Abstract
Plasmid pTC-F14 is a 14.2-kb plasmid isolated from Acidithiobacillus caldus that has a replicon that is closely related to the promiscuous, broad-host-range IncQ family of plasmids. The region containing the mobilization genes was sequenced and encoded five Mob proteins that were related to those of the DNA processing (Dtr or Tra1) region of IncP plasmids rather than to the three-Mob-protein system of the IncQ group 1 plasmids (e.g., plasmid RSF1010 or R1162). Plasmid pTC-F14 is the second example of an IncQ family plasmid that has five mob genes, the other being pTF-FC2. The minimal region that was essential for mobilization included the mobA, mobB, and mobC genes, as well as the oriT gene. The mobD and mobE genes were nonessential, but together, they enhanced the mobilization frequency by approximately 300-fold. Mobilization of pTC-F14 between Escherichia coli strains by a chromosomally integrated RP4 plasmid was more than 3,500-fold less efficient than the mobilization of pTF-FC2. When both plasmids were coresident in the same E. coli host, pTC-F14 was mobilized at almost the same frequency as pTF-FC2. This enhanced pTC-F14 mobilization frequency was due to the presence of a combination of the pTF-FC2 mobD and mobE gene products, the functions of which are still unknown. Mob protein interaction at the oriT regions was unidirectionally plasmid specific in that a plasmid with the oriT region of pTC-F14 could be mobilized by pTF-FC2 but not vice versa. No evidence for any negative effect on the transfer of one plasmid by the related, potentially competitive plasmid was obtained.
Plasmid pTC-F14 was recently isolated from the moderately thermophilic (50°C), acidophilic, sulfur-oxidizing bacterium Acidithiobacillus caldus (11). The strain of A. caldus in which the plasmid was found was one of two dominant organisms in a bacterial consortium undergoing pilot-scale testing for the commercial extraction of nickel from ores (17). The 14.2-kb plasmid pTC-F14 was shown to have an IncQ-like replicon that was closely related to, but compatible with, the broad-host-range 12.2-kb plasmid pTF-FC2 (9, 10). Plasmid pTF-FC2 had been previously isolated from a different, but related, iron- and sulfur-oxidizing bacterium, Acidithiobacillus ferrooxidans (6, 16, 19).
Although IncQ and IncQ-like plasmids are not self-transmissible, they are efficiently mobilized by conjugative plasmids of the Escherichia coli IncPα and IncPβ groups. By using IncP plasmids or IncP-based helper plasmids, the IncQ plasmids have been successfully mobilized to a large number of hosts, including a wide range of gram-negative bacteria; several gram-positive bacteria, including Arthrobacter spp., Streptomyces lividans, and Mycobacterium smegmatis; and cyanobacteria, such as Synechococcus, as well as being mobilized into plant and animal cells (reviewed in reference 18). Likewise, pTF-FC2 has been mobilized from E. coli to several gram-negative bacteria and from Agrobacterium tumefaciens to plant cells (D. E. Rawlings, unpublished observations). This, together with the broad-host-range properties of their replicons, makes these plasmids highly promiscuous and interesting to study from the fundamental biology, ecology, and applied biology points of view.
There are two major groups of IncQ and IncQ-like plasmids, the most distinguishing characteristic between the groups being whether they possess a three-gene, IncQ-like mobilization system or a five-gene, IncP-like mobilization system (18). Examples of the three-mob-gene plasmid family are the IncQ plasmids RSF1010, R1162, and R300B and the IncQ-like plasmids pIE1107, pIE1115, pIE1130, and pDN1 (25, 27). Plasmid pTF-FC2 was the only example of an IncQ-like plasmid with a five-mob-gene system (19) until the discovery of plasmid pTC-F14. Recently, the sequence of another IncQ-like plasmid, pRAS3, with a five-mob-gene system has been observed (14), although no biology of this system has been reported. The amino acid sequences of the Mob proteins from the two groups of plasmids belonging to the IncQ family are not related to each other.
As part of this study, we report that like pTF-FC2, the mobilization genes of pTC-F14 are of the IncP type. Because pTC-F14 and pTF-FC2 are promiscuous plasmids that were isolated from acidiphilic, iron- and/or sulfur-oxidizing, chemolithotrophic bacteria that share a similar habitat, it is not unlikely that the plasmids may come into contact with each other. Plasmids pTC-F14 and pTF-FC2 have diverged sufficiently for their replicons to be compatible, which should allow them to coexist in the same host cell (9, 10). This raised questions such as have the mob genes diverged sufficiently to be plasmid specific, or will they complement the mobilization activity of each other? Was there competition between plasmids at the level of mobilization? That is, had one of the plasmids evolved a mobilization system that would allow it to dominate the horizontal transfer process, thereby giving it a selective advantage in preference to the other? Here we characterize the mobilization genes of pTC-F14 and report on the ability of the mobilization systems of pTC-F14 and pTF-FC2 to interact with each other.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The strains, plasmids, and primers used in this study are listed in Table 1.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | F′/endA1 hsdR17(rK− mK+) supE44 thi-1 reacA1 gyrA(Nalr) relA1 Δ(lacZYA-argF)U169 (φ80dlacΔ(lacZ)M15) | Promega Corp., Madison, Wis. |
| S17.1 | recA pro hsdR (RP4-2 Tc::Mu Km::Tn7) | 23 |
| CSH56 | F−ara Δ(lac pro) supD nalA thi | Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. |
| HB101 | F− Δ(mcrC-mrr) hsdS20 recA13 ara-14 proA2 lacY1 λ−galK2 rpsL20(Smr) xyl-5 mtl-1 supE44 | 3 |
| Plasmids | ||
| pUC19 | AmprlacZ′; ColE1 replicon, cloning vector | 28 |
| pACYC184 | Tcr Cmr; p15A replicon, cloning vector | 5 |
| pBR322 | Ampr Tcr; ColE1 replicon, cloning vector | 2 |
| pKK223-3 | Ampr; ColE1 replicon, expression vector | 4 |
| pTC-F14Cm | Cmr; natural pTC-F14 plasmid with a chloramphenicol resistance gene inserted at the single BamHI site | 9 |
| pTC-F14Km | Kmr; pTC-F14Cm with the chloramphenicol resistance gene replaced by the kanamycin resistance gene from Tn5 | This study |
| pDER412 | Cmr; natural pTF-FC2 plasmid with chloramphenicol resistance gene cloned into the Tn5467 transposon | 16 |
| pMmob | Ampr, 5,554-bp BamHI-XbaI fragment of pTC-F14 containing all mobilization genes and the repB primase, cloned into pUC19 | This study |
| pMmob1-pMmob9 | Ampr; PCR-based deletions of the mobilization region of pTC-F14 cloned into pUC19; refer to Fig. 1 | This study |
| pMmob1184 | Cmr; minimum mobilization region one of pTC-F14 cloned into the tetracycline resistance marker of pACYC184 | This study |
| pMmob1322 | Ampr; minimum mobilization region one of pTC-F14 cloned into pBR322 | This study |
| pmobE | Ampr; PCR product of mobE gene of pTF-FC2 cloned into pKK223-3 | This study |
| pmobDE | Ampr; PCR product of mobDE genes of pTF-FC2 cloned into pKK223-3 | This study |
| pmobCDE | Ampr; PCR product of mobCDE genes of pTF-FC2 cloned into pKK223-3 | This study |
| pAC105 | Cmr; exonuclease III shortening of the pTF-FC2 mobilization region containing mobC, -D, and -E cloned into pACYC184 | 20 |
| pAC209 | Cmr; exonuclease III shortening of the pTF-FC2 mobilization region containing mobA, -B, -C, and -D and a truncated mobE cloned into pACYC184 | 20 |
| pAC218 | Cmr; exonuclease III shortening of the pTF-FC2 mobilization region containing mobA, -B, and -C with mobD and -E removed also in pACYC184 | 20 |
| pAC221 | Cmr; exonuclease III shortening of the pDER412 mobilization region containing mobA and -B and the oriT cloned into pACYC184 | 20 |
| pOriTF14 | Ampr; a 203-bp HindIII-NcoI fragment of pTC-F14 containing the oriT cloned into pUC19 | This study |
| pOriTFC2 | Ampr; the oriT of pTF-FC2 cloned into pUC19 | This study |
| Primers | ||
| mobEF2 | (EcoRI) 5′-TACAGAATTCAGCAAGCGCATGAGC-3′ | This study |
| mobDEF2 | (EcoRI) 5′-TACAGAATTCCCAAAACCCGACAGC-3′ | This study |
| mobCDEF2 | (EcoRI) 5′-TATAGAATTCCCACGTGGCGAAGCC-3′ | This study |
| mobER2 | (XbaI) 5′-TACATCTAGAATGTTGAGCGCGTCG-3′ | This study |
| mobAR14 | (EcoRI) 5′-TACAGAATTCCGGTCCATGTCGTCG-3′ | This study |
| repBR14 | (EcoRI) 5′-TACAGAATTCGGGTAATCGGATGGC-3′ | This study |
| mobC′R14 | (PstI) 5′-TATACTGCAGCTTTCCCGCCTTTGC-3′ | This study |
| mobCR14 | (PstI) 5′-TATACTGCAGTTGCCACCACCGACG-3′ | This study |
| mobDR14 | (PstI) 5′-TATACTGCAGTCGGGTGTCGGTTCC-3′ | This study |
| mobER14 | (PstI) 5′-TACACTGCAGCTGTCCGAAAGTAGG-3′ | This study |
| mobAR14#2 | 5′-TGGCGTCGCTTGTTTGGTTC-3′ | 10 |
Media and growth conditions.
E. coli strains were grown in either Luria-Bertani broth or on LA plates (21) at 37°C, supplemented as required with antibiotics at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 34 μg/ml; streptomycin, 50 μg/ml; kanamycin, 50 μg/ml; and nalidixic acid, 50 μg/ml.
Mating assays.
Donor and recipient (CSH56) cells were cultured separately overnight with appropriate antibiotic selection. Cells were washed three times in 0.85% (wt/vol) NaCl solution and mixed in a donor/recipient ratio of 1:10. An LA plate was spotted with 100 μl of this mixture and incubated at 37°C for 1 h. The agar plug was excised, suspended in 5 ml of 0.85% NaCl solution, and vigorously shaken to dislodge mating cells. Cells were pelleted by a 2-min spin in a microcentrifuge and resuspended in 1 ml of 0.85% NaCl solution. Serial dilutions were then plated onto media that selected for donor and transconjugant cells. The transfer frequency was calculated as the number of transconjugants per donor during the 1-h mating period.
DNA manipulations, sequencing, and bioinformatics.
General techniques were performed according to standard procedures (21) or the manufacturers' recommendations. DNA was digested to give fragments with sizes of 700 bp to 1.2 kbp and cloned into the pUC19 vector. The DNA sequence was determined by a combination of sequencing from the ends of a number of subclones and synthesis of specific primers to obtain overlapping sequence from both strands. Sequencing was performed by the dideoxy chain termination method with an ABI PRISM 377 automated DNA sequencer, and the sequence was analyzed with a variety of software programs (mainly the PC-based DNAMAN [version 4.1] package from Lynnon Biosoft). Searches for sequences related to Mob proteins were performed by using the gapped-BLAST program of the National Center for Biotechnology Information at www.ncbi.nih.nlm.gov (1). Sequence alignments (based on CLUSTAL W) were carried out with the multiple alignment program, and amino acid sequence homology trees were constructed with the tree output program within the DNAMAN package.
PCRs.
PCRs were performed with the Expand high-fidelity Taq DNA polymerase from Roche with a Hybaid PCR Sprint cycler. Plasmid pDER412 was used as template with primers mobEF2, mobDEF2, mobCDEF2, and mobER2 (listed in Table 1) to produce pmobCDE, pmobDE, and pmobE, respectively. Primers mobAR14, mobAR14#2, repBR14, mobC′R14, mobCR14, mobDR14, and mobER14 (Table 1 and Fig. 1) with pMmob as a template were used to give the mobilization region fragments (pMmob1 to pMmob9). After an initial denaturation of 60 s at 94°C, 25 cycles of 30 s at 55 to 60°C (depending on primer set) and an elongation step of up to 4 min (approximately 1 min per 1,000 bp) at 72°C were performed. A final extension step of 120 min at 72°C before cooling to 4°C completed the reaction.
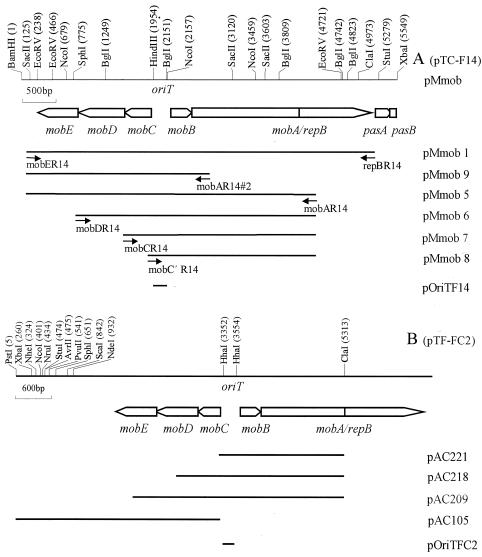
FIG. 1.
Genetic and restriction endonuclease cleavage maps of the mobilization regions of plasmids pTC-F14 and pTF-FC2. (A) The 5.55-kb BamHI-XbaI region of pTC-F14 showing the locations of the mob, repB, and pas genes as well as the oriT gene. The positions of the primers used to amplify and construct certain subclones are shown by short horizontal arrows. (B) The previously reported mob region of pTF-FC2 and subclones (20) used in this study.
Nucleotide sequence accession number.
The nucleotide sequence of the 5.5-kbp region sequenced has been submitted to the EMBL-GenBank database under accession no. NC_004734/AF325537.
RESULTS
Mobilization of pTC-F14.
Selectable chloramphenicol and kanamycin resistance genes were cloned into plasmid pTC-F14 to produce plasmids pTC-F14Cm and pTC-F14Km, respectively (Table 1). These plasmids were transformed into an E. coli S17-1 donor strain that has an RP4 plasmid derivative integrated into the chromosome to provide the conjugative functions required for plasmid mobilization. Both pTC-F14Cm and pTC-F14Km were mobilized to an E. coli CSH56 recipient strain at similar frequencies of approximately 2.8 × 10−3 transconjugants per donor. To determine whether the type of conjugative plasmid affected the mobilization frequency, we compared mobilization frequencies by using two self-transmissible plasmids different from the RP4 (IncPα) that was integrated into the chromosome of E. coli S17-1. Plasmid pTC-F14Cm was mobilized by R751 (IncPβ) from E. coli HB101 at a frequency about 100-fold lower than that by the RP4 derivative in E. coli S17-1, while mobilization by R388 (IncW) was not detectable (Table 2). A 5.55-kb BamHI-XbaI fragment from pTC-F14 was subcloned into the nonmobilizable vector pUC19 and was found to be mobilized by E. coli S17-1 at frequencies that approached saturation. Saturation indicates that after 1 h of mating at a donor/recipient ratio of 1:10, the number of transconjugants was approximately equal to the number of recipients. This 5.55-kb fragment therefore contained all of the components needed for mobilization and was sequenced.
TABLE 2.
Mobilization frequency of plasmids and constructs
| Test plasmida | Plasmid present in transa | Mobilization frequency of test plasmidb |
|---|---|---|
| pTC-F14Km | (2.7 ± 1.5) × 10−3 | |
| pTC-F14Cm | (2.8 ± 1.8) × 10−3 | |
| pTC-F14Cmc | R751 | 1.3 × 10−5 |
| pTC-F14Cmc | R388 | <10−6 |
| pTF-FC2 (pDER412) | ≥10 | |
| pTF-FC2 (pDER412) | pTC-F14Km | ≥10 |
| pTC-F14Km | pTF-FC2 (pDER412) | 8.4 ± 0.52 |
| pMmob (F14, mobEDCBA repB pasA) | ≥10 | |
| pMmob1 (F14, mobEDCBA repB) | ≥10 | |
| pMmob5 (F14, mobEDCBA) | (3.3 ± 3.1) × 10−1 | |
| pMmob6 (F14, mobDCBA) | (1.2 ± 1.0) × 10−1 | |
| pMmob7 (F14, mobCBA) | (5.4 ± 3.3) × 10−3 | |
| pMmob8 (F14, mobBA) | <10−6 | |
| pMmob9 (F14, mobEDCB) | <10−6 | |
| pMmob8 (F14, mobBA) | pTC-F14Cm | 2.4 ± 1.4 |
| pMmob1184 (as for pMmob1) | (2.1 ± 0.8) × 10−1 | |
| pMmob1322 (as for pMmob1) | (2.3 ± 1.6) × 10−1 | |
| pTC-F14Cm | pAC105 (FC2, mobEDC) | (7.9 ± 2.1) × 10−1 |
| pTC-F14Cm | pAC209 (FC2, mobDCBA) | (2.3 ± 3.5) × 10−3 |
| pTC-F14Cm | pAC221 (FC2, mobBA) | (8.9 ± 2.4) × 10−4 |
| pTC-F14Cm | pmobE (FC2) | (4.4 ± 2.1) × 10−3 |
| pTC-F14Cm | pmobDE (FC2) | ≥10 |
| pTC-F14Cm | pmobCDE (FC2) | (4.7 ± 2.9) × 10−1 |
| pTC-F14Cm | pAC218 (FC2 mobC) + pmobE (FC2) | (1.1 ± 2.6) × 10−3 |
| pOriTF14 | <10−6 | |
| pOriTF14 | pTC-F14Cm | 1.5 ± 1.0 |
| pOriTF14 | pTF-FC2 (pDER412) | (3.5 ± 0.1) × 10−2 |
| pOriTF14 | pTC-F14Km + pTF-FC2 (pDER412) | ≥10 |
| pOriTFC2 | <10−6 | |
| pOriTFC2 | pTC-F14Cm | <10−6 |
| pOriTFC2 | pTF-FC2 (pDER412) | ≥10 |
| pOriTFC2 | pTC-F14Km + pAC105 (FC2, mobEDC) | (3.5 ± 4.7) × 10−1 |
| pOriTFC2 | pTC-F14Km + pAC221 (FC2, mobBA) | (1.7 ± 0.8) × 10−2 |
Where relevant, genes or plasmids are indicated in parentheses.
Mobilization frequency is the number of transconjugants per donor during a 60-min mating with a donor/recipient ratio of 1:10 using E. coli S17-1 as the donor and E. coli CSH56 as the recipient. A mating frequency of ≥10 is indicated when the number of transconjugants equaled the number of recipients. Mating frequencies were the average of at least three independent experiments, and standard deviations are indicated.
E. coli HB101 was used as the donor strain.
Analysis of the mobilization region of pTC-F14 and comparison with related plasmids.
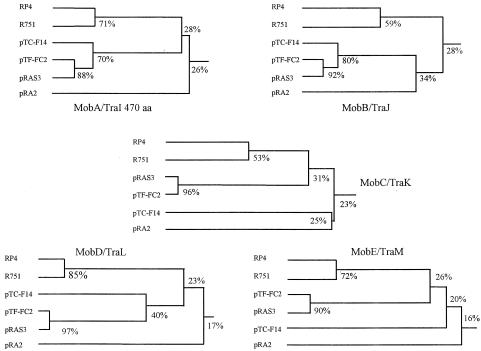
Five open reading frames were identified, arranged in a manner similar to those of pTF-FC2 (Fig. 1). However, some of the predicted amino acid sequences and characteristics of the mobilization proteins of pTC-F14 differed substantially from their counterparts in pTF-FC2. The MobA-RepB fusion and MobB proteins were the best conserved at 75.4 and 77.8% amino acid sequence identity, while the MobC, MobD, and MobE proteins were poorly conserved at 26.5, 39.8, and 21.2% amino acid sequence identity, respectively. Although all pairs of Mob proteins were of comparable sizes, the predicted pI values of the MobE proteins differed by almost 3 pH units. Surprisingly, plasmid pRAS3.1, isolated from Aeromonas salmonicida in Norway, has Mob proteins (GenBank accession no. AY043299.1/NC_003124.1) that are considerably more closely related to pTF-FC2 than pTC-F14 is to pTF-FC2. The sequences of the MobA, MobB, MobC, MobD, and MobE proteins of pRAS3.1 are 93.8, 88.8, 94.1, 97.4, and 88.8% identical to that of pTF-FC2, respectively, whereas they are only 75.0, 74.5, 25.8, 40.7, and 20.8% identical to that of pTC-F14. Three of the Mob proteins (MobA, MobB, and MobC) of pTC-F14, pTF-FC2, and pRAS3 had a greater than 20% amino acid sequence identity to the N-terminal 400-amino-acid portion of TraI and the complete TraJ and TraK proteins of the IncPα plasmid RP4 and the IncPβ plasmid R751, respectively. MobD and MobE had weaker but detectable sequence identity (17 to 18%) to TraL and TraM of RP4 and R751. These Mob proteins clearly belong to the IncP-like family of conjugation-associated, DNA processing proteins (Dtr), and a dendrogram showing the relationship between proteins of this family is presented in Fig. 2 (15, 24).
FIG. 2.
Phylogenetic relationship between the MobA/TraI, MobB/TraJ, MobC/TraK, MobD/TraL, and MobE/TraM proteins of the IncP and IncQ group 2 plasmids as well as pRA2. Since in the IncQ-related plasmids the MobA plasmids exist as a MobA-RepB fusion, only the N-terminal 470 amino acids were considered for comparison. The percentages shown represent amino acid sequence identities. Accession numbers are as follows: RP4, L27758; R751, U67194; pTC-F14, AF325537; pTF-FC2, M57717; pRAS3, AY043298; and pRA2, U88088.
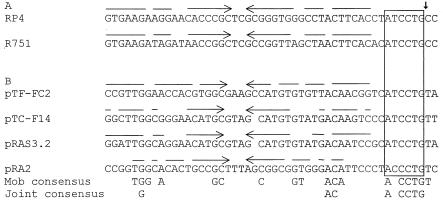
The oriT region of pTC-F14 was identified by sequence analysis and was found to be located on a 203-bp NcoI-HindIII fragment. This fragment was cloned into the nonmobilizable pUC19 vector (pOriT-F14) and transformed into E. coli S17-1, which contained a resistant pTC-F14Cm. pOriT-F14 was mobilized at a frequency that was about 500-fold greater than that of pTC-F14Cm. The oriT regions of IncPα and IncPβ plasmids as well as the four plasmids that have mobilization regions related to the IncP plasmids are compared in Fig. 3. The four mobilizable plasmids each contained an inverted repeat sequence that has been shown to be the site at which the relaxosome of plasmid RP4/RK2 binds prior to nicking at the oriT gene (26, 29). The highly conserved nucleotide hexamer that immediately precedes the nic site is also shown. In contrast to the mobilization proteins, for which plasmids pTF-FC2 and pRAS3 were the most closely related, the oriT regions of pTC-F14 and pRAS3 were considerably more closely related (matches at 42 of 50 bp) than those of pRAS3 and pTF-FC2 (32 of 50 bp) or pTC-F14 and pTF-FC2 (30 of 50 bp) (Fig. 3).
FIG. 3.
Comparison of the oriT regions of the IncP conjugative plasmids (A) and the IncQ group 2 and pRA2 mobilizable plasmids (B). Imperfect inverted repeat sequences are shown by horizontal arrows, while the highly conserved hexamer preceding the nick site is boxed. The small vertical arrow shows the nick site as determined for RP4/RK2.
Determination of which mob genes are essential or nonessential for mobilization.
A series of PCR-based deletions of the pTC-F14 mobilization region were made (Fig. 1). These were designed to test which genes were required for mobilization, as well as to determine the smallest region that is mobilized at the frequency of the intact mobE-repB region. When the entire mobE-repB region of pTC-F14 was cloned into the pUC19 vector (pMmob1), the mobilization frequency was at the level of saturation. This was an increase of more than 3,000-fold relative to the frequency obtained with the mobilization genes linked to its natural replicon (pTC-F14Km or pTC-F14Cm). To test whether this increase in mobilization frequency was due to placement of the mobE-repB region in the pUC19 vector, the mobE-repB region of pMmob1 was cloned into vectors pACYC184 and pBR322. Both of these constructs (pMmob1184 and pMmob1322) had mobilization frequencies ∼50-fold less than that of the pUC19 construct (pMmob1) but still ∼100-fold higher than that of the parent plasmid (Table 2). This suggested that the increase in mobilization frequency was associated with the placement of the mobilization region within the high-copy-number vector pUC19. Deletion of most of repB (pMmob5) reduced the mobilization frequency by about 30-fold. The mobilization frequency was restored to saturation levels by placing a repB-expressing construct in trans with pMmob5 (data not shown). This indicated that the repB gene assisted but was not essential for mobilization. Using the mobE-mobA (pMmob5) construct as a starting point, sequential deletion of the mobE (pMmob6), mobED (pMmob7), and mobEDC (pMmob8) genes was carried out. Deletion of mobE had no discernible effect on the mobilization frequency, while deletion of both mobE and mobD (pMmob7) reduced the mobilization frequency by approximately 600-fold, whereas there was no detectable mobilization of the mobE-mobC deletion (pMmob8). Deletion of most of mobA from pMmob5 (pMmob9) resulted in a construct with a mobilization frequency below the detection limit.
Comparison of the mobilization efficiencies and interaction between the mobilization systems of pTC-F14 and pTF-FC2.
The mobilization frequencies of plasmids containing the mob genes of pTC-F14 and the pTF-FC2 when associated with their natural replicons were compared. Plasmid pTC-F14Km was mobilized from E. coli S17-1 donor cells to CSH56 recipient cells at a frequency of 2.83 × 10−3 transconjugants per donor, which was more than 3,000-fold less than that of plasmid pDER412, which contained the pTF-FC2 mobilization genes (Table 2). To test whether mobilization of one plasmid was affected by coresidence of the other, both pTC-F14Km and pTF-FC2 (pDER412) were placed into E. coli S17-1 cells, and the frequency of transfer was measured. The frequency of mobilization of pTC-F14Km was enhanced almost to saturation in the presence of pTF-FC2, while the presence of pTC-F14 had no discernible effect on the mobilization of pTC-FC2. To determine what property of pTF-FC2 was required for this enhancement of mobilization frequency, plasmid constructs containing combinations of pTF-FC2 mob genes subcloned into the vector pACYC184 were introduced into E. coli S17-1(pTC-F14Cm) cells. Coresident plasmids pAC221 (containing pTF-FC2 mobA and mobB) and pAC209 (containing mobA, mobB, mobC, mobD, and a truncated mobE gene) did not increase the frequency of mobilization. In contrast, pAC105, which contained mobC, mobD, and a complete mobE gene, enhanced the mobilization frequency of pTC-F14 by about 100-fold, although this was about 10-fold less than when the whole of pDER412 was present. To determine whether this result was due to the mobE of pTF-FC2, the gene was amplified by PCR and cloned behind the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible tac promoter in plasmid pKK223-3 (construct pmobE). This construct did not improve the mobilization frequency of pTC-F14Cm. When a combination of pTF-FC2 mobD and mobE genes (pmobDE) were placed in trans and induced with IPTG, mobilization of pTC-F14Cm reached saturation. IPTG induction of a combination of the mobCDE genes (pmobCDE) enhanced pTC-F14Cm mobilization by approximately 150-fold to about the same level as with pAC105. This indicated that it was the combination of mobD and mobE from pTF-FC2 that enhanced pTC-F14 mobilization.
Interaction at the oriT regions.
To test whether the mobilization proteins of the two plasmids could act specifically on the oriT regions of each other, plasmids containing one of the cloned oriT fragments (pOriT-F14 and pOriT-FC2) were transformed into E. coli S17-1 containing either pTC-F14Km or pDER412. Both cloned oriT regions were functional, because they were mobilized by their respective parent plasmids at a frequency comparable to or greater than that of the parent plasmid (Table 2). The construct containing the oriT region of pTC-F14 (pOriT-F14) was mobilized at a frequency of 1.48 transconjugant per donor when pTC-F14 was placed in trans, but only at 3.48 × 10−2 with pDER412 in trans. With both pTC-F14 and pDER412 in trans, the mobilization frequency of pOriT-F14 reached saturation.
In the pTF-FC2 oriT complementation experiments, pDER412 was able to mobilize a construct containing its own oriT gene (pOriT-FC2) at a saturation frequency, while mobilization by pTC-F14 was below the limit of detection. Complementation of the oriT regions was therefore unidirectional, with pTF-FC2 able to mobilize the oriT of pTC-F14, but not vice versa. We tested whether there was any detectable interaction between the mob genes of pTC-F14 and the oriT of pTF-FC2 by providing two subsets of the mob genes of pTF-FC2 in trans. Plasmid pTC-F14Km was able to mobilize pOriT-FC2 at a frequency of 3.54 × 10−1 when in the presence of the pTF-FC2 mobCDE genes (pAC105) and at a frequency of 1.67 × 10−2 when the mobAB genes (pAC221) were present. This result was surprising and suggested that at least one of the pTF-FC2 mobCDE gene products, as well as one of the mobAB gene products, is able to independently assist in the recognition of the heterologous pTF-FC2 oriT by pTC-F14.
DISCUSSION
Plasmid pTC-F14 is the second member of the five-mob-gene, IncQ-like plasmid family to have its mobilization system investigated. Two other members of this IncQ-like plasmid family, now designated IncQ group 2, are pTF-FC2 and pRAS3, although no report on the biology of pRAS3 mobilization has been published. The observation that the amino acid sequence relationship between the mobilization proteins of plasmids pTF-FC2 and pRAS3 is much closer than that between the proteins of pTF-FC2 and pTC-F14 is remarkable (Fig. 2). The implication is that all three mob regions shared the same common ancestor, but pTF-FC2 and pRAS3 diverged more recently than pTF-FC2 and pTC-F14. Since divergence, pTF-FC2 and pRAS3 are now found in bacteria as different as the obligately acidophilic chemolithotrophic A. ferrooxidans strain FC isolated in South Africa and the neutrophilic A. salmonocida strain isolated in Norway. This serves to illustrate the highly promiscuous nature of the IncQ plasmid family. The 32.7-kb plasmid pRA2 is a fourth example of a plasmid containing a set of five mob genes that are related to the Tra1 system of the IncP plasmids (13). Plasmid pRA2 has a unique replicon with no similarity to those of the IncQ-like plasmids, and this suggests that the five-mob-gene system is a mobilization module that may also be acquired by different, otherwise unrelated plasmids.
To facilitate an investigation into the minimum region required for mobilization, the mobE-repB region of pTC-F14 was cloned into the nonmobilizable vector pUC19. The mobilization frequency of this construct (pMmob1) was at saturation level, which was about 104-fold higher than when the mobilization system was linked to its natural IncQ replicon (pTC-F14Km). An increase in mobilization frequency was not likely to be due to derepression of the IncP helper plasmid, because the mobilization frequency of the pTC-F14Km plasmid was not increased (data not shown). We considered whether this increase in mobilization frequency was associated with the high copy number of the pUC19 vector (up to 500 copies) and therefore transferred the mobE-repB region into the lower-copy-number vectors pACYC184 (20 to 30 copies) and pBR322 (25 to 50 copies). The mobilization frequency of these constructs (pMmob1184 and pMmob1322) was reduced 20- to 30-fold but was still 100-fold higher than that of pTC-F14Km (copy number 12 to 16). The increase in mobilization frequency was consistent with an increase in vector copy number, although this observation cannot be taken as proof that the mobilization frequency was affected by copy number.
We wished to detect whether all five of the mob genes were required for mobilization and whether the presence of the repB gene affected the mobilization frequency of pTC-F14. A characteristic of all IncQ and IncQ-like plasmids is that the mobA and repB genes are fused in such a way that the MobA (nickase) and the RepB (primase) may be synthesized as separate proteins or as MobA-RepB fusion protein. All three polypeptides have been detected for plasmids RSF1010 (22) and pTF-FC2 (7; Rawlings, unpublished). A fortuitous ClaI site was present in plasmid pTF-FC2 that allowed deletion of the repB primase gene (Fig. 1). This deletion did not affect the frequency of mobilization between E. coli strains. In the case of pTC-F14, a PCR-generated fragment was used to delete repB (pMmob5), and this reduced the mobilization frequency about 30-fold compared with that of a plasmid containing an intact repB (pMmob1). Placement of a repB-expressing construct in trans with the RepB-truncated MobA restored mobilization frequencies to levels similar to those when MobA-RepB was present, indicating that the decrease in mobilization was not due to an increase in structural instability of the truncated MobA. The repB gene was therefore not essential for mobilization of pTC-F14, although unlike pTF-FC2, the presence of repB did enhance mobilization. This is in sharp contrast with the three-mob-gene IncQ plasmid R1162, in which the MobA-linked RepB primase was essential for the recovery of plasmids in recipient cells (12). These authors argue that the mobA-repB gene fusion of R1162 most likely occurred after the IncQ replicon acquired mobilization genes and may be unique among IncQ-like plasmids. Deletion of the pTC-F14 mobE gene had no noticeable effect on plasmid mobilization, while the additional deletion of mobD reduced mobilization (600-fold) and the further deletion of mobC abolished mobilization. This is in contrast to pTF-FC2, in which deletion of mobE reduced mobilization 150-fold, with no mobilization detected on deletion of both mobE and mobD.
The discovery that when pTF-FC2 was coresident with pTC-F14, the mobilization of the latter plasmid was increased by about 3,000-fold was unexpected. We further discovered that the presence of the combination of the pTF-FC2 mobD and mobE genes, but not the individual mobD and mobE genes, was responsible for this increase. This suggests that the apparently dispensable pTC-F14 mobE gene does play a role in mobilization, but the pTC-F14 mobE gene is not optimally functional in the mating system used (described below). The functions of MobD and MobE proteins are unknown, and the same applies to the related TraL and TraM proteins of the IncPα and IncPβ plasmids. TraL has been found to have an ATP- or GTP-binding Walker A box (24), and this box is present and highly conserved in the MobD proteins of the IncQ-like plasmids (data not shown). The role of MobD and MobE in facilitating the mobilization of one plasmid by another found in this study emphasizes the need to discover the function of the proteins.
In the present study, plasmid pTF-FC2 was clearly much more readily mobilized between E. coli strains than plasmid pTC-F14. Furthermore, a coresident pTF-FC2 could mobilize a plasmid containing the oriT of pTC-F14 (although not as efficiently as pTC-F14), while a coresident pTC-F14 could not mobilize a plasmid containing the oriT gene of pTF-FC2. Based on these results, plasmid pTF-FC2 might be expected to be a more promiscuous plasmid than pTC-F14. However, the fact that mobilization studies were carried out between E. coli strains by using the chromosomally located IncP plasmid RP4 as a conjugative helper plasmid must be taken into account. It is possible that pTF-FC2 is more suited to mobilization by this plasmid than pTC-F14, while there may be an as yet unknown helper plasmid that mobilizes pTC-F14 better than pTF-FC2. The reason for the unexpected observation that the mobD and mobE genes of pTF-FC2 were better able to assist pTC-F14 mobilization than its own genes could be because the MobD and MobE proteins are better suited to work with RP4, while the equivalent proteins of pTC-F14 may be better suited to function with a different conjugative plasmid.
The interpretation of experiments on the ability of plasmids containing the cloned oriT regions to be mobilized by the mob genes of the other plasmid is not fully clear. The oriT of pTC-F14 could be mobilized by its own mob proteins, and this mobilization frequency was enhanced in the presence of pTF-FC2. This result was consistent with the ability of pTF-FC2 to enhance the mobilization frequency of pTC-F14. Plasmid pTC-F14 was not able to mobilize a plasmid containing the oriT of pTF-FC2, unless some of the pTF-FC2 genes were present. What was surprising is that when we attempted to determine which of the pTF-FC2 mobAB or mobCDE genes were required, we found that either set of genes partly enhanced mobilization. A possible explanation is that more than one of the products of the pTF-FC2 mob genes are likely to enhance binding of the mobilization complex to the oriT of pTF-FC2. The nicking and processing of DNA prior to plasmid transfer by conjugation are frequently plasmid specific. For example, despite the high degree of similarity between the DNA-processing transfer proteins and the oriT regions of the IncP plasmids RP4/RK2 and R751, the oriT of RP4/RK2 cannot be transferred by R751 (8). Transfer of the RP4/RK2 oriT took place only when the specific traJ and traK genes of RK2/RP4 were present, with traI also being required, although this was not plasmid specific. Plasmid RP4 TraJ (29) and TraK (30) proteins bind specifically to different features of the oriT region. In the case of pTF-FC2, it is likely that MobB (related to TraJ) and MobC (related to TraK) of pTF-FC2 could bind to its own oriT and thereby assist the otherwise oriT-specific proteins of pTC-F14 to recognize the oriT of pTF-FC2.
Part of the motivation for this study was to gain an understanding of the evolution of mobilization systems. The sequence similarity between the proteins associated with plasmid replication and mobilization suggests that plasmids pTC-F14 and pTF-FC2 share a common ancestor. It has been reported that plasmids pTC-F14 and pTF-FC2 are compatible in E. coli, and this indicates that replicons of the plasmids have diverged sufficiently for them to function as independent units. Pressure for the replicons to diverge may have arisen because the two broad-host-range, promiscuous plasmids occur in bacteria that share a similar ecological niche. This means they may have frequently encountered each other, and divergence to the point of compatiblity would mean that the plasmids will not exclude each other from the same host cell and thereby would each have an increased “replication space.” It was of interest to discover whether the mobilization systems of these related plasmids would compete with each other. If one plasmid has a more dominant mobilization system, it would presumably be transferred horizontally to more host cells than the competing plasmid and thereby dominate an ecosystem. No reduction in mobilization frequency of one plasmid when coresident with the other plasmid was detected. In contrast, a coresident pTF-FC2 appeared to assist the mobilization of pTC-F14. Similarly, the cloned oriT region of pTF-FC2 could be mobilized by a coresident pTC-F14 when some but not all of the pTF-FC2 mob genes were present.
Acknowledgments
This work was funded by grants from the National Research Foundation (Pretoria), The Human Resources for Industry Program (THRIP; Pretoria), the University of Stellenbosch, and the BHP-Billiton Johannesburg Technology Centre.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3308-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 3.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 4.Brosius, J., and A. Holy. 1984. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl. Acad. Sci. USA 81:6929-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorrington, R. A., and D. E. Rawlings. 1990. Characterization of the minimum replicon of the broad-host-range plasmid pTF-FC2 and similarity between pTF-FC2 and the IncQ plasmids. J. Bacteriol. 172:5697-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorrington, R. A., S. Bardien, and D. E. Rawlings. 1991. The broad-host-range plasmid pTF-FC2 requires a primase-like protein for autonomous replication in Escherichia coli. Gene 108:7-14. [DOI] [PubMed] [Google Scholar]
- 8.Fürste, J. P., W. Pansegrau, G. Zieglin, M. Kröger, and E. Lanka. 1989. Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid encoded products with the transfer origin. Proc. Natl. Acad. Sci. USA 86:1771-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner, M. N., S. M. Deane, and D. E. Rawlings. 2001. Isolation of a new broad-host-range IncQ-like plasmid, pTC-F14, from the acidophilic bacterium Acidithiobacillus caldus and analysis of the plasmid replicon. J. Bacteriol. 183:3303-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner, M. N. 2003. An investigation into the replicon of a broad host range mobilizable plasmid isolated from the moderately thermophilic bacterium Acidithiobacillus caldus. Ph.D. thesis. University of Stellenbosch, Stellenbosch, South Africa.
- 11.Hallberg, K. B., and E. B. Lindström. 1994. Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology 140:3451-3456. [DOI] [PubMed] [Google Scholar]
- 12.Henderson, D., and R. J. Meyer. 1999. The MobA-linked primase is the only replication protein of R1162 required for conjugal mobilization. J. Bacteriol. 181:2973-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong, S. M., C. C. Yeo, A. Suwanto, and C. L. Poh. 2000. Characterization of the endogenous plasmid from Pseudomonas alcaligenes NCIB 9867: DNA sequence and mechanism of transfer. J. Bacteriol. 182:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.L'Abée-Lund, T. M., and H. Sørum. 2002. A global non-conjugative Tet C plasmid pRAS3, from Aeromonas salmonicida. Plasmid 47:172-181. [DOI] [PubMed] [Google Scholar]
- 15.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncPα plasmids, compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 16.Rawlings, D. E., I. M. Pretorius, and D. R. Woods. 1984. Expression of a Thiobacillus ferrooxidans origin of replication in Escherichia coli. J. Bacteriol. 158:737-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rawlings, D. E., N. J. Coram, M. N. Gardner, and S. M. Deane. 1999. Thiobacillus caldus and Leptospirillum ferrooxidans are widely distributed in continuous-flow biooxidation tanks used to treat a variety of metal-containing ores and concentrates, p. 777-786. In R. Amils and A. Ballester (ed.), Biohydrometallurgy and the environment toward the mining of the 21st century. Part A. Elsevier Press, Amsterdam, The Netherlands.
- 18.Rawlings, D. E., and E. Tietze. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65:481-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohrer, J., and D. E. Rawlings. 1992. Sequence analysis and characterization of the mobilization region of a broad-host-range plasmid, pTF-FC2, isolated from Thiobacillus ferrooxidans. J. Bacteriol. 174:6230-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohrer, J., and D. E. Rawlings. 1993. Regulation of mobilization of the broad-host-range plasmid pTF-FC2. Mol. Microbiol. 9:1051-1059. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Scholtz, P., V. Haring, B. Wittmann-Liebold, K. Ashman, M. Bagdasarian, and E. Scherzinger. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271-288. [DOI] [PubMed] [Google Scholar]
- 23.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 24.Thorsted, P. B., D. P. Macartney, P. Aktar, A. S. Haines, N. Ali, P. Davidson, T. Stafford, M. J. Pocklington, W. Pansegrau, B. M. Wilkins, E. Lanka, and C. M. Thomas. 1998. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282:969-990. [DOI] [PubMed] [Google Scholar]
- 25.Tietze, E. 1998. Nucleotide sequence and genetic characterization of the novel IncQ-like plasmid pIE1107. Plasmid 39:165-181. [DOI] [PubMed] [Google Scholar]
- 26.Waters, V. L. 1993. Processes at the nick region link conjugation, T-DNA transfer and rolling circle replication. Mol. Microbiol. 9:1123-1130. [DOI] [PubMed] [Google Scholar]
- 27.Whittle, G., M. E. Katz, E. H. Clayton, and B. F. Cheetham. 2000. Identification and characterization of a native Dichelobacter nodosus plasmid, pDN1. Plasmid 43:230-234. [DOI] [PubMed] [Google Scholar]
- 28.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 29.Ziegelin, G., J. P. Fürste, and E. Lanka. 1989. TraJ protein of plasmid RP4 binds to a 19-base pair invert sequence repetition within the transfer origin. J. Biol. Chem. 264:11989-11994. [PubMed] [Google Scholar]
- 30.Ziegelin, G., W. Pansegrau, R. Lurz, and E. Lanka. 1992. TraK protein of conjugative plasmid RP4 forms a specialized nucleoprotein complex with the transfer origin. J. Biol. Chem. 267:17279-17286. [PubMed] [Google Scholar]