Abstract
Escherichia coli responses to four inhibitors that interfere with translation were monitored at the transcriptional level. A DNA microarray method provided a comprehensive view of changes in mRNA levels after exposure to these agents. Real-time reverse transcriptase PCRanalysis served to verify observations made with microarrays, and a chromosomal grpE::lux operon fusion was employed to specifically monitor the heat shock response. 4-Azaleucine, a competitive inhibitor of leucyl-tRNA synthetase, surprisingly triggered the heat shock response. Administration of mupirocin, an inhibitor of isoleucyl-tRNA synthetase activity, resulted in changes reminiscent of the stringent response. Treatment with kasugamycin and puromycin (targeting ribosomal subunit association as well as its peptidyl-transferase activity) caused accumulation of mRNAs from ribosomal protein operons. Abundant biosynthetic transcripts were often significantly diminished after treatment with any of these agents. Exposure of a relA strain to mupirocin resulted in accumulation of ribosomal protein operon transcripts. However, the relA strain's response to the other inhibitors was quite similar to that of the wild-type strain.
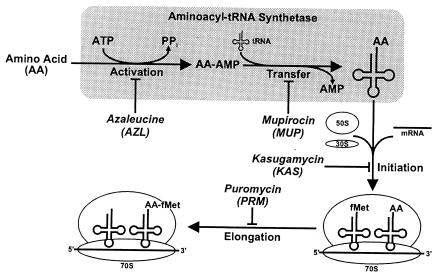
Protein synthesis is a multistep process (Fig. 1). Free amino acids are selected with high specificity byaminoacyl-tRNA synthetases and charged to tRNA. The resulting aminoacyl-tRNAs are essential components in the initiation and elongation steps of protein synthesis that take place on the ribosome. In Escherichia coli these processes are integrated through the stringent response (9). When uncharged tRNA occupies the ribosomal A site, the alarmones ppGpp and pppGpp accumulate due to the action of the relA gene product, the stringent factor. These alarmones encourage transcription of amino acid biosynthetic genes (41) and discourage initiation at the strong promoters that drive formation of the translational apparatus (3, 4).
FIG. 1.
Scheme of protein synthesis showing the sites of action of the inhibitors used. The indicated sites of action were deduced from references cited in the introduction to this article. Inhibitor abbreviations are indicated.
Over the years, many herbicides and antibiotics have been developed that target these processes (reviewed in references 11, 19, and 20). Rhodius et al. have recently reviewed evidence that DNA microarray-mediated gene expression profiling sheds light on the consequences of administration of inhibitors to E. coli (35). Cellular responses to both the glutamine mimic acivicin (39) and the DNA-damaging agent mitomycin C (53) have been investigated. Such success encouraged us to consider the consequences of inhibiting protein biosynthesis at a variety of steps (Fig. 1).
Inhibitors exist that interfere with the synthesis of branched-chain amino acids and their utilization in translation. Sulfometuron methyl (SM), a slow tight-binding inhibitor of the branched-chain amino acid biosynthetic enzyme acetolactate synthase (21, 38), has been extensively studied from many perspectives. These include an E. coli gene expression-profiling study with a set of gene fusions (47) estimated to be about 30% complete (48). SM administration results in starvation for the branched-chain amino acids and pantothenate as well as 2-ketoacid imbalances (22, 24, 25). Moreover, the global transcriptional response to this inhibitor is characterized by a strong rpoS regulon (14) signature, as if SM triggers a premature conversion from exponential- to stationary-phase growth (47).
Two inhibitors of branched-chain aminoacyl-tRNA formation have been studied in some detail. One, 4-azaleucine (AZL), is a competitive inhibitor of leucine binding to E. coli leucyl-tRNA synthetase (LeuRS) that does not progress to the azaleucyl-adenylate in vitro (40). AZL differs from leucine by having a tertiary, titratable N at the branch point of the R group (42). E. coli mutations conferring resistance to AZL have been identified in a variety of genes, including genes encoding amino acid transport and LeuRS (19). The second inhibitor, mupirocin (MUP), a mimic of isoleucyl-adenylate also known as pseudomonic acid (15), is utilized as a topical antibiotic. Resistance to MUP is caused by alterations in IleRS structure (7, 57). Thus, it is expected that SM treatment causes a paucity of branched-chain amino acids and 2-ketoacid imbalance and that AZL treatment saturates LeuRS with an amino acid analog that cannot condense with ATP. In addition it is speculated that MUP binding to IleRS yields an enzyme with an activated amino acid analog incapable of being condensed with tRNA. That is, MUP treatment mimics what happens when a supply of uncharged tRNA is inaccessible.
Kasugamycin (KAS) inhibits initiation of polypeptide synthesis (32). Resistance mutations map to several loci in E. coli that encode either structural components or modifying activities of the ribosome (19). Thus, we expect that branched-chain and other aminoacyl-tRNAs accumulate upon KAS treatment since aminoacyl-tRNA synthesis is unrestrained while the major route of aminoacyl-tRNA consumption is blocked. Puromycin (PRM) is an analog of aminoacyl-tRNA that binds to the acceptor site of the ribosome, blocking elongation and causing premature release of the growing polypeptide chain (11). Thus, PRM elevates the titer of unfolded proteins in the cytoplasm, triggering the heat shock response (13).
Here we investigate the action of AZL, MUP, KAS, and PRM through the use of DNA microarray-mediated gene expression profiling of stringent and relaxed derivatives of E. coli K-12 and compare this work to recent studies of antibiotic treatment of the pathogen Streptococcus pneumoniae (31) and of E. coli growth transitions (10).
MATERIALS AND METHODS
Chemicals.
Kanamycin, PRM, and 4-aza-d,l-leucine were purchased from Sigma (St. Louis, Mo.). KAS was obtained from Calbiochem (San Diego, Calif.). MUP was a gift from Smith Kline Beecham Pharmaceuticals (West Sussex, United Kingdom).
Strains, growth conditions, and materials.
The E. coli K-12 derivatives (obtained from M. Cashel [National Institutes of Health]) used were CF1943 (W3110) and CF1944 (W3110 ΔrelA251::kan) (56). Strains used in other microarray experiments were the near-wild-type strain MG1655 (2), the tolC mutant DE 112 (49), and the ilvB mutant DPD1675 (47). Prototrophic E. coli strain DPD3084 harbors a grpE::luxCDABE fusion at the chromosomal lac locus (51).
Aerobic growth was carried out at 37°C in M9 minimal medium (28) supplemented with 0.4% glucose and treated with shaking (250 rpm) unless otherwise specified. The concentration of each inhibitor of translation used to treat cultures prior to RNA isolation was determined by dilution of overnight cultures into fresh M9 glucose medium. At an A600 of 0.2, individual cultures were split before treatment with various concentrations of each inhibitor and growth was monitored for 4 h after the inhibitor addition.
Range finding determined that the concentration of each of the inhibitors AZL, MUP, KAS, and PRM needed to decrease the growth rate (increase the doubling time) by a factor of 5 for the relA+ strain CF1943 was 750, 8.5, 750, and 818 μg/ml, respectively, while a concentration of each inhibitor of only 375, 4.3, 750, and 546 μg/ml, respectively, was needed to retard growth of the isogenic relA strain CF1944 to the same extent. These inhibitor concentrations were later used to determine gene expression changes after a chronic, 30-min administration of the antibacterial agents.
RNA isolation.
Total RNA used in microarray procedures and real-time PCR measurements was extracted (following established methods) from cultures. Overnight cultures of each strain were diluted into fresh M9 glucose medium and incubated at 37°C. Once the cultures had reached an A600 of 0.4, they were split into two portions. One portion was treated with the inhibitor at the previously determined concentration, while the other was left unchallenged. The cells were pelleted after 30 min of shaking at 37°C, and the total RNA was immediately extracted using RNeasy minicolumns (Qiagen, Inc., Valencia, Calif.) as previously described (54).
DNA microarray experiments.
The procedures used to generate fluorescently labeled cDNA for microarray experiments, as well as those used in hybridization, data acquisition, and analysis, have been extensively described (55) and reviewed (34) previously. A total of 4,290 distinct open reading frames (ORFs) were spotted in duplicate on each glass slide (55).
Measurement of relative transcript levels using real-time PCR.
Bulk RNA isolated as described above was used as a template for cDNA production using random primers (Invitrogen, Carlsbad, Calif.) and Superscript II RNaseH− reverse transcriptase (Invitrogen). These cDNA samples were diluted 20-fold and used as the template for real-time PCRs. Primer pairs specific to several genes of interest (Table 1) were designed using software on the Primer3 website (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) (36). The PCR was carried out using QuantiTect SYBR Green PCR Master Mix (Qiagen), and the thermal cycling and real-time monitoring of SYBR green fluorescence were performed on an iCycler (Bio-Rad, Hercules, Calif.) according to the protocol supplied with the master mix. Data were collected in duplicate (using cDNA prepared from two independently isolated bulk RNA samples) for each gene in each data set. The severalfold changes in expression levels were calculated using a ΔCt method described elsewhere (54).
TABLE 1.
Primer pairs used in real-time PCR
| Gene name | B number | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|
| argA | b2818 | GCCAAAACACTGGAACTGGT | ATCGACAGGCGAGCAGTAAT |
| argB | b3959 | CGGCAAATAAAACCCTGTTG | CGTCACCGAGAAACAAACCT |
| aroF | b2601 | CATTGAGCCTGCAACAAGAA | CCCGGCGATAATATCTGAAA |
| cysK | b2414 | GGTATTGGCGCTGGTTTTAT | TTGGTGATGCCAATGACTT |
| dnaK | b0014 | TCGTATGCCAATGGTTCAGA | TCCGGGTTAACGTCTTTACG |
| glnA | b3870 | TCCGCTGAACACGTACTGA | TGAAGCGCAAATCAACAAA |
| ilvCa | b3774 | TGTACGAAATGAACGTGGTTATC | CACCAAAGCGTAAGAGAACAGAT |
| ilvCb | b3774 | CCGCAGTATGAAGGCAAAAT | TCACCATCGCAATCATCAGT |
| lacZ | b0344 | CACCCGAGTGTGATCATCTG | GATACAGCGCGTCGTGATTA |
| leuS | b0642 | AAACCGACACTTTCGACACC | TGTACTGCGGGCAAGTGTAG |
| livJ | b3460 | GAAAGCGAACTCCGTGGATA | AAGCCTTTCAGATCGCCTTT |
| metE | b3829 | GTACATAATCCGGCGGTAGAA | CAGCACGCACTTCATAGACAT |
| mopA | b4143 | AAAGATGGTGTTTCCGTTGC | ATCTGCGCACCCATATTTTC |
| ompA | b0957 | ACGGTGCATACAAAGCTCAG | GTCCAGGTCGTCAGTGATTG |
| rplV | b3315 | CATGCTCGTTCTTCTGCTCA | GCCTGCGACACTTTCTTACC |
| rpmD | b3302 | CTGCGTCGTATTGGTCACAC | TGATCATACCGCGAATAGCA |
| rpsL | b3342 | TGCGTAAAGTATGCCGTGTT | TGACCTTCACCACCGATGTA |
| sfsA | b0146 | GTTAGCGGAGAACGAACAGG | TCAACTCCCGAAGGTGTTTC |
| soxS | b4062 | ACCAGCCGCTTAACATTGAT | CGGAACATTCGTTGCAAGT |
| trmD | b2607 | CCGAAATTGACGAAGAATGG | GCGTCATTGCTGGTAACTCA |
| trpC | b1262 | TATCGATACGCTGCGTGAAG | GGTTTCACCGACGCTTAATG |
| trxA | b3781 | AACTGACCGTTGCAAAACTG | TACCACGGATGCCATATTTC |
| wrbA | b1004 | AAAACTGGCGAGCGTCTTTA | CAGGTGGATGTGATGGTTTG |
Used in measures of RNA decay.
Used in microarray verification.
Confirmation of rel phenotypes.
Upon amino acid starvation, stringent (relA+) strains greatly reduce incorporation of precursor into RNA while the bulk incorporation of UTP into macromolecules proceeds at an unaffected pace in a relaxed (relA) strain (9). The rel phenotype of each strain tested was determined by monitoring the incorporation of [14C]uracil into macromolecules during isoleucine starvation induced by addition of l-valine to the culture medium (27). Overnight cultures were diluted into M9 glucose medium and incubated at 30°C. Upon reaching an A600 of 0.4, the culture was split into two portions and [14C]uracil (0.55 mCi/mmol) was added to achieve a final concentration of 20 μg/ml. l-valine was added to 0.2 mg/ml to one sample, while the other remained untreated. At various times before and after the addition of valine, 0.5 ml was taken from each culture and added to 2.5 ml of cold 6% trichloroacetic acid. Samples were incubated on ice for 20 min before being collected on Whatman GF/A filters. Each sample was washed three times with 5% trichloroacetic acid and twice with 70% ethanol and allowed to dry. The extent of incorporation of [14C]uracil into macromolecules was measured by scintillation counting.
Bioluminescence of grpE::lux operon fusion.
A 10-ml culture of strain DPD3084 was incubated with shaking (250 rpm) in a 50-ml Erlenmeyer flask at 37°C in minimal medium E (12) supplemented with 0.4% glucose and thiamine until early exponential-phase growth was achieved. At that point, 50-μl samples of the culture were exposed in microtiter plates to twofold dilution series with inhibitors in the same medium (49). Light production was monitored as a function of time as described previously (49), with updated instrumentation (Labsystems Lunminoskan Ascent). The response ratio and maximal-response ratio for such a time course study have been defined previously (5); the minimal-response ratio is the smallest fraction of light emitted by a treated culture relative to that emitted by an untreated control over the duration of an experiment. Experiments were performed at 37°C because the gene products of the luxCDABE operon utilized (47) (derived from Photorhabdus luminescens [50]) are thermotolerant.
RESULTS
Expression profile of wild-type E. coli.
Preliminary characterization determined the inhibitor concentrations used to treat each E. coli strain (see Materials and Methods). The stringent and relaxed phenotypes of the wild-type (CF1943) and relA (CF1944) strains were verified by monitoring the bulk incorporation of uracil into RNA (data not shown). E. coli strain CF1943 was then treated with each of the four protein synthesis inhibitors at the concentrations specified above.
AZL.
Expression profiling of transcripts corresponding to the 4,290 E. coli ORFs indicated that the response to AZL was quite broad. Expression of 134 genes was elevated >2-fold, while expression of 33 genes was elevated >4-fold and transcript levels of 16 ORFs were elevated >8-fold in response to the challenge. Examination of the 58 genes encoding transcripts whose levels were elevated >3-fold was revealing (Table 2 and supplementary material [Table 1A; http://trna.chem.yale.edu/supdata/sup001/]). Of the 58 genes, 12 were heat shock genes (dnaK, dnaJ, htrA, htpG, htpX, clpB, grpE, ibpA, hslU, hslV, mopB, and mopA), another 4 were genes induced by other stresses (suhB, mazE, soxS, and nrdG), and 12 specified proteins of uncharacterized function. Elevated expression of a limited subset of amino acid biosynthesis genes was noted; it was striking that of the 11 genes in this class whose expression was elevated, 9 were arg and 2 were met. Similarly, unanticipated elevation of arg and met transcripts has been noted in a microarray study of the E. coli response to the histidine biosynthesis inhibitor acivicin (39).
TABLE 2.
Transcripts accumulated after inhibitor challenge of a relA+ straina
| Inhibitor and gene | b no. | Expression ratio | Inhibitor and gene | b no. | Expression ratio | |
|---|---|---|---|---|---|---|
| AZL | ||||||
| ibpA | b3687 | 260 | ||||
| argA | b2818 | 39 | ||||
| b1455 | b1455 | 35 | ||||
| argI | b4254 | 20 | ||||
| mgtA | b4242 | 14 | ||||
| dnaK | b0014 | 13 | ||||
| argF | b0273 | 12 | ||||
| celA | b1738 | 12 | ||||
| clpB | b2592 | 12 | ||||
| argC | b3958 | 11 | ||||
| b0235 | b0235 | 10 | ||||
| htpG | b0473 | 9.7 | ||||
| argB | b3959 | 9.2 | ||||
| argG | b3172 | 9.1 | ||||
| celB | b1737 | 8.9 | ||||
| sdaA | b1814 | 8.9 | ||||
| mopA | b4143 | 7.9 | ||||
| htpX | b1829 | 7.1 | ||||
| ybdO | b0603. | 6.9 | ||||
| dnaJ | b0015 | 6.7 | ||||
| mopB | b4142 | 6.7 | ||||
| chaA | b1216 | 6.2 | ||||
| pstS | b3728 | 6.0 | ||||
| grpE | b2614 | 5.9 | ||||
| ycjW | b1320 | 5.6 | ||||
| hisJ | b2309 | 5.2 | ||||
| htrA | b0161 | 4.4 | ||||
| ybdQ | b0607 | 4.4 | ||||
| dadX | b1190 | 4.3 | ||||
| dadA | b1189 | 4.2 | ||||
| MUP | ||||||
| ycjF | b1322 | 7.6 | ||||
| b1976 | b1976 | 6.2 | ||||
| cpsG | b2048 | 6.1 | ||||
| leuC | b0072 | 5.9 | ||||
| b4103 | b4103 | 4.5 | ||||
| yagU | b0287 | 4.3 | ||||
| yaiM | b0355 | 4.3 | ||||
| ilvG | b3767 | 3.8 | ||||
| b1433 | b1433 | 3.6 | ||||
| mukF | b0922 | 3.4 | ||||
| rnc | b2567 | 3.4 | ||||
| yagR | b0284 | 3.1 | ||||
| thrA | b0002 | 3.0 | ||||
| hemN | b3867 | 2.9 | ||||
| ycjI | b1326 | 2.9 | ||||
| b1762 | b1762 | 2.8 | ||||
| molR | b2115 | 2.8 | ||||
| xseB | b0422 | 2.8 | ||||
| mhpT | b0353 | 2.6 | ||||
| b1513 | b1513 | 2.5 | ||||
| KAS | ||||||
| yjfI | b4181 | 52 | ||||
| b1629 | b1629 | 25 | ||||
| b1644 | b1644 | 21 | ||||
| ycjX | b1321 | 19 | ||||
| glvG | b3681 | 16 | ||||
| mviM | b1068 | 15 | ||||
| yceO | b1058 | 15 | ||||
| yjcB | b4060 | 15 | ||||
| ydgO | b1630 | 14 | ||||
| yhaL | b3107 | 14 | ||||
| ttdB | b3062 | 13 | ||||
| yfiK | b2578 | 13 | ||||
| ycjG | b1325 | 12 | ||||
| ylcB | b0572 | 12 | ||||
| sgbE | b3583 | 11 | ||||
| nikA | b3476 | 10 | ||||
| rpsF | b4200 | 9.7 | ||||
| caiT | b0040 | 9.4 | ||||
| yeeT | b2003 | 9.3 | ||||
| b2460 | b2460 | 9.0 | ||||
| ybgG | b0732 | 8.9 | ||||
| rpsR | b4202 | 8.7 | ||||
| priB | b4201 | 8.1 | ||||
| sgaB | b4194 | 7.6 | ||||
| ycjT | b1316 | 7.5 | ||||
| rplI | b4203 | 7.1 | ||||
| elaC | b2268 | 7.0 | ||||
| hyfH | b2488 | 6.4 | ||||
| yjfJ | b4182 | 6.2 | ||||
| yraJ | b3144 | 6.2 | ||||
| PRM | ||||||
| metA | b4013 | 82 | ||||
| ycdU | b1029 | 77 | ||||
| chpR | b2783 | 59 | ||||
| ybeK | b0651 | 28 | ||||
| b1832 | b1832 | 27 | ||||
| phoQ | b1129 | 24 | ||||
| uhpT | b3666 | 24 | ||||
| b1444 | b1444 | 20 | ||||
| b1601 | b1601 | 19 | ||||
| kdpC | b0696 | 17 | ||||
| arp | b4017 | 16 | ||||
| mesJ | b0188 | 15 | ||||
| yfiK | b2578 | 13 | ||||
| xthA | b1749 | 12 | ||||
| yhcR | b3242 | 12 | ||||
| yhaI | b3104 | 11 | ||||
| celC | b1736 | 9.6 | ||||
| rpmJ | b3299 | 9.4 | ||||
| b0011 | b0011 | 8.3 | ||||
| b1447 | b1447 | 8.1 | ||||
| rpmD | b3302 | 7.9 | ||||
| b1451 | b1451 | 7.8 | ||||
| menC | b2261 | 7.5 | ||||
| yagS | b0285 | 7.1 | ||||
| b1600 | b1600 | 6.9 | ||||
| yaeS | b0174 | 6.9 | ||||
| b2534 | b2534 | 6.5 | ||||
| hemL | b0154 | 6.5 | ||||
| rpmC | b3312 | 6.2 | ||||
| b1400 | b1400 | 6.1 |
Additional data can be found at http://trna.chem.yale.edu/supdata/sup001.
Down-regulation was also observed. Of the 34 genes whose transcripts were decreased >4-fold (Table 3 and supplementary material [Table 2A; http://trna.chem.yale.edu/supdata/sup001/]), functions cannot yet be assigned to 22. Similarities among members of this group of down-regulated genes were not discerned.
TABLE 3.
Transcripts diminished after inhibitor challenge of a relA+ straina
| Inhibitor and gene | b no. | Expression ratio | Inhibitor and gene | b no. | Expression ratio | |
|---|---|---|---|---|---|---|
| AZL | ||||||
| b2880 | b2880 | 0.04 | ||||
| b2879 | b2879 | 0.07 | ||||
| yhaM | b3108 | 0.08 | ||||
| b2635 | b2635 | 0.09 | ||||
| b1445 | b1445 | 0.10 | ||||
| b2274 | b2274 | 0.10 | ||||
| b2450 | b2450 | 0.10 | ||||
| farR | b0730 | 0.10 | ||||
| b1310 | b1310 | 0.11 | ||||
| hmpA | b2552 | 0.12 | ||||
| yehT | b2125 | 0.13 | ||||
| kdpD | b0695 | 0.14 | ||||
| ypfH | b2473 | 0.15 | ||||
| caiC | b0037 | 0.16 | ||||
| yiaB | b3563 | 0.16 | ||||
| yjfK | b4183 | 0.17 | ||||
| yjfL | b4184 | 0.17 | ||||
| b2460 | b2460 | 0.19 | ||||
| nuoK | b2279 | 0.19 | ||||
| yhaB | b3120 | 0.20 | ||||
| yibG | b3596 | 0.20 | ||||
| b2666 | b2666 | 0.21 | ||||
| b0359 | b0359 | 0.22 | ||||
| gapC_2 | b1416 | 0.22 | ||||
| wrbA | b1004 | 0.22 | ||||
| apbA | b0425 | 0.23 | ||||
| wcaI | b2050 | 0.23 | ||||
| yadN | b0141 | 0.23 | ||||
| ytfA | b4205 | 0.23 | ||||
| gadB | b1493 | 0.24 | ||||
| MUP | ||||||
| yhjG | b3524 | 0.03 | ||||
| aroF | b2601 | 0.11 | ||||
| trpB | b1261 | 0.13 | ||||
| trpE | b1264 | 0.16 | ||||
| gadB | b1493 | 0.17 | ||||
| gadA | b3517 | 0.18 | ||||
| rplC | b3320 | 0.21 | ||||
| trpD | b1263 | 0.21 | ||||
| tyrA | b2600 | 0.21 | ||||
| trpA | b1260 | 0.22 | ||||
| b1973 | b1973 | 0.23 | ||||
| trpC | b1262 | 0.23 | ||||
| fusA | b3340 | 0.24 | ||||
| gapA | b1779 | 0.25 | ||||
| rpsJ | b3321 | 0.25 | ||||
| rplE | b3308 | 0.26 | ||||
| rplR | b3304 | 0.26 | ||||
| tdh | b3616 | 0.27 | ||||
| tufA | b3339 | 0.27 | ||||
| cirA | b2155 | 0.29 | ||||
| metE | b3829 | 0.29 | ||||
| rplB | b3317 | 0.29 | ||||
| rpsD | b3296 | 0.29 | ||||
| serA | b2913 | 0.29 | ||||
| ybeD | b0631 | 0.29 | ||||
| ptsI | b2416 | 0.30 | ||||
| rplF | b3305 | 0.3 | ||||
| rpsE | b3303 | 0.3 | ||||
| csgC | b1043 | 0.31 | ||||
| hdeB | b3509 | 0.31 | ||||
| KAS | ||||||
| gadA | b3517 | 0.04 | ||||
| aceA | b4015 | 0.05 | ||||
| gadB | b1493 | 0.05 | ||||
| gapA | b1779 | 0.05 | ||||
| aceK | b4016 | 0.06 | ||||
| livJ | b3460 | 0.06 | ||||
| hdeB | b3509 | 0.08 | ||||
| aroF | b2601 | 0.09 | ||||
| dps | b0812 | 0.09 | ||||
| b1513 | b1513 | 0.11 | ||||
| ilvC | b3774 | 0.11 | ||||
| osmE | b1739 | 0.11 | ||||
| aroG | b0754 | 0.12 | ||||
| otsB | b1897 | 0.12 | ||||
| serA | b2913 | 0.12 | ||||
| yeaG | b1783 | 0.12 | ||||
| asd | b3433 | 0.13 | ||||
| pflB | b0903 | 0.13 | ||||
| icdA | b1136 | 0.14 | ||||
| ompC | b2215 | 0.14 | ||||
| serC | b0907 | 0.14 | ||||
| pgi | b4025 | 0.15 | ||||
| ppc | b3956 | 0.15 | ||||
| aceB | b4014 | 0.16 | ||||
| gatY | b2096 | 0.16 | ||||
| wrbA | b1004 | 0.16 | ||||
| eno | b2779 | 0.17 | ||||
| fba | b2925 | 0.17 | ||||
| hdeA | b3510 | 0.17 | ||||
| leuB | b0073 | 0.18 | ||||
| PRM | ||||||
| ais | b2252 | 0.02 | ||||
| b1973 | b1973 | 0.02 | ||||
| atoC | b2220 | 0.03 | ||||
| livJ | b3460 | 0.03 | ||||
| nikE | b3480 | 0.03 | ||||
| nuoL | b2278 | 0.03 | ||||
| ycdY | b1035 | 0.03 | ||||
| gadA | b3517 | 0.05 | ||||
| gadB | b1493 | 0.05 | ||||
| b1284 | b1284 | 0.06 | ||||
| gapA | b1779 | 0.06 | ||||
| hmpA | b2552 | 0.06 | ||||
| yjfQ | b4191 | 0.06 | ||||
| apbA | b0425 | 0.07 | ||||
| b1604 | b1604 | 0.07 | ||||
| hdeB | b3509 | 0.07 | ||||
| ibpB | b3686 | 0.07 | ||||
| livK | b3458 | 0.07 | ||||
| uhpB | b3668 | 0.07 | ||||
| yhaB | b3120 | 0.07 | ||||
| yiaA | b3562 | 0.07 | ||||
| b1045 | b1045 | 0.08 | ||||
| serC | b0907 | 0.08 | ||||
| yeaG | b1783 | 0.08 | ||||
| ynfM | b1596 | 0.08 | ||||
| b3254 | b3254 | 0.09 | ||||
| mraY | b0087 | 0.09 | ||||
| sucA | b0726 | 0.09 | ||||
| udp | b3831 | 0.09 | ||||
| b1057 | b1057 | 0.10 |
Additional data can be found at http://trna.chem.yale.edu/supdata/sup001.
MUP.
The response to MUP was quite different from that caused by AZL (for up-regulation, see Table 2 and supplementary material [Table 1B; http://trna.chem.yale.edu/supdata/sup001/]; for down-regulation, see Table 3 and supplementary material [Table 2B; http://trna.chem.yale.edu/supdata/sup001/]). Only 20 mRNA levels were modestly elevated (between 2.5- and 7.5-fold), and 10 of the corresponding genes have no assigned function. It was comforting that ilv, leu, and thr transcription was elevated by MUP treatment, since the ilvGMEDA, thr, and leu operons are thought to be responsive to isoleucyl-tRNA limitation due to the presence of isoleucine codons in each operon's attenuation leader genes (18, 33, 45).
Transcription reduced by MUP treatment was mostly associated with known genes; only 7 of the 64 genes with reduced expression had an unassigned role. Moreover, many of these genes were previously noted to be among the 50 most abundantly expressed ORFs when E. coli was cultured in minimal medium to either the exponential (22 of the 64 genes) or transitional (21 of the 64) phase of a typical growth curve (55). Of the 64 genes displaying reduced transcript abundance, 27 were in the list of 73 genes most highly expressed during the exponential or transition phase of growth in minimal medium. Moreover, 33 of these 64 genes showed at least twofold-reduced expression levels after challenge with acivicin, a glutamine analog (39).
KAS.
Compared to interference with tRNA aminoacylation, inhibition of translation initiation led to accumulation of more than 200 transcripts (Table 2 and supplementary material [Table 1C; http://trna.chem.yale.edu/supdata/sup001/]). Among those were mRNAs derived from ribosomal protein genes organized into operons and loci encoding transporters for diverse polyvalent molecules such as citrate, taurine, and carnitine. Many of the 150 down-regulated genes (Table 3 and supplementary material [Table 2C; http://trna.chem.yale.edu/supdata/sup001/]) were also familiar. Among them were genes (previously identified to be highly expressed when cells are grown in minimal medium [55]) specifying the glyoxylate bypass (aceABK), glycolysis (ptsHI, pgi, pgk, fba, tpiA, eno, and gapA), the pentose shunt (talA and tktB), the TCA cycle (gltA and icdA), and other central carbon pathways (zwf, pflB, and adhE) as well as biosynthesis (aroF, cysK, folE, ilvC, and metE). Levels of transcripts normally associated with growth cessation either through membership in the rpoS regulon (cbpA, dps, hdeA, hdeB, osmY, otsA, otsB, poxB, wrbA, and yeaG) (14) or by empirical study (rmf, gadA, and gadB) (55) were counterintuitively found not to be elevated after the KAS challenge. Taking these data together, it appears that E. coli was responding to KAS by attempting to preserve or even enhance its protein biosynthetic capacity while jettisoning its ability to produce energy.
PRM.
The expression of 119 genes was enhanced by treatment with the inhibitor PRM (Table 2 and supplementary material [Table 1D; http://trna.chem.yale.edu/supdata/sup001/]), including that of 34 genes that encode ribosomal proteins as well as that of 6 translation-related genes embedded in ribosomal protein operons (infB, nusA, prlA, rimM [yfjA], rnpA, and trmD) (16). Transcript levels of 383 genes were found to be decreased after this challenge (Table 3 and supplementary material [Table 2D; http://trna.chem.yale.edu/supdata/sup001/]). As with KAS treatment, exposure to PRM decreased expression of a subset of the rpoS regulon (14) and stationary-phase stimulon (55) genes such as gadAB, hdeABD, osmEY, otsAB, treAR, wrbA, and xasA as well as genes involved in a much broader array of energy metabolism functions, ranging from glycolysis and associated functions (dld, eno, gapA, gapC, pgi, pgk, and ptsHI), the TCA cycle (gltA, icdA, sdhBC, sucACD, and aceA), and the pentose shunt (talA and tktB) to respiration (cydA, cyoD, fdhE, fdnI, hyfGHI, narVW, nikBDE, nrdEFHI, and pflB). Normally highly expressed genes (55), including aroF, folE, ilvC, metE, and livJK, were subjected to an apparent down-regulation. Transcripts of genes of biosynthetic pathways (aroAG, asd, glnBEGL, gltD, guaC, hisBDFIL, leuB, lysC, serAC, thrAC, trpABC, and tyrB), catabolism of alternative carbon sources (ebgAC, gatYZ, tauABCD, malGKM, and manXYZ), and iron metabolism (cirA, entABC, fecBR, and fhuBF) also accumulated to a substantially lower level.
Differential responses of a relA mutant to the four inhibitors.
The absence of an intact RelA protein in the cell interfered with its ability to mount an effective response to a limited supply of amino acid. Differences in the transcriptional patterns in the presence and absence of the relA gene product, possibly revealing response elements under stringent control, are highlighted below.
AZL.
Transcription of about 9% of E. coli genes (381 genes) increased >2.5-fold after AZL treatment of the relA mutant (Table 4 and supplementary material [Table 3A; http://trna.chem.yale.edu/supdata/sup001/]); for 41 of the genes, transcription was induced >8-fold. Of this subset of 41 genes, 22 encoded products of unknown function, 3 specified amino acid biosynthetic genes (argA, argG, and metA), and nine (clpB, dnaK, grpE, htpG, htpX, htrA, ibpA, ibpB, and mopA) were involved in the heat shock response to unfolded cytoplasmic proteins (58). Moreover, this group included 12 ribosomal protein genes (16).
TABLE 4.
Transcripts accumulated after inhibitor challenge of a relA straina
| Inhibitor and gene | b no. | Expression ratio | Inhibitor and gene | b no. | Expression ratio | |
|---|---|---|---|---|---|---|
| AZL | ||||||
| ibpB | b3686 | 210 | ||||
| ibpA | b3687 | 160 | ||||
| yfiE | b2577 | 35 | ||||
| metA | b4013 | 33 | ||||
| ybfB | b0702 | 30 | ||||
| clpB | b2592 | 25 | ||||
| mgtA | b4242 | 22 | ||||
| cpsB | b2049 | 22 | ||||
| b2451 | b2451 | 20 | ||||
| b1776 | b1776 | 17 | ||||
| b2656 | b2656 | 17 | ||||
| b2681 | b2681 | 16 | ||||
| yhcN | b3238 | 15 | ||||
| yshA | b3875 | 14 | ||||
| yfiA | b2597 | 14 | ||||
| ygfS | b2886 | 14 | ||||
| b2680 | b2680 | 13 | ||||
| ybfH | b0691 | 13 | ||||
| cyoD | b0429 | 13 | ||||
| ybcU | b0557 | 13 | ||||
| argA | b2818 | 12 | ||||
| b1627 | b1627 | 12 | ||||
| sdaA | b1814 | 12 | ||||
| htpG | b0473 | 12 | ||||
| yiaB | b3563 | 11 | ||||
| htpX | b1829 | 11 | ||||
| htrA | b0161 | 11 | ||||
| dnaK | b0014 | 10 | ||||
| yfiD | b2579 | 10 | ||||
| ylcB | b0572 | 9.9 | ||||
| MUP | ||||||
| sdaB | b2797 | 9.7 | ||||
| sdaC | b2796 | 8.3 | ||||
| mhpC | b0349 | 6.9 | ||||
| ybfA | b0699 | 6.3 | ||||
| b1759 | b1759 | 5.8 | ||||
| rplK | b3983 | 5.7 | ||||
| yafU | b0218 | 5.6 | ||||
| rplA | b3984 | 5.4 | ||||
| yhbE | b3184 | 5.4 | ||||
| ylcD | b0574 | 5.2 | ||||
| b1445 | b1445 | 5.1 | ||||
| ycdY | b1035 | 5.1 | ||||
| yjeR | b4162 | 5.1 | ||||
| rplL | b3986 | 4.8 | ||||
| rpsS | b3316 | 4.8 | ||||
| rplJ | b3985 | 4.7 | ||||
| rplP | b3313 | 4.4 | ||||
| rpsC | b3314 | 4.4 | ||||
| ybgD | b0719 | 4.4 | ||||
| rplB | b3317 | 4.3 | ||||
| rplV | b3315 | 4.3 | ||||
| rplW | b3318 | 4.3 | ||||
| pin | b1158 | 4.2 | ||||
| rplD | b3319 | 4.1 | ||||
| b2667 | b2667 | 3.9 | ||||
| rplC | b3320 | 3.9 | ||||
| rplY | b2185 | 3.9 | ||||
| rpmC | b3312 | 3.9 | ||||
| rpsJ | b3321 | 3.9 | ||||
| trmD | b2607 | 3.9 | ||||
| KAS | ||||||
| yaiL | b0354 | 46 | ||||
| mobA | b3857 | 29 | ||||
| wbbI | b2034 | 27 | ||||
| yeeT | b2003 | 27 | ||||
| b1983 | b1983 | 26 | ||||
| yjfM | b4185 | 23 | ||||
| rpsR | b4202 | 23 | ||||
| fadA | b3845 | 20 | ||||
| b2447 | b2447 | 19 | ||||
| rpsM | b3298 | 18 | ||||
| b1601 | b1601 | 18 | ||||
| yfbM | b2272 | 17 | ||||
| rpsF | b4200 | 17 | ||||
| rplP | b3313 | 17 | ||||
| bacA | b3057 | 16 | ||||
| ybdJ | b0580 | 16 | ||||
| rpmC | b3312 | 15 | ||||
| b0833 | b0833 | 15 | ||||
| glvC | b3683 | 15 | ||||
| yjeP | b4159 | 14 | ||||
| priB | b4201 | 14 | ||||
| uhpC | b3667 | 14 | ||||
| b2451 | b2451 | 12 | ||||
| rpsU | b3065 | 11 | ||||
| rpmJ | b3299 | 11 | ||||
| wbbJ | b2033 | 11 | ||||
| rplO | b3301 | 10 | ||||
| b1593 | b1593 | 10 | ||||
| gapC_2 | b1416 | 9.9 | ||||
| dadA | b1189 | 9.5 | ||||
| PRM | ||||||
| yehQ | b2122 | 85 | ||||
| b0011 | b0011 | 16 | ||||
| ylcB | b0572 | 16 | ||||
| dksA | b0145 | 14 | ||||
| wecD | b3790 | 13 | ||||
| yfhB | b2560 | 13 | ||||
| ygfR | b2885 | 12 | ||||
| yfiD | b2579 | 12 | ||||
| purL | b2557 | 12 | ||||
| ygaA | b2709 | 11 | ||||
| yfiK | b2578 | 11 | ||||
| metA | b4013 | 11 | ||||
| seqA | b0687 | 11 | ||||
| sgaB | b4194 | 11 | ||||
| ybcU | b0557 | 11 | ||||
| ycjF | b1322 | 10 | ||||
| yjfR | b4192 | 10 | ||||
| phoQ | b1129 | 10 | ||||
| malG | b4032 | 9.6 | ||||
| b1628 | b1628 | 8.8 | ||||
| yciR | b1285 | 8.3 | ||||
| b1445 | b1445 | 8.2 | ||||
| b1045 | b1045 | 8.1 | ||||
| priB | b4201 | 8.1 | ||||
| ybdM | b0601 | 7.9 | ||||
| rplO | b3301 | 7.7 | ||||
| rpsR | b4202 | 7.6 | ||||
| trmD | b2607 | 7.5 | ||||
| glnB | b2553 | 7.4 | ||||
| insA_1 | b0022 | 7.4 |
Additional data can be found at http://trna.chem.yale.edu/supdata/sup001.
However, levels of transcripts of 61 ORFs decreased after AZL treatment (Table 5 and supplementary material [Table 4A; http://trna.chem.yale.edu/supdata/sup001/]). Among those were many biosynthetic genes, including several (aroF, folE, ilvC, and metE) that are known to be quite highly expressed during normal exponential growth in minimal medium. Levels of transcripts of genes known to accumulate during acidification (1) and entry into stationary phase (55) (dps, gadA, gadB, hdeA, hdeB, osmC, osmY, wrbA, xasA, and yeaG) were also found to be lowered.
TABLE 5.
Transcripts diminished after inhibitor challenge of a relA straina
| Inhibitor and gene | b no. | Expression ratio | Inhibitor and gene | b no. | Expression ratio | |
|---|---|---|---|---|---|---|
| AZL | ||||||
| uhpB | b3668 | 0.06 | ||||
| aroF | b2601 | 0.07 | ||||
| trpE | b1264 | 0.07 | ||||
| gadB | b1493 | 0.08 | ||||
| gadA | b3517 | 0.10 | ||||
| ilvC | b3774 | 0.11 | ||||
| ycjD | b1289 | 0.13 | ||||
| glnB | b2553 | 0.15 | ||||
| kdpA | b0698 | 0.15 | ||||
| wrbA | b1004 | 0.15 | ||||
| lpxD | b0179 | 0.16 | ||||
| tyrA | b2600 | 0.16 | ||||
| trpA | b1260 | 0.17 | ||||
| hdeB | b3509 | 0.18 | ||||
| trpB | b1261 | 0.18 | ||||
| b2342 | b2342 | 0.20 | ||||
| fucA | b2800 | 0.20 | ||||
| livJ | b3460 | 0.22 | ||||
| livK | b3458 | 0.22 | ||||
| trpD | b1263 | 0.23 | ||||
| yeaG | b1783 | 0.23 | ||||
| serC | b0907 | 0.24 | ||||
| ybaJ | b0461 | 0.24 | ||||
| talA | b2464 | 0.25 | ||||
| dps | b0812 | 0.26 | ||||
| pflB | b0903 | 0.26 | ||||
| trpC | b1262 | 0.26 | ||||
| yadF | b0126 | 0.26 | ||||
| aceA | b4015 | 0.27 | ||||
| nrdI | b2674 | 0.28 | ||||
| MUP | ||||||
| aroF | b2601 | 0.04 | ||||
| aceA | b4015 | 0.07 | ||||
| b1973 | b1973 | 0.08 | ||||
| gapA | b1779 | 0.08 | ||||
| ilvC | b3774 | 0.08 | ||||
| trpE | b1264 | 0.09 | ||||
| aceB | b4014 | 0.11 | ||||
| eno | b2779 | 0.11 | ||||
| trpB | b1261 | 0.11 | ||||
| b2350 | b2350 | 0.14 | ||||
| gadB | b1493 | 0.14 | ||||
| livJ | b3460 | 0.14 | ||||
| hisD | b2020 | 0.15 | ||||
| lysC | b4024 | 0.15 | ||||
| gadA | b3517 | 0.16 | ||||
| hdeB | b3509 | 0.16 | ||||
| hisG | b2019 | 0.16 | ||||
| mobB | b3856 | 0.16 | ||||
| trpA | b1260 | 0.16 | ||||
| leuA | b0074 | 0.17 | ||||
| metE | b3829 | 0.17 | ||||
| pheA | b2599 | 0.17 | ||||
| ompC | b2215 | 0.18 | ||||
| yrbG | b3196 | 0.18 | ||||
| cycA | b4208 | 0.19 | ||||
| cyoB | b0431 | 0.19 | ||||
| trpD | b1263 | 0.19 | ||||
| aceK | b4016 | 0.20 | ||||
| gltA | b0720 | 0.20 | ||||
| livK | b3458 | 0.20 | ||||
| KAS | ||||||
| ybeK | b0651 | 0.01 | ||||
| ynfM | b1596 | 0.02 | ||||
| b0263 | b0263 | 0.02 | ||||
| ycjW | b1320 | 0.04 | ||||
| b1644 | b1644 | 0.06 | ||||
| gapA | b1779 | 0.07 | ||||
| ilvC | b3774 | 0.09 | ||||
| b1565 | b1565 | 0.09 | ||||
| lysC | b4024 | 0.11 | ||||
| b2653 | b2653 | 0.11 | ||||
| b1513 | b1513 | 0.15 | ||||
| yagY | b0292 | 0.15 | ||||
| yagU | b0287 | 0.15 | ||||
| ymfM | b1148 | 0.15 | ||||
| b0165 | b0165 | 0.15 | ||||
| b1153 | b1153 | 0.16 | ||||
| ycgX | b1161 | 0.16 | ||||
| aceA | b4015 | 0.18 | ||||
| gloB | b0212 | 0.19 | ||||
| b1462 | b1462 | 0.19 | ||||
| glnA | b3870 | 0.19 | ||||
| yrfD | b3395 | 0.19 | ||||
| serA | b2913 | 0.19 | ||||
| gltA | b0720 | 0.20 | ||||
| malS | b3571 | 0.21 | ||||
| trpB | b1261 | 0.21 | ||||
| gadA | b3517 | 0.21 | ||||
| aroF | b2601 | 0.22 | ||||
| smpA | b2617 | 0.22 | ||||
| trpD | b1263 | 0.22 | ||||
| PRM | ||||||
| yhcO | b3239 | 0.01 | ||||
| yi81_1 | b0016 | 0.02 | ||||
| b1547 | b1547 | 0.03 | ||||
| yfaE | b2236 | 0.03 | ||||
| b2386 | b2386 | 0.03 | ||||
| ykgG | b0308 | 0.04 | ||||
| eutB | b2441 | 0.04 | ||||
| glpR | b3423 | 0.04 | ||||
| b1172 | b1172 | 0.05 | ||||
| cadA | b4131 | 0.05 | ||||
| ybhI | b0770 | 0.06 | ||||
| flhB | b1880 | 0.06 | ||||
| rfe | b3784 | 0.06 | ||||
| aceA | b4015 | 0.06 | ||||
| yhcK | b3226 | 0.08 | ||||
| b1759 | b1759 | 0.09 | ||||
| aceK | b4016 | 0.09 | ||||
| ybcK | b0558 | 0.10 | ||||
| b1503 | b1503 | 0.11 | ||||
| celF | b1734 | 0.11 | ||||
| cirA | b2155 | 0.11 | ||||
| b2361 | b2361 | 0.11 | ||||
| b2650 | b2650 | 0.11 | ||||
| ybbT | b0505 | 0.12 | ||||
| ybeJ | b0655 | 0.13 | ||||
| sucA | b0726 | 0.13 | ||||
| eno | b2779 | 0.13 | ||||
| atpD | b3732 | 0.14 | ||||
| sucB | b0727 | 0.16 | ||||
| modA | b0763 | 0.16 |
Additional data can be found at http://trna.chem.yale.edu/supdata/sup001.
MUP.
The relA mutant's response to the MUP inhibitor was quite different from that observed for the parental strain. Transcripts of 105 genes attained moderately (between 2.5- and 10-fold) elevated levels (Table 4 and supplementary material [Table 3B; http://trna.chem.yale.edu/supdata/sup001/]). One-third of the elevated transcripts emanated from ribosomal protein operons (16). Thus, the relA mutation was responsible for an apparent inversion in the expression of ribosomal protein gene transcripts; these mRNAs accumulated after MUP treatment of the relA strain, while they did not do so after inhibition of the parental strain.
Transcripts of 94 ORFs were down-regulated >2.5-fold by MUP treatment of the relA strain, while the expression levels of 43 were decreased >4-fold (Table 5 and supplementary material [Table 4B; http://trna.chem.yale.edu/supdata/sup001/]). Included in this more stringent subset were nine genes (aceA, aceB, aceK, eno, gapA, gltA, icdA, pflB, and ptsI) of central carbon metabolism and two genes (cycA and cyoB) of respiration as well as many amino acid biosynthetic genes. Four genes (aroF, cysK, ilvC, and metE) classified as amino acid biosynthetic geneshave been previously noted to be highly expressed (55), while eight others (aroF, pheA, trpA, trpB, trpC, trpD, trpE, and tyrA) belong to the family of genes specifying synthesis of the aromatic amino acids. Thus, the patterns of reduced transcript accumulation were similar for the relA and relA+ pair challenged with MUP; the striking though expected difference (9) was that expression of the genes specifying the translational machinery did not cease in the relA mutant.
KAS.
Challenge with the KAS inhibitor resulted in at least a 2.5-fold-increased abundance of transcripts corresponding to 339 ORFs in the relA strain (Table 4 and supplementary material [Table 3C; http://trna.chem.yale.edu/supdata/sup001/]), echoing the broad impact of this inhibitor upon the relA+ strain reported above. Of these genes, 32 specify ribosomal proteins (16). Further inspection revealed that 10 of the 63 ORFs (overexpressed >7-fold) encoded ribosomal proteins. This challenge also reduced transcript levels for 120 ORFs (Table 5 and supplementary material [Table 4C; http://trna.chem.yale.edu/supdata/sup001/]), including genes involved in stress responses (mopA, mopB, hdeB, gadA, gadB, and otsB) (43), biosynthetic genes (both those previously noted to be highly expressed [aroF, cysK, folE, glnA, and ilvC, and metE] [55] and those that do not appear to be extraordinary [aroB, aroG, cysA, cysN, ilvE, ilvG, ilvI, lysC, serA, thrB, thrC, trpA, trpB, trpC, trpD, and trpE]), and genes of central carbon metabolism (aceA, fba, gapA, gltA, icdA, pgk, ppc, ptsI, sucA, and tpiA).
PRM.
Transcripts of 259 ORFs accumulated in the relA strain after PRM administration (Table 4 and supplementary material [Table 3D; http://trna.chem.yale.edu/supdata/sup001/]). The pattern seen was reminiscent of that observed with the parental strain. The accumulated transcripts were produced from 45 genes specifying ribosomal proteins and 17 genes (cmk, dnaA, dnaG, dnaN, himD, infB, insA_1, insB_1, mviM, nusA, priB, prlA, rnpA, rpoA, rpoD, trmD, and tsf) that were adjacent to ribosomal protein-specifying genes (16). Down-regulated transcripts fell into the same general classes seen with the parental strain; transcripts from genes involved in energy metabolism (aceA, aceK, adhP, atpA, atpD, atpG, eno, fba, gapA, pflB, pgk, ppc, ptsI, sdhB, sucA, sucB, and sucC), iron metabolism (entB, entE and fur), stress responses (cadA, gadA, gadB, and hdeA) (43), and biosynthesis (aroF, aroG, ilvC, leuA, leuB, lysC, metE, serA, thrB, thrC, trpA, and trpB) and highly expressed genes (aroF, ilvC, and metE) (55) were less abundant in the mutant after PRM incubation (Table 5 and supplementary material [Table 4D; http://trna.chem.yale.edu/supdata/sup001/]).
Independent measurements of inhibitor effects.
Microarray data can reveal an organism's transcriptional response to stimuli on a genomic scale. However, to reinforce this system-wide profile, two alternative measures of monitoring a gene's transcription were used: real-time PCR and assays of an E. coli strain harboring the lux operon fused to the grpE promoter.
Real-time PCR.
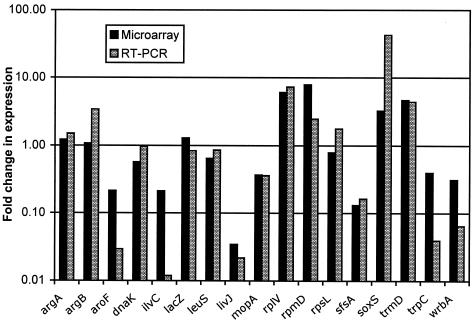
Verification of the major trends suggested by the microarray data was sought, using real-time PCR to measure relative changes in the abundance of selected transcripts (Table 6) between different RNA preparations. The direction of change, or lack thereof, observed with this technique generally confirmed results obtained by global profiling with microarrays, although variations in magnitude between the two technologies were observed (see results for aroF in Table 6). Overall, there was more than 80% concordance of the data generated by the microarray with that generated by the real-time PCR procedure with regard to the direction of the expression level change. Transcripts of genes involved in amino acid biosynthesis and uptake, a major functional group involved in the cell's responses to all inhibitors tested, showed consistent agreement between the trends suggested by both techniques (see results for argA, argB, aroF, ilvC, livJ, and trpC in Fig. 2). Also, the data from genes tested, which are involved in the cell's response to stresses and entry into stationary phase, exhibited firm agreement (see results for dnaK, mopA, soxS, and wrbA in Fig. 2). Disagreement was apparent between the microarray and real-time reverse transcriptase PCR (RT-PCR) data for two genes involved in sugar metabolism (lacZ and sfsA) and the gene coding for ribosomal protein S12 (rpsL). While the reasons for the discrepancies between the two data sets for these three genes are not clear, the overall trends suggested by the real-time PCR data for the set of genes shown in Table 6 were in good agreement with expression changes inferred from the genome-wide data set (see above).
TABLE 6.
Transcript changes measured by RT-PCR and microarrays
| Gene | b no. | Expression ratio for indicated
E. coli K-12 strain and
treatment
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT +
AZL
|
relA +
AZL
|
WT +
MUP
|
relA +
MUP
|
WT +
KAS
|
relA +
KAS
|
WT +
PRM
|
relA + PRM
|
||||||||||
| Array | RT-PCR | Array | RT-PCR | Array | RT-PCR | Array | RT-PCR | Array | RT-PCR | Array | RT-PCR | Array | RT-PCR | Array | RT-PCR | ||
| argA | b2818 | 38 | 64 | 12 | 16 | 1.4 | 1.0 | 0.68 | 1.5 | 0.95 | 1.6 | 1.9 | 0.21 | 1.2 | 1.5 | 0.72 | 0.28 |
| argB | b3959 | 9.2 | 17 | 3.5 | 7.2 | 0.87 | 0.90 | 0.83 | 1.0 | 0.99 | 4.6 | 0.60 | 0.54 | 1.1 | 3.4 | 0.82 | 0.15 |
| aroF | b2601 | 0.38 | 0.063 | 0.065 | 0.015 | 0.11 | 0.59 | 0.041 | 0.75 | 0.093 | 0.016 | 0.21 | 0.0041 | 0.21 | 0.029 | 0.26 | 0.0063 |
| dnaK | b0014 | 13 | 12 | 10 | 7.1 | 1.2 | 0.95 | 0.30 | 1.8 | 0.30 | 0.15 | 0.43 | 0.26 | 0.57 | 0.98 | 1.1 | 2.2 |
| ilvC | b3774 | 0.48 | 0.013 | 0.11 | 0.0042 | 0.40 | 0.86 | 0.078 | 0.72 | 0.11 | 0.0035 | 0.088 | 0.0062 | 0.21 | 0.012 | 0.16 | 0.068 |
| lacZ | b0344 | 1.3 | 0.046 | 0.75 | 0.59 | 1.0 | 0.56 | 1.2 | 0.84 | 1.2 | 0.22 | 0.36 | 0.70 | 1.3 | 0.83 | 0.85 | 1.2 |
| leuS | b0642 | 1.0 | 1.0 | 1.0 | 1.6 | 0.85 | 0.52 | 0.98 | 0.83 | 0.94 | 0.85 | 0.94 | 0.58 | 0.64 | 0.86 | 0.97 | 0.70 |
| livJ | b3460 | 0.4 | 0.20 | 0.22 | 0.19 | 0.37 | 1.6 | 0.14 | 0.97 | 0.062 | 0.017 | 0.34 | 0.029 | 0.035 | 0.022 | 0.26 | 0.016 |
| mopA | b4143 | 7.9 | 25 | 8.7 | 14 | 0.45 | 0.93 | 0.31 | 1.1 | 0.24 | 0.10 | 0.35 | 0.25 | 0.37 | 0.36 | 0.54 | 0.64 |
| rplV | b3315 | 3.0 | 4.6 | 4.0 | 2.3 | 0.37 | 0.70 | 4.3 | 0.93 | 3.0 | 5.1 | 7.5 | 2.1 | 6.1 | 7.3 | 4.1 | 4.2 |
| rpmD | b3302 | 2.0 | 2.4 | 2.0 | 5.4 | 0.59 | 0.32 | 2.9 | 1.1 | 3.1 | 1.7 | 4.1 | 2.0 | 7.9 | 2.5 | 3.9 | 3.2 |
| rpsL | b3342 | 0.80 | 2.5 | 0.62 | 2.5 | 0.88 | 0.62 | 0.82 | 1.4 | 0.64 | 1.6 | 1.1 | 1.7 | 0.79 | 1.8 | 0.88 | 1.6 |
| sfsA | b0146 | 1.5 | 0.36 | 1.2 | 0.064 | 0.83 | 0.47 | 0.21 | 0.65 | 0.49 | 0.16 | 7.7 | 0.79 | 0.13 | 0.16 | 4.2 | 0.79 |
| soxS | b4062 | 4.0 | 45 | 6.4 | 15 | 1.1 | 0.94 | 1.3 | 1.3 | 1.6 | 1.7 | 1.8 | 1.2 | 3.3 | 42 | 1.5 | 13 |
| trmD | b2607 | 1.2 | 0.95 | 1.3 | 0.94 | 0.35 | 0.24 | 3.9 | 0.60 | 2.4 | 0.97 | 1.1 | 1.0 | 4.7 | 4.4 | 7.5 | 3.1 |
| trpC | b1262 | 0.39 | 0.056 | 0.26 | 0.023 | 0.23 | 0.86 | 0.20 | 0.51 | 0.37 | 0.040 | 0.34 | 0.025 | 0.40 | 0.039 | 0.50 | 0.013 |
| wrbA | b1004 | 0.22 | 0.61 | 0.15 | 2.1 | 0.85 | 1.1 | 0.42 | 1.0 | 0.16 | 0.032 | 0.28 | 0.12 | 0.31 | 0.065 | 0.81 | 0.14 |
FIG. 2.
Severalfold changes in levels of selected mRNAs for relA+ strain CF1943 treated with PRM, as determined by two distinct methods (RT-PCR and hybridization to DNA microarrays). The strain was challenged, RNA was prepared from challenged and control cultures, and the RNA samples were used to determine the changes in RNA level after challenge. Distinct cultures were challenged for the RT-PCR and DNA microarray analyses.
Bioluminescence of the grpE::lux operon fusion strain.
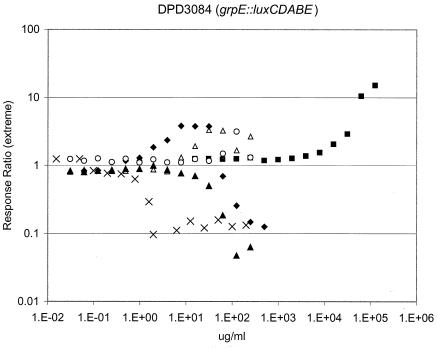
The heat shock response (58) is thought to be triggered by the presence of unfolded proteins in the cytoplasm (23). To determine whether such a response can be triggered by the translation inhibitors studied here, we used E. coli strain DPD3084, harboring the rpoH-controlled grpE promoter fused to the lux operon. As seen in Fig. 3, the expected result (that exposure to high concentrations of ethanol elevated expression from this fusion) was revealed by bioluminescent monitoring. At moderate concentrations (16 to 250 μg/ml), AZL treatment elevated grpE::luxCDABE expression. At higher concentrations (data not shown), light emission was lowered by this treatment, presumably because protein synthesis needed to make Lux polypeptides was blocked. Concentrations lower than 16 μg/ml had no effect on light output. MUP concentrations of more than 0.4 μg/ml compromised light emission, while the strain was unresponsive to lower concentrations. Treatment with KAS lowered bioluminescence at concentrations higher than 63 μg/ml. A robust bioluminescent response indicative of heat shock was observed at 2 to 31 μg/ml of the antibiotic, amounts well below that needed for growth inhibition, while lower doses were ineffectual. The response to PRM was more complicated. Reduced light emission was seen over a wide range of concentrations (from 8 to 250 μg/ml); at lower concentrations, DPD3084 appeared indifferent to the challenge. During the time course studied, increased heat shock expression was seen in a narrow window of PRM concentrations of 63 to 125 μg/ml (consistent with a previously reported modest response to a related grpE::lux fusion at 200 μg/ml [52]).
FIG. 3.
Dose responses obtained with a bioluminescent sensor that detects both induction of a heat shock-regulated promoter and compromise of a “healthy” metabolism. An exponential-phase culture of E. coli strain DPD3084 was challenged individually with the indicated amount of ethanol (▪), PRM (▴ and ○), KAS (♦), AZL (▵), or MUP (×), and bioluminescence was recorded as a function of time after treatment. Maximal (▪, ○, ♦, ▵, and ×) and minimal (▴) response ratios observed during the period following exposure to each concentration of the different chemicals are reported as extreme response ratios.
Characterization of abundant biosynthetic transcripts often lost after inhibitor challenge.
Several such transcripts were lost after inhibitor challenges (Table 3 and Table 5) (39). To examine this phenomenon more thoroughly, cysK, glnA, ilvC, and metE transcripts were studied.
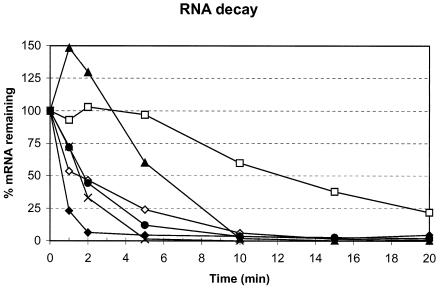
RT-PCR was used to measure the chemical half-lives of several transcripts in an early-exponential-phase culture (A600 = 0.4) of E. coli strain MG1655 subjected to shaking at 37°C in Luria-Bertani medium after blocking of transcription initiation with rifampin (0.15 mg/ml) (Fig. 4). As controls, the long-lived ompA transcript and the labile trxA mRNA (17) were also examined. The half-life of the trxA mRNA was found to be 1.9 min, while that of the ompA transcript was 8.8 min. Those for cysK, glnA, ilvC, and metE were determined to be 1.8, 0.5, 1.4, and 2.3 min, respectively. Thus, the chemical half-lives of these transcripts were not remarkable.
FIG. 4.
mRNA decay after rifampin inhibition. The inhibitor was added to exponential-phase cultures at the initiation of the experiment. At the indicated times, RNA was prepared prior to being subjected to RT-PCR analyses to determine the amounts of trxA (⋄), ompA (□), metE (▴), ilvC (×), glnA (♦), and cysK (•) mRNA that remained.
The loss of ilvC, glnA, and metE mRNAs after different inhibitor challenges was determined by microarray analyses (see Table 7 for details). RT-PCR analysis of the same mRNA preparations confirmed the observations. Treatment with SM, 2,4-dinitrophenol, p-hydroxybenzoate, and acivicin was shown by both methodologies to cause substantial loss of these mRNAs consistent with the RT-PCR studies of inhibitors arresting translation (Table 6).
TABLE 7.
Loss of abundant transcripts after distinct challenges
| Transcript | Expression
ratio after challenge
with:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| SMa
|
Acivicinb
|
p-Hydroxybenzoatec
|
2,4-Dinitrophenold
|
|||||
| Array | RT-PCR | Array | RT-PCR | Array | RT-PCR | Array | RT-PCR | |
| cysK | 0.5 | 1.4 | 0.39 | 0.19 | 1 | 1 | 0.56 | 0.71 |
| glnA | 0.14 | 0.12 | 0.15 | 0.01 | 0.14 | 0.001 | 0.1 | 0.015 |
| ilvC | 0.21 | 0.03 | 0.04 | 0.001 | 0.06 | 0.003 | 0.05 | 0.014 |
| metE | 0.08 | 0.0031 | 0.13 | 0.033 | 0.3 | 0.067 | 0.13 | 0.13 |
RNA isolated from strain DPD1675 challenged with inhibitor (8 μg/ml) for 45 min with shaking (250 rpm) at 37°C.
RNA isolated from an exponential-phase culture of strain MG1655 treated with inhibitor (2 μg/ml) for 60 min with shaking (250 rpm) at 37°C in medium E (12) supplemented with thiamine and 0.4% glucose.
RNA isolated from an exponential-phase culture of strain DE112 (49) incubated with inhibitor (3,453 μg/ml) for 60 min with shaking (250 rpm) at 37°C.
RNA isolated from an exponential-phase culture of strain DE112 (49) treated with inhibitor (75 μg/ml) for 60 min with shaking (250 rpm) at 37°C in medium E (12) supplemented with thiamine and 0.4% glucose.
DISCUSSION
RNA polymerase activity is limiting in E. coli. The 4,641 predicted promoters responsible for expression of the 2,326 transcription units (37) are serviced by approximately 2,800 molecules of RNA polymerase in a cell that doubles once every hour, a rate typical of growth using glucose as a carbon source in minimal medium (the standard, uninhibited conditions used in this study). Of these enzymes, only 500 are actively engaged in transcription at any instant, with 300 producing mRNA and 200 synthesizing tRNA and rRNA (8). Even when E. coli is growing at its maximal rate, only about 700 RNA polymerase molecules are engaged in the production of mRNA at any one time, with 2,600 RNA polymerase molecules devoted to stable RNA synthesis.
We have previously used a parts-per-million scale (55) to estimate mRNA content in E. coli. Under standard conditions, at any instant >300 RNA polymerase molecules are engaged in mRNA synthesis, with a transit time of 21 s (8) for an average 951-bp gene (6). Thus, transcription across some ORF occurs about 55,000 times per generation. This is equivalent to between 10 and 15 transcription events per generation for a gene whose expression is average (223 ppm [0.0223%]). Deviations from this value are large; many ORFs may be transcribed less than once per cell division (18 ppm) while others are estimated to be transcribed about 900 times per doubling (16,200 ppm) (55). This range is consistent with other thoughts on gene expression, including that the range of protein expression is from <1 to 105 molecules per cell (29, 46).
These experiments provide an approximation of the distribution of RNA polymerase among those promoters that produce mRNA. It is conceivable, however, that distribution of RNA polymerase between mRNA and stable RNA synthesis can change greatly upon inhibition (3, 4); that would have caused us to underestimate induction of gene expression in response to some inhibitors.
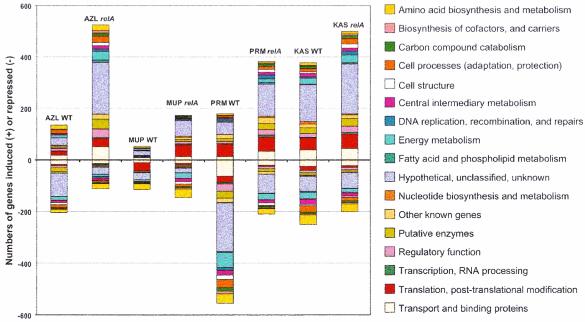
The broad effects of each inhibitor on gene expression levels are evident when the distribution of induced and repressed genes over the spectrum of functional classifications in the cell is examined. Major trends and large effects become easily discernible using this approach. One can immediately see that the total number of gene transcripts affected varies greatly with the inhibitor used (for an example, see the results of MUP inhibition of the wild-type relA strain versus those of KAS inhibition of the wild-type relA strain [Fig. 5]). This property of the response may illustrate the inherent differential complexity of the inhibitors' targets. In the case of MUP, the primary effect is that of blocking one amino acid's incorporation into protein (i.e., MUP inhibits Ile charging onto tRNAIle). KAS, on the other hand, targets a process involving a higher order of complexity, interfering with the central machinery of protein synthesis itself (i.e., KAS blocks formation of the 70S ribosomal particle). The relA-dependent inversion of the expression of ribosomal proteins and those associated with the translation apparatus is easily recognizable for MUP treatment as discussed above (Fig. 5). Another striking feature of the various expression patterns is that a large fraction of the cell's response to each inhibitor is derived from hypothetical or unknown ORFs (Fig. 5). It is therefore clear that a large part of any inhibitor's effects on cellular physiology cannot yet be rationalized.
FIG. 5.
A global view of changes in mRNA distribution. Each bar represents a separate inhibitor-strain combination. The number of genes whose expression was elevated or diminished is represented by the height of each bar. The contribution of each functional class of genes to the observed change is signified by the area of the colored block within each bar. WT, wild-type strain W3110; relA, W3110 ΔrelA251::kan strain.
The four inhibitors elicited different patterns of elevated gene expression from the relA+ strain. AZL caused elevated expression of many stress-induced genes (including those of the heat shock regulon) as well as increased content of amino acid, though not branched-chain amino acid, biosynthetic transcripts. The thought that AZL is not charged to tRNA and incorporated into protein within E. coli (40) needs to be reconsidered in light of these results and those found with Salmonella enterica serovar Typhimurium (42) and E. coli (26). Those mRNAs elevated after MUP treatment were quite different, and the increased amount of thr, leu, and ilv operon mRNAs detected conformed to expectations consistent with deattenuation caused by a limitation for isoleucyl-tRNA (18, 45). That the responses to these two inhibitors differed from those caused by treatment with agents acting on ribosomes was not surprising. Blocking initiation of translation with KAS and causing premature release of polypeptides with PRM resulted in elevated expression of ribosomal protein and other mRNAs specifying the translational machinery, as if the cell sensed that there was insufficient protein synthesis relative to other cellular activities. Surprisingly, PRM treatment did not cause an elevated heat shock response.
Shared patterns of reduced gene expression after inhibition of the relA+ strain were also observed. MUP, KAS, and PRM treatment elicited reduced accumulation of transcripts previously identified as being highly expressed in exponentially growing cells or cells transitioning to the stationary phase (55). In addition, KAS and PRM, but not AZL or MUP, treatment resulted in diminished titers of transcripts specifying several enzymes involved in central carbon metabolism. MUP treatment resulted in a decrease in mRNAs specifying ribosomal proteins, as would be expected for the stringent response (9).
The relA mutation changed the observed pattern of gene expression in several ways. Most notably, each of the four treatments of a relA mutant resulted in elevated levels of ribosomal protein mRNAs; AZL and MUP did not cause elevated titers in the relA+ strain. These observations reinforce the concept of RelA acting as a “brake” to subvert the inclination to increase the titer of the translational machinery when protein synthesis is limited. Such a hypothesis could not be reached without comparisons of the various responses of a mutant strain challenged with a series of inhibitors blocking various points in the process of translation (Fig. 1). While a more restricted set of translational inhibitors has been used to generate transcriptional profiles with another species (31), a broader set of chemicals, a global regulatory mutant, and prior quantitative knowledge of steady-state mRNA levels (55) have contributed to the concept suggested here.
The stringent response is complicated and has been operationally linked to relA function. Others have presented an expanded stringent response model (10) and have linked ppGpp, lrp, and rpoS to adaptation to famine (44). Proper utilization of terminology and concepts surrounding the words stimulon, regulon, and modulon (30) may reveal considerable commonality among those results, the work reported in this article, and the results of studies presented elsewhere (39, 55). Such convergence suggests that systems biology will contribute a great deal to the understanding of microbial physiology.
Acknowledgments
We thank Jordan S. Pober (Yale University) for use of real-time PCR equipment.
Research in the laboratory of D.G.S. was supported by grants from the National Institute of General Medical Sciences, the Department of Energy, and the National Aeronautics and Space Administration.
Footnotes
This work is dedicated to the memory of Professor Philip E. Hartman, Department of Biology, The Johns Hopkins University.
REFERENCES
- 1.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance stress response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 3.Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305:689-702. [DOI] [PubMed] [Google Scholar]
- 4.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. [DOI] [PubMed] [Google Scholar]
- 5.Belkin, S., T. K. Van Dyk, A. C. Vollmer, D. R. Smulski, and R. A. LaRossa. 1996. Monitoring subtoxic environmental hazards by stress-responsive luminous bacteria. Environ. Toxicol. Water Qual. 11:179-185. [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 7.Bocazzi, P., J. K. Zhang, and W. W. Metcalf. 2000. Generation of dominant selectable markers for resistance to pseudomonic acid by cloning and mutagenesis of the ileS gene from the archaeon Methanosarcina barkeri Fusaro. J. Bacteriol. 182:2611-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bremer, H., and P. P. Dennis. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p. 1553-1569. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 9.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 10.Chang, D. E., D. J. Smalley, and T. Conway. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289-306. [DOI] [PubMed] [Google Scholar]
- 11.Davis, B. D., P.-C. Tai, and B. J. Wallace. 1974. Complex interactions of antibiotics with the ribosome, p. 771-789. In M. Nomura, A. Tissieres, and P. Lengyel (ed.), Ribosomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 12.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 13.Goff, S. A., and A. L. Goldberg. 1985. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock genes. Cell 41:587-595. [DOI] [PubMed] [Google Scholar]
- 14.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 15.Hughes, J., and G. Mellows. 1980. Interaction of pseudomonic acid A with Escherichia coli B isoleucyl-tRNA synthetase. Biochem. J. 191:209-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keener, J., and M. Nomura. 1996. Regulation of ribosome synthesis, p. 1417-1431. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 17.Kushner, S. R. 1996. mRNA decay, p. 849-860. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 18.Landick, R., C. L. Turnbough, Jr., and C. Yanofsky. 1996. Transcription attenuation, p. 1263-1286. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 19.LaRossa, R. A. 1996. Mutant selections linking physiology, inhibitors, and genotypes, p. 2527-2587. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 20.LaRossa, R. A., and S. C. Falco. 1984. Amino acid biosynthetic enzymes as targets of herbicide action. Trends Biotechnol. 2:158-161. [Google Scholar]
- 21.LaRossa, R. A., and J. V. Schloss. 1984. The sulfonylurea herbicide sulfometuron methyl is an extremely potent and selective inhibitor of acetolactate synthase in Salmonella typhimurium. J. Biol. Chem. 259:8753-8757. [PubMed] [Google Scholar]
- 22.LaRossa, R. A., and T. K. Van Dyk. 1987. Metabolic mayhem caused by 2-ketoacid imbalances. Bioessays 7:125-130. [DOI] [PubMed] [Google Scholar]
- 23.LaRossa, R. A., and T. K. Van Dyk. 1991. Physiological roles of the dnaK and groE stress proteins: catalysts of protein folding or macromolecular sponges? Mol. Microbiol. 5:529-534. [DOI] [PubMed] [Google Scholar]
- 24.LaRossa, R. A., T. K. Van Dyk, and D. R. Smulski. 1990. A need for metabolic insulation: lessons from sulfonylurea genetics, p. 109-121. In Z. Barak, D. M. Chipman, and J. V. Schloss (ed.), Biosynthesis of branched chain amino acids. VCH and Balaban Publishers, New York, N.Y.
- 25.LaRossa, R. A., T. K. Van Dyk, and D. R. Smulski. 1987. Toxic accumulation of α-ketobutyrate caused by inhibition of the branched-chain amino acid biosynthetic enzyme acetolactate synthase in Salmonella typhimurium. J. Bacteriol. 169:1372-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemeignan, B., P. Sonigo, and P. Marliere. 1993. Phenotypic suppression by incorporation of an alien amino acid. J. Mol. Biol. 231:161-166. [DOI] [PubMed] [Google Scholar]
- 27.Lund, E., and N. O. Kjeldgaard. 1972. Metabolism of guanosine tetraphosphate in Escherichia coli. Eur. J. Biochem. 28:316-326. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Inc., Sunderland, Mass.
- 30.Neidhardt, F. C., and M. F. Savageau. 1996. Regulation beyond the operon, p. 1310-1324. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 31.Ng, W.-L., K. M. Kazmierczak, G. T. Robertson, R. Gilmour, and M. E. Winkler. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 185:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okuyama, A., M. N., T. Kinoshita, and N. Tanaka. 1971. Inhibition by kasugamycin of initiation complex formation on 30S ribosomes. Biochem. Biophys. Res. Commun. 43:196-199. [DOI] [PubMed] [Google Scholar]
- 33.Patte, J.-C. 1996. Biosynthesis of threonine and lysine, p. 528-541. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 34.Picataggio, S. K., L. J. Templeton, D. R. Smulski, and R. A. LaRossa. 2002. Comprehensive transcript profiling of Escherichia coli using high-density DNA microarrays, p. 177-188. In V. L. Clark and P. M. Bavoil (ed.), Bacterial pathogenesis, vol. 358, part C. Academic Press, San Diego, Calif. [DOI] [PubMed]
- 35.Rhodius, V., T. K. Van Dyk, C. Gross, and R. A. LaRossa. 2002. Impact of genomic technologies on the study of bacterial gene expression. Annu. Rev. Microbiol. 56:599-624. [DOI] [PubMed] [Google Scholar]
- 36.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 37.Salgado, H., A. Santos-Zavaleta, S. Gama-Castro, D. Millán-Zárate, E. Díaz-Peredo, F. Sánchez-Solano, E. Pérez-Rueda, C. Bonavides-Martínez, and J. Collado-Vides. 2001. RegulonDB (version 3.2): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 29:72-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schloss, J. V. 1989. Modern aspects of enzyme inhibition with particular emphasis on reaction-intermediate analogs and other potent, reversible inhibitors, p. 165-245. In P. Boger and G. Sandman (ed.), Target sites of herbicide action. CRC Press, Boca Raton, Fla.
- 39.Smulski, D. R., L. L. Huang, M. P. McCluskey, M. J. G. Reeve, A. C. Vollmer, T. K. Van Dyk, and R. A. LaRossa. 2001. Combined, functional genomic-biochemical approach to intermediary metabolism: interaction of acivicin, a glutamine amidotransferase inhibitor, with Escherichia coli K-12. J. Bacteriol. 183:3353-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soll, D., and P. R. Schimmel. 1974. Aminoacyl-tRNA synthetases, p. 489-538. In P. D. Boyer (ed.), The enzymes, 3rd ed., vol. 10. Academic Press, New York, N.Y.
- 41.Stephens, J. C., S. W. Artz, and B. N. Ames. 1975. Guanosine 5′-diphosphate 3′-diphosphate (ppGpp): positive effector for histidine operon transcription and general signal for amino acid deficiency. Proc. Natl. Acad. Sci. USA 72:4389-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stieglitz, B. 1971. Ph.D. dissertation. Cornell University, Ithaca, N.Y.
- 43.Storz, G., and R. Hengge-Aronis (ed.). 2000. Bacterial stress responses. ASM Press, Washington, D.C.
- 44.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Umbarger, H. E. 1996. Biosynthesis of the branched-chain amino acids, p. 442-457. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 46.VanBogelen, R. A., K. Z. Abshire, A. Pertsemlidis, R. L. Clark, and F. C. Neidhardt. 1996. Gene-protein database of Escherichia coli K-12, edition 6, p. 2067-2117. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Resnikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 47.Van Dyk, T. K., B. L. Ayers, R. W. Morgan, and R. A. LaRossa. 1998. Constricted flux through the branched-chain amino acid biosynthetic enzyme acetolactate synthase triggers elevated expression of genes regulated by rpoS and internal acidification. J. Bacteriol. 180:785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Dyk, T. K., E. J. DeRose, and G. E. Gonye. 2001. LuxArray, a high-density, genomewide transcription analysis of Escherichia coli using bioluminescent reporter strains. J. Bacteriol. 183:5496-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Dyk, T. K., W. R. Majarian, K. B. Konstantinov, R. M. Young, P. S. Dhurjati, and R. A. LaRossa. 1994. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl. Environ. Microbiol. 60:1414-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Dyk, T. K., and R. A. Rosson. 1998. Photorhabdus luminescens luxCDABE promoter probe vectors. Methods Mol. Biol. 102:85-95. [DOI] [PubMed] [Google Scholar]
- 51.Van Dyk, T. K., D. R. Smulski, D. A. Elsemore, R. A. LaRossa, and R. W. Morgan. 2000. A panel of bioluminescent biosensors for characterization of chemically induced bacterial stress responses, p. 167-184. In A. Mulcandani and O. A. Sadik (ed.), Chemical and biological sensors for environmental monitoring. American Chemical Society Symposium Series, no. 762. American Chemical Society, Washington, D.C.
- 52.Van Dyk, T. K., D. R. Smulski, T. R. Reed, S. Belkin, A. C. Vollmer, and R. A. LaRossa. 1995. Responses to toxicants of an Escherichia coli strain carrying a uspA′::lux genetic fusion and an E. coli strain carrying a grpE′::lux genetic fusion are similar. Appl. Environ. Microbiol. 61:4124-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Dyk, T. K., Y. Wei, M. K. Hanafey, M. Dolan, M. J. G. Reeve, J. A. Rafalski, L. B. Rothman-Denes, and R. A. LaRossa. 2001. A genomic approach to gene fusion technology. Proc. Natl. Acad. Sci. USA 98:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei, Y., J.-M. Lee, and R. A. LaRossa. 2001. The global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J. Bacteriol. 183:2265-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei, Y., J.-M. Lee, C. Richmond, F. R. Blattner, J. A. Rafalski, and R. A. LaRossa. 2001. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 183:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao, H., M. Kalman, I. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 57.Yanagisawa, T., J. T. Lee, H. C. Wu, and M. Kawakami. 1994. Relationship of protein structure of isoleucyl-tRNA synthetase with pseudomonic acid resistance of Escherichia coli. A proposed mode of action of pseudomonic acid as an inhibitor of isoleucyl-tRNA synthetase. J. Biol. Chem. 269:24304-24309. [PubMed] [Google Scholar]
- 58.Yura, T., M. Kanemori, and M. T. Morita. 2000. The heat shock response: regulation and function, p. 3-18. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.