Abstract
Archaeal protein trafficking is a poorly characterized process. While putative type I signal peptidase genes have been identified in sequenced genomes for many archaea, no biochemical data have been presented to confirm that the gene product possesses signal peptidase activity. In this study, the putative type I signal peptidase gene in Methanococcus voltae was cloned and overexpressed in Escherichia coli, the membranes of which were used as the enzyme source in an in vitro peptidase assay. A truncated, His-tagged form of the M. voltae S-layer protein was generated for use as the substrate to monitor the signal peptidase activity. With M. voltae membranes as the enzyme source, signal peptidase activity in vitro was optimal between 30 and 40°C; it was dependent on a low concentration of KCl or NaCl but was effective over a broad concentration range up to 1 M. Processing of the M. voltae S-layer protein at the predicted cleavage site (confirmed by N-terminal sequencing) was demonstrated with the overexpressed archaeal gene product. Although E. coli signal peptidase was able to correctly process the signal peptide during overexpression of the M. voltae S-layer protein in vivo, the contribution of the E. coli signal peptidase to cleavage of the substrate in the in vitro assay was minimal since E. coli membranes alone did not show significant activity towards the S-layer substrate in in vitro assays. In addition, when the peptidase assays were performed in 1 M NaCl (a previously reported inhibitory condition for E. coli signal peptidase I), efficient processing of the substrate was observed only when the E. coli membranes contained overexpressed M. voltae signal peptidase. This is the first proof of expressed type I signal peptidase activity from a specific archaeal gene product.
The export of proteins and their arrival at the correct final destinations are vital cellular processes. To achieve proper recognition and targeting, many proteins that reside beyond the cytoplasm are synthesized as preproteins, with an N-terminal signal peptide that acts as a targeting signal. In bacteria, most exported proteins pass through the Sec secretory machinery, followed by processing by a signal peptidase that removes the signal peptide (11). Signal peptidase I is the principal peptidase responsible for processing most exported proteins in bacteria, while signal peptidase II is responsible for processing lipoprotein precursors (8, 27). In many gram-negative bacteria, a third signal peptidase, termed the prepilin peptidase, is required for processing of type IV prepilin subunits and related pseudopilins (18).
To date, the archaeal type I signal peptidase is still poorly characterized. Limited information has come from genome analysis and sequence annotation, which seems to suggest several similarities between the archaeal signal peptidase and its eukaryal counterpart (13). Bacterial signal peptidase I is active as a monomer and is composed of two structural components referred to as domain I and domain II (20). Within domain I, serine 90 and lysine 145 are the two critical amino acids identified; they are believed to form the catalytic dyad responsible for enzymatic activity and are conserved among bacteria. Domain II folds as a β-sheet, and while it is found in all bacterial type I signal peptidases, its role in the enzymatic activity remains unclear. Unlike those in bacteria, eukaryotic signal peptidases do not contain domain II regions. Also, unlike their bacterial counterparts, eukaryotic signal peptidases function as part of a multisubunit signal peptidase complex, which in Saccharomyces cerevisiae is composed of the Sec11, Spc1, Spc2, and Spc3 proteins (31). In terms of the catalytic dyad, serine 90 remains conserved while lysine 145 found in bacteria is replaced by a conserved histidine residue in eukaryotes (13). Site-directed mutagenesis of conserved and nonconserved lysine residues present in Sec11 as well as in Spc3 do not hinder enzymatic activity, suggesting that eukaryal signal peptidases possess a very different catalytic mechanism (30). Archaeal signal peptidases appear to resemble those in eukarya in terms of catalysis. Like those in eukarya, archaeal signal peptidases lack the conserved lysine of the bacterial serine/lysine catalytic dyad; it is replaced by a histidine residue. Also, most archaeal signal peptidases lack a significant domain II region, and even in the species that do possess domain II regions, these regions appear to be quite different from those identified in bacteria (13).
Thus far the only signal peptidase in archaea for which there are biochemical data is the preflagellin peptidase in methanococci encoded by flaK, which is responsible for the removal of the short atypical signal peptide found on archaeal flagellins (3, 7). In other archaea, a limited number of other substrates, often sugar-binding proteins, are also processed by the same enzyme (2). While putative type I signal peptidase genes have been identified in annotated complete sequenced genomes for many archaea, no biochemical data for any archaea have yet been presented to confirm that the annotated gene possesses signal peptidase activity. Recent studies of complete genomes of various archaea have identified between 8 and 32% of the proteomes predicted to be exported or secreted (22, 24), the majority of which would be expected to have signal peptides processed by the type I signal peptidase. Clearly the lack of data on type I signal peptidase in archaea represents a serious deficiency in our knowledge of protein secretion in the domain Archaea. In this paper, we report the cloning and overexpression of the type I signal peptidase of Methanococcus voltae in Escherichia coli as a His-tagged protein. Signal peptidase activity of this enzyme against the M. voltae S-layer protein as the substrate was demonstrated in vitro by employing a high-salt-concentration buffer to inhibit background E. coli enzymatic activity. This is the first report to demonstrate expressed type I signal peptidase activity from a specific archaeal gene product.
(Portions of this work have been presented previously [S. L. Bardy, S. Y. Ng, L. Noad, and K. F. Jarrell, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. I-26, 2002].)
MATERIALS AND METHODS
Microbial strains and growth conditions.
M. voltae PS was grown in Balch medium III at 37°C under an atmosphere of CO2 and H2 (20:80) as previously described (14). E. coli strains were grown at 37°C in Luria-Bertani medium supplemented with ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), and streptomycin (50 μg/ml) when necessary. E. coli DH5α (Novagen) was used for all in vitro cloning. E. coli BL21(DE3)/pLysS (Novagen) and E. coli BL21(DE3)/pSJS1240 (obtained from S. Sandler, University of California, Berkeley) were used as expression strains.
Cloning of the truncated S-layer gene.
Although the signal peptide of the S-layer protein was originally predicted to be 12 amino acids in length (10) (S-layer protein initially reported as an ATPase [GenBank accession no. M59200]), new evidence has suggested that the translation start codon lies further upstream (1), extending the signal peptide to 28 amino acids in length. The cloning in this study was done based on this information.
Due to the size of the natural S-layer protein (75 kDa) in M. voltae (16), it was reasoned that the processed and unprocessed forms would be difficult to discern by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting. Therefore, a truncated version of the S-layer protein was produced for use in the peptidase assay. Primers were designed with NdeI and XhoI restriction sites (boldface type) incorporated into the forward (5′-GGAATTCCATATGGCAATGAGCTTAAAAAAAATCGGTGC −3′) and reverse (5′-CCGCTCGAGTTGGAAGTCTTCGTTATCGTCATCG-3′) primers, respectively. The amplified fragment, when expressed with the His tag, would generate a predicted protein of approximately 24.1 kDa, a size that has been proven to be effective for analyzing preflagellin cleavage (5). PCR was performed on a MiniCycler (MJ Research, Inc., Waltham, Mass.) using Pwo DNA polymerase under the following conditions: 95°C for 5 min; 29 cycles of 94°C for 45 s; 50°C for 45 s, and 72°C for 1 min; and a final cycle with an extension time of 5 min at 72°C. The amplified product was purified using a PCR purification column (Qiagen, Chatsworth, Calif.) and cloned into the multiple cloning site of the pET23a(+) vector via the NdeI and XhoI restriction sites to produce plasmid pKJ341. In the PCR product, there is no stop codon, and when cloned into pET23a, an in-frame fusion with a His-tag sequence at the C terminal of the protein was created, allowing for its subsequent detection by immunoblotting using anti-His antibodies.
Cloning of the M. voltae signal peptidase gene.
A signal peptidase gene (MJ0260) was identified in the complete genomic sequence of Methanococcus jannaschii (6). From this, forward (5′-GGAATTCCATATGGTTGTTTTGTTTTTAATTTGG-3′) and reverse (5′-GGAATTCCATATGTTATTTTCTCCCTCCTTTAAGATAATTG-3′) PCR primers were used to amplify, from M. jannaschii genomic DNA, a 609-bp fragment which was subsequently used to generate a digoxigenin-labeled probe by using a DNA labeling kit (Boehringer Mannheim, Mannheim, Germany). A 2.3-kb HindIII fragment that bound this probe was identified in a Southern blot with M. voltae genomic DNA. To clone the corresponding gene, M. voltae genomic DNA was digested with HindIII and the fragments in the size range of 2 to 2.5 kb were excised, purified, and ligated into pUC21, followed by transformation into E. coli (DH5α) and plating on Luria-Bertani plates containing ampicillin (100 μg/ml) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml). White colonies were screened, and a probable positive clone was detected by Southern hybridization. Subsequent sequencing revealed the presence of a putative signal peptidase gene (GenBank accession no. AF395308) as well as its surrounding genes.
Based on the sequence of this clone, primers were designed with NdeI and XhoI restriction sites (boldface type) incorporated into the forward (5′-GGAATTCCATATGGATAATAAAGATAATAATC-3′) and reverse (5′-CCGCTCGAGTTTACTAAACATTTTAAATATGC-3′) primers, respectively. PCR was performed as described above, and the entire amplified M. voltae signal peptidase gene product was cloned into pET23a to generate an in-frame fusion with the C-terminal His-tag sequences, generating plasmid pKJ385.
Isolation of M. voltae membranes.
Six milliliters of late-exponential-phase M. voltae cells was harvested by aerobic centrifugation at 16,000 × g for 5 min. The pellets were resuspended in 100 μl of medium and diluted with sterile distilled H20 to a final volume of 1.5 ml, resulting in lysis of the osmotically fragile cells. The lysate was centrifuged at 16,000 × g for 10 min to pellet the membranes, which were resuspended in 100 μl of distilled H20 for use in the peptidase assay.
Overexpression and isolation of the truncated S-layer protein and signal peptidase.
E. coli BL21(DE3) harboring pSJS1240 was used as the expression host for the plasmid pKJ341 (truncated S-layer protein gene cloned into pET23a). Protein expression was induced in log-phase cells with the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) (Life Technologies, Gibco-BRL, Mississauga, Ontario, Canada) to a concentration of 0.4 mM. At 20-min postinduction, when the unprocessed S-layer protein was the dominant expressed form, the cells were heat treated at 80°C for 30 min to inactivate the E. coli signal peptidase I. Cells were harvested by centrifugation at 5,000 × g for 10 min, and the pellet was kept frozen overnight. The pellet was resuspended in 20 mM Tris-HCl buffer (pH 8.0), and the viscous solution was sonicated twice for 30 s each time on ice to aid cell lysis. Unbroken cells and potential inclusion bodies were removed by centrifugation at 5,000 × g for 5 min. A crude membrane fraction containing the S-layer protein was then obtained by centrifugation of the supernatant at 20,000 × g for 30 min. The pellet was resuspended in 5 ml of buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, 0.75 M urea [pH 7.9]) and dialyzed against sterile distilled H2O overnight at 4°C. This material was used in subsequent peptidase assays as a substrate source.
Membranes containing overexpressed M. voltae signal peptidase were obtained from E. coli cells harboring pKJ385 in a similar fashion except that the expression host was E. coli BL21(DE3)/pLysS, the induction time was 90 min, and the cells were not heat treated prior to membrane isolation.
Signal peptidase assay using S-layer protein as the substrate.
The assay for S-layer signal peptidase activity in M. voltae was based on the conditions determined as being optimal for in vitro activity of the preflagellin peptidase from M. voltae (7), which in turn had been based on the assay for prepilin peptidase developed for Pseudomonas aeruginosa (25). The substrate for the assay was the E. coli membrane preparation containing the overexpressed S-layer protein, isolated as described above. The enzyme source was an M. voltae membrane preparation. The assay was performed with 72 μl of E. coli membranes containing the overexpressed S-layer protein (approximately 40 μg) and 12 μl of M. voltae membranes (approximately 30 μg of protein) in the presence of 12 μl of buffer A (125 mM HEPES [pH 8.5], with 1.25% Triton X-100), 12 μl of buffer B (125 mM HEPES, 4 M KCl [pH 8.5], with 1.25% Triton X-100), and 12 μl of sterile distilled H2O. The reaction was started by the addition of M. voltae membranes, and the reaction mixture was kept at 37°C in a water bath. Samples (10-μl aliquots) were taken from the reaction mixture at various time points, mixed into 2× ESB (0.0625 M Tris-HCl [pH 6.8], 1% [wt/vol] SDS, 10% glycerol, 2% 2-mercaptoethanol, 0.001% bromophenol blue), and immediately boiled for 5 min prior to SDS-PAGE and Western blot analysis using anti-His antibodies (26).
The peptidase assay using the cloned M. voltae signal peptidase was performed in a similar manner, with 12 μl of E. coli membrane preparation containing the M. voltae signal peptidase (approximately 40 and 0.4 μg of protein, respectively, for the high- and low-enzyme-concentration reactions) substituting as the enzyme source.
The cation concentrations of the peptidase assay were varied by using different amounts of buffer A, buffer B, and buffer C (125 mM HEPES, 5 M NaCl [pH 8.5], with 1.25% Triton X-100).
N-terminal sequence analysis.
The purified S-layer proteins (processed and unprocessed forms) were resolved by SDS-PAGE and transferred onto an Immobilon-P nitrocellulose membrane (Millipore, Bedford, Mass.) as previously described (28). The membrane was briefly stained with Coomassie blue R250 (0.1% Coomassie blue R250, 40% methanol, 1% acetic acid), destained in 50% methanol, and rinsed thoroughly in distilled water. Protein bands were excised, extensively washed to remove glycine, and dried. Sequencing was performed by David Watson, National Research Council of Canada, Ottawa, Ontario, Canada.
RESULTS
Cloning, overexpression, and isolation of the truncated, His-tagged M. voltae S-layer protein.
The only likely substrate for processing by a type I signal peptidase in M. voltae that has been studied is the S-layer protein. It has a predicted signal peptide that is 28 amino acids in length (1), and the protein has an apparent molecular mass of 75 kDa (16). In order to have a readily discernible difference between the processed and unprocessed forms of the protein in SDS-PAGE and immunoblotting analyses, a truncated version of the S-layer protein was produced for use in the peptidase assay. Although the truncated S-layer protein with the attached His tag was predicted to be 24.1 kDa, the unprocessed form of the polypeptide ran with an apparent molecular mass of 35.5 kDa, suggesting that it is somehow modified, possibly through glycosylation—a common feature of archaeal S-layer proteins (23). The induced expression of the truncated S-layer in E. coli was monitored by Western blotting (Fig. 1). It was observed that one protein band (apparent molecular mass of 35.5 kDa) was present at early time points up to 30 min postinduction. This protein appeared to undergo a gradual conversion through time, resulting in the appearance of a smaller protein (approximate apparent molecular mass of 31.5 kDa), with the difference being approximately equal to that which is expected if the 28-amino-acid signal peptide were removed. Subsequent N-terminal sequencing confirmed the two bands as being the unprocessed and processed forms of the S-layer protein, respectively.
FIG. 1.
Overexpression of the truncated S-layer protein. The cloned truncated S-layer protein was overexpressed in E. coli BL21(DE3)/pLysS. Following induction with 0.4 mM IPTG, 1.5-ml samples were taken at various time points and centrifuged and the pellet was resuspended in 100 μl of 2× ESB and boiled for 5 min. Ten-microliter aliquots were examined by immunoblotting using a primary anti-His antibody at a dilution of 1:20,000. Lanes (from left): 1, immediately prior to IPTG addition; 2 to 8: 0.5, 1, 1.5, 2, 2.5, 3, and 4 h postinduction, respectively; 9, uninduced cells after 4-h mock induction. The corresponding N-terminal sequence determined for each band is indicated.
Due to different codon preferences in methanococci, the overexpression of Methanococcus proteins in E. coli hosts is sometimes problematic, and an effective method used to circumvent this is to have multiple plasmid-borne copies of the tRNA genes corresponding to the rare codons present in the expressing host (15). A comparison of S-layer overexpression was done with pKJ341 in E. coli BL21(DE3)/pLysS and E. coli BL21(DE3) harboring pSJS1240. Plasmid pSJS1240 contains genes for several tRNAs recognizing rare codons in E. coli and has previously been used successfully to help overexpress methanococcal proteins in E. coli (15). It was found that expression in the latter strain was more reproducible with a greater yield (data not shown), and subsequently it was chosen as the overexpression host used for substrate isolation.
Detection of peptidase activity against the truncated S-layer protein.
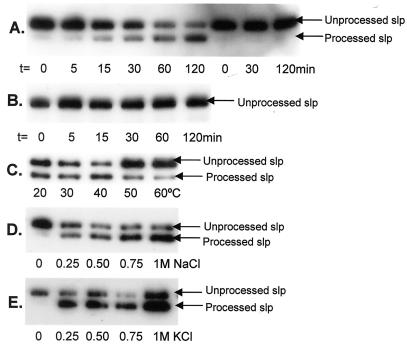
A signal peptidase assay was performed to evaluate the peptidase activity in crude M. voltae membrane preparations against the truncated S-layer protein (Fig. 2A). As revealed by Western blotting, the unprocessed S-layer band was found to decrease in intensity with time and a band with gradually increasing intensity was seen where the processed S-layer protein was expected. In control experiments where no M. voltae membranes or heat-treated (95°C for 5 min) M. voltae membranes were present (Fig. 2A and B), no evidence for processing of the S-layer was detected by Western blotting, indicating that the processing observed in the experimental set is directly attributable to the enzymatic activity present in the M. voltae membranes. These controls also indicate that the heat treatment used to prepare the S-layer-containing membranes effectively inactivated all E. coli signal peptidase I enzymatic activity against the archaeal substrate while leaving the S-layer substrate still amenable to processing.
FIG. 2.
Detection of peptidase activity against the truncated S-layer protein. (A) The truncated S-layer protein preparation was used as the substrate in the in vitro peptidase assay with M. voltae membranes as the enzyme source (lanes 1 to 6 from left). Controls with sterile distilled H2O substituting for M. voltae membranes were also performed (lanes 7 to 9 from left). Samples were taken from the reaction mixture at the various indicated time points. Peptidase activity against the His-tagged S-layer protein was detected via Western blotting with anti-His antibodies. (B) The peptidase assay described for panel A was repeated, with heat-treated (95°C for 5 min) M. voltae membranes substituting as the enzyme source. The next three panels show optimization of the signal peptidase assay. The peptidase assays were performed under standard conditions with one changed variable in each case, as indicated. Time courses up to 120 min were performed in each case, but only samples at t = 120 min, at which the reaction appeared to be complete, are shown for comparison. (C) Optimization with respect to temperature. The peptidase assay was performed at 0.4 M KCl with different incubation temperatures as indicated. (D) Optimization with respect to [NaCl]. The peptidase assay was performed at 37°C with different final [NaCl] as indicated. (E) Optimization with respect to [KCl]. The peptidase assay was performed at 37°C with different final [KCl] as indicated.
Characterization of the S-layer protein peptidase assay.
To characterize the range of in vitro working conditions for the M. voltae signal peptidase, the signal peptidase assay was run in conditions with varying [NaCl], [KCl], and temperature. Optimal activity was detected between 30 and 40°C, which was as expected since M. voltae is a mesophile (Fig. 2C). Processing of the S-layer in the in vitro assay was dependent on a low concentration of either NaCl or KCl but was then effective over a broad salt concentration range up to 1 M (Fig. 2D and E). No preference for either monovalent cation was observed.
Cloning and overexpression of the M. voltae type I signal peptidase.
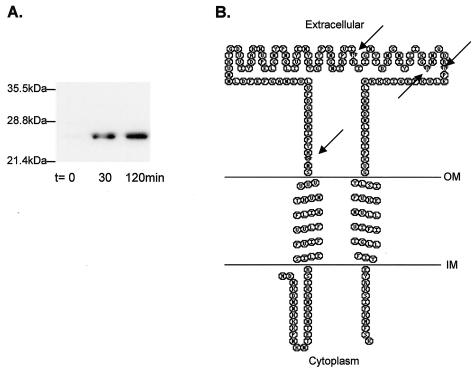
Using a heterologous probe from M. jannaschii followed by PCR, the putative type I signal peptidase gene of M. voltae was identified. It is 630 bp in length and encodes a protein of 210 amino acids with a predicted molecular mass of 24.6 kDa. When the His-tagged version of the protein was overexpressed and detected by Western blotting in E. coli, it had an apparent molecular mass of 21.5 kDa (Fig. 3A). The gene is apparently transcribed as a monocistronic mRNA, as the gene for histone B is upstream and transcribed in the opposite direction while the gene for geranylgeranyl hydrogenase is downstream and also transcribed in the opposite direction. The intergenic region between the histone B gene and the signal peptidase gene is 222 bp, while that between the signal peptidase gene and the geranylgeranyl hydrogenase gene is 83 bp. The neighboring genes of the signal peptidase are different from those in the related methanogen M. jannaschii (6). The secondary structure predicted for the M. voltae signal peptidase was examined by using the TMHMM program, version 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (17). This program predicts that there are two transmembrane segments, one located at the very N terminus (amino acids 27 to 49) and the other located at the very C terminus (amino acids 176 to 198) of the protein, with the intervening sequence (amino acids 50 to 175) predicted to be located outside the cell (Fig. 3B).
FIG. 3.
(A) Overexpression of the His-tagged M. voltae signal peptidase. The cloned M. voltae signal peptidase was overexpressed in E. coli BL21(DE3)/pLysS. Samples (1.5 ml) were taken preinduction (t = 0) and at 30 and 120 min postinduction and centrifuged, and the pellet was resuspended in 100 μl of ESB and boiled for 5 min. Ten-microliter aliquots were examined by immunoblotting using a primary anti-His antibody at a dilution of 1:20,000. (B) Secondary structure of M. voltae type I signal peptidase, as predicted by TMHMM (17). Arrows indicate the amino acids (Ser-52, His-122, Asp-142, and Asp-148) predicted to be important in activity based on sequence similarity to Sec11. This panel was generated using TOPO transmembrane protein display software (S. J. Johns and R. C. Speth) available online at http://www.sacs.ucsf.edu/TOPO/. OM, outer membrane; IM, inner membrane.
S-layer protein signal peptidase assay using cloned M. voltae signal peptidase as the enzyme source.
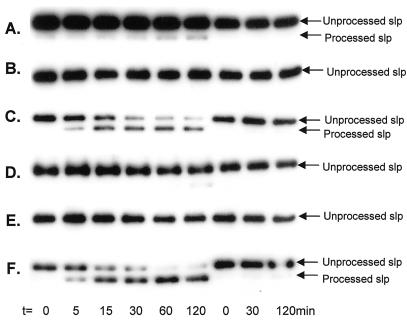
The enzymatic activity of the cloned M. voltae putative type I signal peptidase was examined using the in vitro peptidase assay. The assay of the cloned M. voltae peptidase activity was complicated by the demonstrated activity of the host E. coli cells alone against the substrate (Fig. 1). While this can be eliminated in the E. coli membranes containing the S-layer substrate by heating the membranes (Fig. 2B), this is not possible in the E. coli membranes containing the overexpressed M. voltae signal peptidase. To circumvent this problem, the in vitro assay was run in both low (50 mM) and high (1 M) NaCl concentrations, since the M. voltae enzyme showed high activity under both conditions while the E. coli enzyme has been reported to be inhibited above 0.16 M NaCl (32). Control experiments showed that membranes from E. coli BL21(DE3)/pLysS harboring pET23a alone, when used in high concentration (40 μg), possessed weak peptidase activity against the S-layer substrate at an NaCl concentration of 50 mM (Fig. 4A) but were completely inactive in assays conducted in 1 M NaCl (Fig. 4D). When approximately 100-fold-less protein (0.4 μg) was added to the assays as the enzyme source, no processing of the substrate was apparent (Fig. 4B and E). When 0.4 μg of the membrane preparation of the BL21(DE3)/pLysS strain carrying the M. voltae signal peptidase gene was added to the assays, significant enzymatic activity was observed compared to that seen with the same strain carrying the pET23a vector alone in both 50 mM and 1 M NaCl (Fig. 4C and F).
FIG. 4.
Comparison of E. coli signal peptidase and M. voltae signal peptidase activities. The signal peptidase assay was performed as described in the legend to Fig. 2A but with different enzyme sources and NaCl concentrations. The three rightmost lanes in each panel represent the corresponding controls with sterile distilled H2O added in place of membrane preparations. (A) 50 mM NaCl and membrane preparation (40 μg) from E. coli BL21(DE3)/pLysS harboring pET23 alone. (B) 50 mM NaCl and membrane preparation (0.4 μg) from E. coli BL21(DE3)/pLysS harboring pET23 alone. (C) 50 mM NaCl and membrane preparation (0.4 μg) from E. coli BL21(DE3)/pLysS harboring the cloned M. voltae signal peptidase. (D to F) Same conditions as for panels A to C, respectively, except that the NaCl concentration was 1 M. The incubation time of each sample is indicated.
DISCUSSION
Information from completed archaeal genome sequences indicates that the vital processes of protein trafficking and secretion are functionally conserved in all three domains of life (21). While some variations exist between the three domains (12), signal peptides and membrane-targeting and translocation systems, as well as putative signal peptidases, are all present in archaeal species, although protein trafficking in archaea is still poorly understood. The actual demonstration of type I signal peptidase activity attributed to any archaeal gene has not been reported until this study.
The substrate used in the in vitro assays for archaeal signal peptidase activity was the M. voltae S-layer protein, truncated to a size of 240 amino acids and His tagged at the C-terminal end, facilitating the detection of both the unprocessed and unprocessed forms with anti-His antibodies. When the cloned S-layer protein was overexpressed in E. coli, it was observed that the S-layer protein was recognized and properly processed at the native cleavage site (Fig. 1). Around the S-layer signal sequence cleavage site are amino acids which are permissible for E. coli signal peptidase I recognition and cleavage (29). When the truncated S-layer sequence was subjected to analysis by SignalP version 1.1, an online server that predicts the presence and location of signal peptide cleavage sites in amino acid sequences from different organisms (http://www.cbs.dtu.dk/services/SignalP/) (19), it was found that the signal peptide predicted for the M. voltae S-layer protein would be recognized by gram-positive and gram-negative bacteria as well as eukaryotes.
It was thus not surprising that when the truncated S-layer substrate was expressed in E. coli, both the unprocessed and the correctly processed forms were observed. However, at early time points following induction, the processed form was barely detectable in Western blots. Consequently, induction of S-layer synthesis for 30 min followed by a heating step to inactivate the native E. coli signal peptidase I activity was found to be an effective method of generating substrate for in vitro assays.
Direct demonstration of expressed signal peptidase activity from the specific gene was achieved by performing the peptidase assay with the cloned M. voltae signal peptidase as the enzyme source under conditions that are nonpermissive for the E. coli signal peptidase but would still allow M. voltae signal peptidase to efficiently function. Using 1 M NaCl to inhibit the E. coli signal peptidase, significant signal peptidase activity was demonstrated for the membrane preparation of the BL21(De3)/pLysS strain harboring the M. voltae signal peptidase gene. The fact that significant activity was observed at 1 M NaCl clearly indicated that the enzymatic activity was directly attributable to the overexpressed M. voltae signal peptidase, since the E. coli signal peptidase, when used in this concentration, was demonstrated to be completely inhibited at this salt concentration (Fig. 4B and E), in agreement with early work on the salt dependence of the bacterial enzyme (32). In fact, the E. coli signal peptidase demonstrated little to no activity even when the added membrane concentration was increased by 100-fold (40 μg) (Fig. 4A and D). The lack of difference at 50 mM versus 1 M NaCl in the extents of processing of the membrane preparation of the BL21(De3)/pLysS strain harboring the M. voltae signal peptidase gene further reinforced the negligible role of the E. coli signal peptidase in processing the S-layer protein in the assays. The lack of processing observed in low salt concentration for the E. coli control membranes was somewhat surprising given that the S-layer is processed in vivo by E. coli. However, the presence of almost entirely unprocessed S-layer at early time points following induction of synthesis might suggest that the processing in vivo, while correct, is not very efficient. The in vitro assay, in addition, is optimized for the archaeal enzyme and may not be conducive to processing by the E. coli enzyme.
There is a paucity of biochemically determined signal peptides reported for archaea in general and for M. voltae in particular. This means that the archaeal signal peptidases may have evolved to recognize certain features around the cleavage site that are not typical of bacterial signal peptides, even though in the case of the S-layer protein used here the cleavage sites for the bacterial and archaeal enzymes are the same. There are only limited data and predications of what archaeal signal peptides might look like (2, 4, 19); work on M. jannaschii has suggested that the signal peptides might be a bacterial and eukaryal hybrid.
The mechanism of action of the archaeal signal peptidase has not been reported. The lack of the conserved lysine known to be important for the activity of the bacterial enzyme has been noted and is striking (13). In the yeast Sec11p subunit of signal peptidase, site-directed mutagenesis has determined that a conserved serine, histidine, and two aspartic acids are important for activity, but this enzyme apparently lacks the catalytic lysine found in bacteria (30). Indeed, it has been suggested based on the nonessential nature of all lysines in the eukaryotic signal peptidase that the type I signal peptidase family is actually composed of two groups, one with an essential catalytic lysine and one without (30). Alignments of the M. voltae signal peptidase with the Sec11 subunit of the yeast signal peptidase indicate that the archaeal enzyme shares the four amino acids shown to be important in the eukaryotic enzyme (M. voltae signal peptidase amino acids Ser-52, His-122, Asp-142, and Asp-148 corresponding to Sec11 amino acids Ser-44, His-83, Asp-103, and Asp-109 [Fig. 5 ]). Indeed, all putative archaeal signal peptidases possess the conserved serine and histidine and the large majority also contain the two aspartic acids (Fig. 5) (13). The two essential aspartic acid residues in Sec11 have also been shown to align with aspartic acid residues in the bacterial enzyme (30), and in the bacterial case these aspartic acid residues are thought to play an important structural role in the enzyme (30). It has not yet been determined whether the yeast enzyme uses a serine/histidine dyad for catalysis or whether one of the essential aspartic acids is also catalytic, resulting in a serine/histidine/aspartic acid catalytic site. However, it is clear that eukaryotic signal peptidases employ a different mechanism of action than the serine/lysine catalytic dyad used by bacterial signal peptidases. The conservation of the four essential amino acids in both the archaeal and eukaryotic enzymes, especially the replacement of the bacterial lysine with histidine, strongly suggests that the archaeal enzyme may have a eukaryal enzyme-like mechanism of action.
FIG. 5.
Conserved regions of bacterial, eukaryal, and archaeal type I signal peptidases. Boldface type indicates amino acids important for catalysis in the bacterial and eukaryal enzymes and their conservation in the archaeal enzymes. EC, E. coli; Sec11, yeast; MV, M. voltae; MJ, M. jannaschii; MT, Methanobacterium thermoautotrophicum; MM, Methanosarcina mazei; MA, Methanosarcina acetivorans; MK, Methanothermus kandlerii; AF, Archaeoglobus fulgidus; AP, Aeropyrum pernix; H.NRC-1, Halobacterium NRC-1; PAE, Pyrobaculum aerophilum; PA, Pyrococcus abysii; PF, Pyrococcus furiosus; PH, Pyrococcus horikoshii; SS, Sulfolobus sulfataricus; ST, Sulfolobus tokodaii; TA, Thermoplasma acidophilum; TV, Thermoplasma volcanium.
The secondary structure of the M. voltae signal peptidase as determined by TMHMM version 2.0 (Fig. 3B) is similar to that of type I signal peptidases in that the majority of the protein, including the proposed catalytic-site amino acids, lies outside the cell (9). The M. voltae signal peptidase has two transmembrane segments located at the extreme N and C termini. The same pattern is observed in the signal peptidases of Thermoplasma volcanium and Sulfolobus tokodaii, as well as several nonarchaeal species (data not shown). Other archaeal signal peptidases, for example, those from Pyrococcus horikoshii, Halobacterium halobium, Sulfolobus sulfataricus, Aeropyrum pernix, Methanobacterium thermoautotrophicum, Methanosarcina acetivorans, and Methanosarcina mazei, have predicted secondary structures similar to that of Bacillus subtilis, with a single transmembrane domain located near the N terminus and the majority of the protein, including the putative catalytic amino acids, located external to the cell (data not shown). Leader peptidase I of E. coli, on the other hand, has two transmembrane segments in the N terminus of the protein but still has the majority of the molecule, including the catalytic site, located in the periplasm. Thus, as in other type I signal peptidases, it seems that the archaeal enzyme cleaves the signal peptides from preproteins on the external face of the cytoplasmic membrane, thereby releasing the translocated protein from the membrane.
Acknowledgments
This research was supported by a grant from the Natural Sciences and Engineering Research Council of Canada to K.F.J.
We thank S. Bardy, E. Chao, and N. Thomas for early contributions to this work and A. Kropinski for invaluable advice throughout the course of the work.
REFERENCES
- 1.Akca, E., H. Claus, N. Schultz, G. Karbach, B. Schlott, T. Debaerdermaeker, J. P. Declercq, and H. König. 2002. Genes and derived amino acid sequences of S-layer proteins from mesophilic, thermophilic, and extremely thermophilic methanococci. Extremophiles 6:351-358. [DOI] [PubMed] [Google Scholar]
- 2.Albers, S. V., and A. M. Driessen. 2002. Signal peptides of secreted proteins of the archaeon Sulfolobus solfataricus: a genomic survey. Arch. Microbiol. 177:209-216. [DOI] [PubMed] [Google Scholar]
- 3.Bardy, S. L., and K. F. Jarrell. 2002. FlaK of the archaeon Methanococcus maripaludis possesses preflagellin peptidase activity. FEMS Microbiol. Lett. 208:53-59. [DOI] [PubMed] [Google Scholar]
- 4.Bardy, S. L., J. Eichler, and K. F. Jarrell. 2003. Archaeal signal peptides: a comparative survey at the genome level. Protein Sci. 12:1833-1843. [DOI] [PMC free article] [PubMed]
- 5.Bayley, D. P., and K. F. Jarrell. 1999. Overexpression of Methanococcus voltae flagellin subunits in Escherichia coli and Pseudomonas aeruginosa: a source of archaeal preflagellin. J. Bacteriol. 181:4146-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 7.Correia, J. D., and K. F. Jarrell. 2000. Posttranslational processing of Methanococcus voltae preflagellin by preflagellin peptidases of M. voltae and other methanogens. J. Bacteriol. 182:855-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalbey, R. E., M. O. Lively, S. Bron, and J. M. van Dijl. 1997. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 6:1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalbey, R. E., and G. von Heijne. 1992. Signal peptidases in prokaryotes and eukaryotes—a new protease family. Trends Biochem. Sci. 17:474-478. [DOI] [PubMed] [Google Scholar]
- 10.Dharmavaram, R., P. Gillevet, and J. Konisky. 1991. Nucleotide sequence of the gene encoding the vanadate-sensitive membrane-associated ATPase of Methanococcus voltae. J. Bacteriol. 173:2131-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driessen, A. J., P. Fekkes, and J. P. van der Wolk. 1998. The Sec system. Curr. Opin. Microbiol. 1:216-222. [DOI] [PubMed] [Google Scholar]
- 12.Eichler, J. 2000. Archaeal protein translocation: crossing membranes in the third domain of life. Eur. J. Biochem. 267:3402-3412. [DOI] [PubMed] [Google Scholar]
- 13.Eichler, J. 2002. Archaeal signal peptidases from the genus Thermoplasma: structural and mechanistic hybrids of the bacterial and eukaryal enzymes. J. Mol. E. 54:411-415. [DOI] [PubMed] [Google Scholar]
- 14.Kalmokoff, M. L., K. F. Jarrell, and S. F. Koval. 1988. Isolation of flagella from the archaebacterium Methanococcus voltae by phase separation with Triton X-114. J. Bacteriol. 170:1752-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, R., S. J. Sandler, S. Goldman, H. Yokota, A. J. Clark, and S. Kim. 1998. Overexpression of archaeal proteins in Escherichia coli. Biotechnol. Lett. 20:207-210. [Google Scholar]
- 16.Koval, S. F., and K. F. Jarrell. 1987. Ultrastructure and biochemistry of the cell wall of Methanococcus voltae. J. Bacteriol. 169:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 18.Lory, S. 1994. Leader peptidases of type IV prepilins and related proteins, p. 31-48. In G. von Heijne (ed.), Signal peptidases. R. G. Landes, Austin, Tex.
- 19.Nielsen, H., S. Brunak, and G. von Heijne. 1999. Machine learning approaches for the prediction of signal peptides and other learning sorting signals. Protein Eng. 12:3-9. [DOI] [PubMed] [Google Scholar]
- 20.Paetzel, M., R. E. Dalbey, and N. C. J. Strynadka. 1998. Crystal structure of a bacterial signal peptidase in complex with a β-lactam inhibitor. Nature 396:186-190. [DOI] [PubMed] [Google Scholar]
- 21.Pohlschröder, M., W. A. Prinz, E. Hartmann, and J. Beckwith. 1997. Protein translocation in the three domains of life: variations on a theme. Cell 91:563-566. [DOI] [PubMed] [Google Scholar]
- 22.Saleh, M. T., M. Fillon, P. J. Brennan, and J. T. Belisle. 2001. Identification of putative exported/secreted proteins in prokaryotic proteomes. Gene 269:195-204. [DOI] [PubMed] [Google Scholar]
- 23.Schäffer, C., P. Graninger, and P. Messner. 2001. Prokaryotic glycosylation. Proteomics 1:248-261. [DOI] [PubMed] [Google Scholar]
- 24.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. Mcdougall, G. Himer, A. Goyal, S. Pietrokovski, G. M. Church, C. J. Daniels, J.-I. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strom, M. S., D. N. Nunn, and S. Lory. 1994. Posttranslational processing of type IV prepilin and homologs by PilD of Pseudomonas aeruginosa. Methods Enzymol. 235:527-540. [DOI] [PubMed] [Google Scholar]
- 26.Thomas, N. A., and K. F. Jarrell. 2001. Characterization of flagellum gene families of methanogenic archaea and localization of novel flagellum accessory proteins. J. Bacteriol. 183:7154-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tjalsma, H., V. P. Kontinene, Z. Pragai, H. Wu, R. Meima, G. Venema, S. Bron, M. Sarvas, and J. M. van Dijl. 1999. The role of lipoprotein processing by signal peptidase II in the gram-positive eubacterium Bacillus subtilis. J. Biol. Chem. 274:1698-1707. [DOI] [PubMed] [Google Scholar]
- 28.Towbin, M., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tschantz, W. R., and R. E. Dalbey. 1994. Bacterial leader peptidase I. Methods Enzymol. 244:285-301. [DOI] [PubMed] [Google Scholar]
- 30.van Valkenburgh, C., X. Chen, C. Mullins, H. Fang, and N. Green. 1999. The catalytic mechanism of endoplasmic reticulum signal peptidase appears to be distinct from most eubacterial signal peptidases. J. Biol. Chem. 274:11519-11525. [DOI] [PubMed] [Google Scholar]
- 31.YaDeau, J. T., C. Klein, and G. Blobel. 1991. Yeast signal peptidase contains a glycoprotein and the Sec11 gene product. Prot. Natl. Acad. Sci. USA 88:517-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zwizinski, C., T. Date, and W. Wickner. 1981. Leader peptidase is found in both the inner and outer membranes of Escherichia coli. J. Biol. Chem. 256:3593-3597. [PubMed] [Google Scholar]