5)FF(−41.5) promoters (data not shown).
FIG. 3.
Analysis of promoter binding by FNR D154A. For DNaseI footprint analysis, promoter fragments were labeled with [γ-32P]ATP, were incubated with various concentrations of FNR D154A prior to treatment with DNaseI, and were analyzed on a DNA sequencing gel. The gel was calibrated with Maxam-Gilbert G+A sequencing reactions. Regions of protection due to FNR D154A are indicated with gray rectangles. The samples were loaded as follows: G+A sequencing reaction (M); no DNaseI, 3 μM FNR D154A control (lanes 1, 5, and 9); DNaseI, no protein control (lanes 2, 6, and 10); DNaseI, 0.1 μM FNR D154A (lanes 3, 7, and 11); DNaseI, 3 μM FNR D154A (lanes 4, 8, and 12).
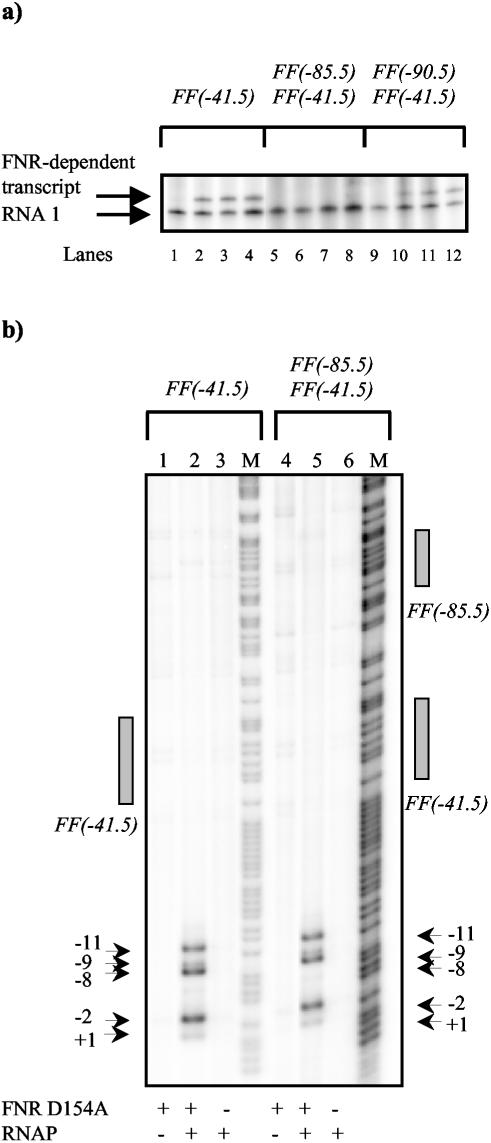
Second, we used an in vitro transcription assay to measure FNR-dependent activation at the different promoters. Labeled RNA products from the assay were analyzed by using a phosphorimager, and the image is shown in Fig. 4a. Bands due to transcripts regulated by the different FNR-dependent promoters and due to the control RNA1 can be distinguished. With the FF(−41.5) and FF(−90.5)FF(−41.5) promoters, transcription is clearly activated by FNR. In contrast, no FNR-dependent transcript was found with the FF(−85.5)FF(−41.5) promoter. This suggests that the suppression of promoter activity by upstream-bound FNR observed with the FF(−85.5)FF(−41.5) promoter in vivo can be reproduced in vitro. In complementary experiments, we checked for promoter opening by using potassium permanganate as a probe. The results in Fig. 4b show clear FNR-dependent unwinding around the transcript start site at both the FF(−41.5) and FF(−85.5)FF(−41.5) promoters. Thus, the defect in FNR-dependent activation of FF(−85.5)FF(−41.5) may not be due to a defect in promoter opening.
FIG. 4.
Analysis of FNR D154A-dependent open complex formation and transcription. (a) In vitro transcription analysis. The FF(−41.5)/pSR, FF(−85.5)FF(−41.5)/pSR, and FF(−90.5)FF(−41.5)/pSR constructs were used as templates for in vitro transcription. Eight nanomolar template plasmid DNA was incubated with 50 nM RNA polymerase, nucleoside triphosphates, and [α-32P]UTP, with or without FNR D154A (0 to 0.5 μM). Transcripts were analyzed on a denaturing polyacrylamide gel. The control RNA1 and the FNR-dependent transcript are marked. Samples were loaded as follows: no FNR D154A (lanes 1, 5, and 9); 0.05 μM FNR D154A (lanes 2, 6, and 10); 0.1 μM FNR D154A (lanes 3, 7, and 11); 0.5 μM FNR D154A (lanes 4, 8, and 12). (b) Permanganate footprint analysis. Shown is a phosphor image of a denaturing polyacrylamide sequencing gel on which DNA cleavage due to attack by permanganate was analyzed. [γ-32P]ATP-labeled promoter fragments were incubated with or without FNR D154A (1 μM) and with or without purified RNA polymerase σ70 holoenzyme (50 nM) prior to treatment with permanganate. The gel is calibrated with Maxam-Gilbert G+A sequencing reactions. The locations of the permanganate-induced cleavage sites and the FNR consensus binding sites are marked. Samples were loaded as follows: G+A sequencing reaction (M); 1 μM FNR D154A (lanes 1 and 4); 1 μM FNR D154A, 50 nM RNAP (lanes 2 and 5); 50 nM RNAP (lanes 3 and 6).
Activity of promoters with tandem sites for FNR: the FF(−n)FF(−61.5) series.
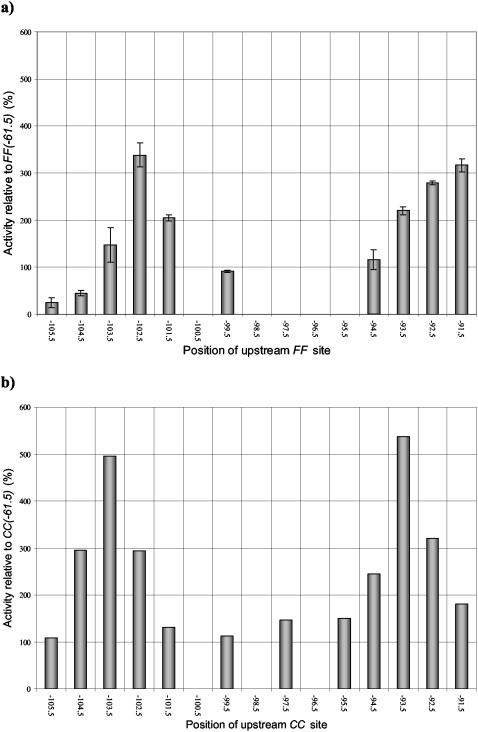
We constructed a second series of related promoters carrying one consensus DNA site for FNR centered at position −61.5 and a second consensus DNA site for FNR located further upstream [the FF(−n)FF(−61.5) series] (Fig. 1b). The starting point for these constructions was the CC(−n)CC(−61.5) promoter series described in a previous study (Table 3) (24). Each new FF(−n)FF(−61.5) promoter was transferred into pRW50, and β-galactosidase activity measurements showed that expression is anaerobically induced and is dependent on FNR (data not shown). Figure 5a illustrates the expression from each promoter in an fnr+ background during anaerobic growth. The results show that expression depends on the spacing between the two DNA sites for FNR.
FIG. 5.
Transcription activation by FF(−n)FF(−61.5) and CC(−n)CC(−61.5) promoters. (a) The anaerobic β-galactosidase activity of M182 (Δlac) cells transformed with each of the FF(−n)FF(−61.5) promoters cloned in pRW50, displayed as a percentage of the activity achieved using the FF(−61.5) promoter. The relative activities are the means of three independent determinations, and error bars depict one standard deviation from the mean. The x axis indicates the location of the center of the upstream 22-bp FF binding site. (b) For comparison, the activities of M182 cells transformed with the CC(−n)CC(−61.5) promoter series reported in Tebbutt et al. (24) are also shown, relative to the activity achieved using the CC(−61.5) promoter. The x axis indicates the location of the center of the upstream 22-bp CC binding site.
For comparison, Fig. 5b illustrates expression from the previously constructed promoters carrying tandem DNA sites for CRP [the CC(−n)CC(−61.5) series] (24). As described before, when the upstream DNA site for CRP is located near position −93.5 or position −103.5, expression is increased up to fivefold. At intermediate locations the upstream-bound CRP has little or no effect, and the observed expression is similar to that found at the CC(−61.5) promoter, with a single DNA site for CRP at position −61.5.
Comparison of data in Fig. 5a and b shows both similarities and differences between the effects of upstream-bound FNR at the FF(−n)FF(−61.5) promoters and the effects of upstream-bound CRP at the CC(−n)CC(−61.5) promoters. The upstream-bound FNR clearly causes increased expression when it is located near positions −92.5 and −102.5. At the intermediate positions, −94.5 and −99.5, the upstream-bound FNR has little or no effect, and the observed expression is similar to that found at the FF(−61.5) promoter, with a single DNA site for FNR at position −61.5. Thus, at these promoters the pattern of expression from the FF(−n)FF(−61.5) and CC(−n)CC(−61.5) series of promoters is at least superficially similar. However, with the FF(−104.5)FF(−61.5) and FF(−105.5)FF(−61.5) promoters, expression is sharply reduced by the upstream-bound FNR. Interestingly, at these promoters the center-to-center distances between the tandem DNA sites for FNR are 43 and 44 bp, which are identical to the distances between the DNA sites for FNR at the FF(−n)FF(−41.5) promoters where greatest repression is observed (Fig. 2a).
DISCUSSION
Tandem DNA sites for CRP or FNR are found at many promoters. At some of these, optimal expression depends on the binding of CRP to both target sites. In a study aimed to investigate this, Belyaeva et al. (3) started with the CC(−41.5) promoter (carrying a single DNA site for CRP) and introduced a second DNA site for CRP at different upstream locations. Belyaeva et al. (3) showed that the tandem-bound CRP molecule could activate transcription synergistically, provided it was located at particular positions. To explain their data, Belyaeva et al. (3) argued that synergy depended on the upstream and downstream CRP molecules being positioned such that they could each make productive interactions with αCTD of RNAP. When the upstream CRP was incorrectly positioned, it had no effect on CRP-dependent activation due to the CRP molecule at position −41.5 (although repression was found when the upstream-bound CRP was positioned at −71.5 or other downstream locations). In this study we performed a parallel experiment, starting with the FF(−41.5) promoter, which is dependent on FNR, and introducing a second DNA site for FNR at different upstream locations. Our results (Fig. 2) show that the pattern of expression from the FF(−n)FF(−41.5) promoter series is very different from that observed by Belyaeva et al. (3) for the CC(−n)CC(−41.5) series. First, we found no promoter where the upstream-bound FNR increased expression by more than twofold, arguing that strong synergy between tandem-bound FNR molecules does not occur (at least in our system). Second, we found that at some promoters, upstream-bound FNR causes a sharp suppression of FNR-dependent transcription. Thus, upstream-bound FNR, located near position −85.5 or position −95.5, down-regulates transcription activation by FNR bound at position −41.5. Interestingly, the repression effect appears to be face-of-the-DNA helix-dependent, suggesting that it results from a particular juxtaposition of bound FNR molecules. Our in vitro studies show that, at least at the FF(−85.5)FF(−41.5) promoter, both FNR molecules bind but that transcription is hindered (Fig. 3 and 4a). Interestingly, according to its reactivity with potassium permanganate, the target promoter can open (Fig. 4b). However, kinetic studies will be needed to pinpoint the precise step that is down-regulated by upstream-bound FNR.
The overall conclusion from our study with the FF(−n)FF(−41.5) series of promoters is that tandem-bound FNR molecules work together to repress rather than to activate transcription. This is consistent with conclusions from studies of the E. coli ndh and yfiD promoters, where repression is dependent on tandem binding of FNR. Expression from the ndh promoter is FNR independent but is repressed by FNR binding at positions −50.5 and −94.5 (21), with efficient repression requiring FNR binding to the more upstream site. Expression from the yfiD promoter is activated by FNR binding at position −40.5, but this activation is suppressed by the binding of upstream FNR at position −93.5 (12). Strikingly, the center-to-center distances between the tandem FNR sites at the ndh and yfiD promoters are 44 and 53 bp, respectively, which correspond to spacings that give sharp repression in the FF(−n)FF(−41.5) promoter series. Working with the yfiD promoter, Green and colleagues (11, 20) have shown that down-regulation is due to specific interactions between the tandem-bound FNR molecules, which interact via a surface-exposed determinant. They have identified residues where substitutions prevent or reduce these interactions. We have studied the effects of one such substitution (G74C), and we found that it relieved, at least partially, the repression at the FF(−n)FF(−41.5) promoters (Table 5).
In the final part of our study we performed a parallel set of constructions to make the FF(−n)FF(−61.5) promoter series. Our results (Fig. 5) show that the pattern of expression of this promoter series is somewhat similar to that observed by Tebbutt et al. (24) for the CC(−n)CC(−61.5) series. At some locations, upstream-bound FNR leads to increased promoter expression, although we can make no conclusion about synergy, since the controls to check that the tandem-bound FNR molecules functioned via the same promoter were not done. The striking result concerns the FF(−104.5)FF(−61.5) and FF(−105.5)FF(−61.5) promoters where expression is sharply reduced by the upstream-bound FNR. At these promoters the center-to-center distances between the tandem DNA sites for FNR are 43 and 44 bp. Taken together with previous results, we can conclude that 43 or 44 bp is a critical spacing for transcription repression by tandem-bound FNR molecules.
Many transcription activators can function as repressors if they are misplaced. Thus, both CRP and FNR can function as simple repressors merely by blocking access of RNAP to a promoter (8, 13). To do this, a single correctly placed FNR or CRP molecule is needed (10, 25). However, as well as using this simple repression mechanism, FNR has evolved a second repression strategy that depends on interactions between tandem-bound FNR molecules. This study shows that these interactions can occur in different contexts and that they are optimal when the spacing between the two FNR molecules is around 44 or 53 bp. These interactions appear to depend solely on FNR, but we are still ignorant of their precise nature. While FNR and CRP appear to have evolved from a common ancestor, CRP seems to have evolved so that tandem-bound CRP molecules can function synergistically to activate transcription at target promoters. In contrast, FNR has evolved so that tandem-bound FNR molecules cooperate in repression (5).
Acknowledgments
This project was generously funded by the UK BBSRC, project grant number G13155. The Birmingham Functional Genomics Laboratory was funded by the BBSRC, grant 6/JIF13209.
We thank Rick Gourse and Andy Bell for providing E. coli strains and Eleanor Chant for assistance with some of the promoter construction and β-galactosidase assays.
REFERENCES
- 1.Bell, A., and S. Busby. 1994. Location and orientation of an activating region in the Escherichia coli transcription factor, FNR. Mol. Microbiol. 11:383-390. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A. I., K. L. Gaston, J. A. Cole, and S. J. W. Busby. 1989. Cloning of binding sequences for the Escherichia coli transcriptional activators FNR and CRP, location of bases involved in discrimination between FNR and CRP. Nucleic Acids Res. 17:3865-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belyaeva, T. A., V. A. Rhodius, C. L. Webster, and S. J. W. Busby. 1998. Transcription activation at promoters carrying tandem DNA sites for the Escherichia coli cyclic AMP receptor protein: organisation of the RNA polymerase α subunits. J. Mol. Biol. 277:789-804. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Browning, D. F., D. Lee, J. Green, and S. J. W. Busby. 2002. Secrets of bacterial transcription initiation taught by the Escherichia coli FNR protein, p. 127-142. In D. Hodgson and C. Thomas (ed.), Signals, switches, regulons, and cascades. SGM symposium. Cambridge University Press, Cambridge, United Kingdom.
- 6.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376-379. [Google Scholar]
- 7.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 8.Busby, S., and A. Kolb. 1996. The CAP modulon, p. 255-279. In E. C. C. Lin and A. S. Lynch (ed.), Regulation of gene expression in Escherichia coli. R. G. Landes & Co., Austin, Tex.
- 9.Busby, S., D. Kotlarz, and H. Buc. 1983. Deletion mutagenesis of the Escherichia coli galactose operon promoter region. J. Mol. Biol. 154:211-227. [DOI] [PubMed] [Google Scholar]
- 10.Cruz-Ramos, H., J. Crack, G. Wu, M. N. Hughes, C. Scott, A. J. Thomson, J. Green, and R. K. Poole. 2002. NO-sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 21:3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green, J., and F. A. Marshall. 1999. Identification of a surface of FNR overlapping activating region 1 that is required for repression of gene expression. J. Biol. Chem. 274:10244-10248. [DOI] [PubMed] [Google Scholar]
- 12.Green, J., M. L. Baldwin, and J. Richardson. 1998. Downregulation of Escherichia coli yfiD expression by FNR occupying a site at −93.5 involves the AR1-containing face of FNR. Mol. Microbiol. 29:1113-1123. [DOI] [PubMed] [Google Scholar]
- 13.Guest, J. R., J. Green, A. S. Irvine, and S. Spiro. 1996. The FNR modulon and FNR-regulated gene expression, p. 317-342. In E. C. C. Lin and A. S. Lynch (ed.), Regulation of gene expression in Escherichia coli. R. G. Landes & Co., Austin, Tex.
- 14.Kelsall, A., C. Evans, and S. Busby. 1985. A plasmid vector that allows fusion of the Escherichia coli galactokinase gene to the translation startpoint of other genes. FEBS Lett. 180:155-159. [Google Scholar]
- 15.Kolb, A., D. Kotlarz, S. Kusano, and A. Ishihama. 1995. Selectivity of the Escherichia coli RNA polymerase Eσ38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 23:819-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazazzera, B. A., D. M. Bates, and P. J. Kiley. 1993. The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 7:1993-2005. [DOI] [PubMed] [Google Scholar]
- 17.Lazazzera, B. A., H. Beinert, N. Khoroshilova, M. C. Kennedy, and P. J. Kiley. 1996. DNA binding and dimerization of the FeS containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 271:2762-2768. [DOI] [PubMed] [Google Scholar]
- 18.Lloyd, G. S., W. Niu, J. Tebbutt, R. H. Ebright, and S. J. W. Busby. 2002. Requirement for two copies of RNA polymerase α subunit C-terminal domain for synergistic transcription activation at complex bacterial promoters. Genes Dev. 16:2557-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lodge, J., J. Fear, S. Busby, P. Gunasekaran, and N.-R. Kamini. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Lett. 95:271-276. [DOI] [PubMed] [Google Scholar]
- 20.Marshall, F. A., S. L. Messenger, N. R. Wyborn, J. R. Guest, H. Wing, S. J. W. Busby, and J. Green. 2001. A novel promoter architecture for microaerobic activation by the anaerobic transcription factor FNR. Mol. Microbiol. 39:747-753. [DOI] [PubMed] [Google Scholar]
- 21.Meng, W., J. Green, and J. R. Guest. 1997. FNR-dependent repression of ndh gene expression requires two upstream FNR-binding sites. Microbiology 143:1521-1532. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Savery, N. J., T. Belyaeva, and S. Busby. 1996. DNaseI footprinting, p. 21-24. In K. Docherty (ed.), Gene transcription: DNA binding proteins. Essential techniques. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 24.Tebbutt, J., V. A. Rhodius, C. L. Webster, and S. J. W. Busby. 2002. Architectural requirements for optimal activation by tandem CRP molecules at a Class I CRP-dependent promoter. FEMS Microbiol. Lett. 210:55-60. [DOI] [PubMed] [Google Scholar]
- 25.Williams, S. M., H. J. Wing, and S. J. W. Busby. 1998. Repression of transcription initiation by Escherichia coli FNR protein: repression by FNR can be simple. FEMS Microbiol. Lett. 163:203-208. [DOI] [PubMed] [Google Scholar]
- 26.Wing, H. J., J. Green, J. R. Guest, and S. J. W. Busby. 2000. Role of activating region 1 of Escherichia coli FNR protein in transcription activation at class II promoters. J. Biol. Chem. 275:29061-29065. [DOI] [PubMed] [Google Scholar]