Abstract
We have isolated 25 MexR mutants that retained their dimerizing ability but were unable to bind mexOP DNA. Surprisingly, 20 mutations were located in the hydrophobic core region at α4, W1, α2, α3, and β2, and only 3 were in positively charged residues. These results verified that DNA binding is mediated by distinct regions of MexR and showed the importance of the hydrophobic core region of the DNA-binding domain.
Wild-type cells of Pseudomonas aeruginosa express a low level of MexAB-OprM transporter, which provides decreased susceptibility to multiple species of antipseudomonal antibiotics (5, 7, 9). Mutations in the mexR (nalB) gene confer high resistance to the same antibiotics (10, 11, 13, 16, 18). It was reported that the MexR repressor, a member of the newly recognized marR family (1, 8), coregulates expression of the divergently transcribed mexA-mexB-oprM and mexR by binding to their shared operator-promoter region (mexOP), located between mexA and mexR (3, 13, 15). MexR, consisting of 147 amino acid residues, forms a homodimer for DNA binding. The three-dimensional structure of MexR has been solved by X-ray crystallography (6), and the structure predicted distinct regions for dimerization (N- and C-terminal helices) and DNA binding (winged helix-turn-helix motif). Since the structure of MexR cocrystallized with MexOP-DNA remains unresolved, the specific details of its interaction with DNA remain unclear. To identify residues in the MexR structure that are important for DNA binding, we isolated mutants that are unable to repress expression of the mexAB-oprM operon and show a dominant-negative phenotype relative to wild-type MexR. Mutation sites were mapped on the structure of MexR, suggesting how the key residues might impair DNA binding.
The strain used for DNA manipulation was Escherichia coli DH5α (Takara). The wild-type P. aeruginosa strain used was PAO4290 (17). Mutant strains used were TNP076 lacking mexA-mexB-oprM (17) and TNP030#10 carrying a mutation in mexR (12). Plasmids used were pET19 (Novagen) and the shuttle vector pMMB67HE (4). Cells were grown in L broth throughout this study except that MICs of antibiotics were determined in Mueller-Hinton agar at 37°C as reported previously (12). The gene encoding MexR was amplified by PCR using the primer pair 5′-GATGCCATGGGCAACTACCCCGTGAATCCCGAC-3′ and 5′-GCGCAACCGCTTGAGGATATTTGGCACCATCACCATCACCATTAAGGATCCCG-3′, and the product was subcloned into pET19 (Novagen). The DNA fragment containing the mexR gene with codons for a six-histidine tag at the carboxyl-terminal end was transferred onto the P. aeruginosa shuttle vector, pMMB67HE, yielding pMEXR-His. Next, we generated random mutations on the plasmid-borne mexR using the mutator strain XL1-Red according to the manufacturer's instructions (Stratagene). Mutants were selected for increased antibiotic resistance due to the production of inactive mexR and consequent production of a derepressed level of the MexAB-OprM efflux pump. P. aeruginosa PAO4290 cells having chromosomal native mexR+ were transformed with the mutant mexR library, and cells with the dominant-negative phenotype relative to native mexR+ were selected for resistance against 6.25 μg of aztreonam/ml and 150 μg of sulbenicillin (for a plasmid marker)/ml on Luria-Bertani agar containing 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
The function of the mutant MexR protein was assessed by determining the MICs of antibiotics. To test the influence of His6 modification, we first determined the antibiotic susceptibility of the cells harboring pMEXR-His6 (wild-type version of MexR with the His6 modification) and found that the MIC of aztreonam was 32-fold lower in the presence of IPTG than that without pMEXR, confirming that the His6 modification did not bother the repressor function. This value was comparable to the MIC of aztreonam in cells lacking MexAB-OprM. In the next experiment, we generated random mutations on the plasmid-borne mexR gene using a mutator strain (Table 1). We reasoned that if mutant MexR-His6 retained its dimer-forming capability but was unable to bind with DNA, its expression in cells with chromosomal mexR+ might result in a dominant-negative phenotype. Consequently, wild-type strains harboring such a plasmid were expected to exhibit increased MICs of antibiotics, to a level close to that for mexR-negative cells (13).
TABLE 1.
Effect of mexR mutation on MICs of antibiotics for P. aeruginosa
| Description of strain(s) | mexR allele on pMEXR-His6a | MIC of antibiotic (μg/ml)b,c
|
No. of isolates | |||
|---|---|---|---|---|---|---|
| Aztreonam
|
Ofloxacin
|
|||||
| −IPTG | +IPTG | −IPTG | +IPTG | |||
| Wild-type, | None | 6.25 | 6.25 | 0.78 | 0.78 | |
| PAO4290 | Wild-type | 3.13 | 0.2 | 0.78 | 0.1 | |
| Dominant- | L45P | 6.25 | 12.5 | 1.56 | 3.13 | 1 |
| negative | I46N | 6.25 | 25 | 1.56 | 3.13 | 1 |
| mutants | L57P | 6.25 | 25 | 1.56 | 3.13 | 1 |
| L57R | 6.25 | 25 | 1.56 | 3.13 | 1 | |
| T69I | 6.25 | 12.5 | 1.56 | 3.13 | 1 | |
| I72N | 6.25 | 25 | 1.56 | 3.13 | 9 | |
| L75P | 6.25 | 25 | 1.56 | 3.13 | 6 | |
| L75R | 6.25 | 25 | 1.56 | 3.13 | 1 | |
| R83C | 6.25 | 6.25 | 1.56 | 3.13 | 1 | |
| R91C | 6.25 | 12.5 | 1.56 | 3.13 | 2 | |
| R91H | 6.25 | 12.5 | 1.56 | 3.13 | 1 | |
| ΔmexAB-oprM | None | 0.2 | 0.2 | 0.1 | 0.1 | |
| mutant, | L57P | 0.2 | 0.39 | 0.1 | 0.1 | |
| TNP076 | R91H | 0.2 | 0.2 | 0.1 | 0.1 | |
| mexR-deficient | None | 25 | 25 | 6.25 | 6.25 | |
| mutant, | L57P | 50 | 50 | 6.25 | 6.25 | |
| TNP030#10 | R91H | 50 | 50 | 6.25 | 6.25 | |
Number refers to amino acid residue of the mutation site.
MIC was determined by the agar dilution method with Mueller-Hinton agar (Becton-Dickinson).
MexR-His6 was induced in the presence (+) or absence (−) of 1 mM IPTG.
We selected 25 transformants on aztreonam-impregnated plates, extracted the plasmid DNA, and identified the mutation by sequencing the entire mexR gene by the dideoxy chain termination method (10, 14). We found a single amino acid substitution for all 25 plasmid-borne mutant mexR genes, which occurred in eight different sites (Table 1). Upon IPTG induction, all MexR-His6 mutants showed dominant-negative behavior relative to native MexR, increasing resistance to aztreonam and ofloxacin to a level close to that in the mexR-deficient strain, except for aztreonam susceptibility of the Arg83Cys mutant (Table 1). These results suggested that the mutant proteins might have an impaired DNA-binding capability but could still form a complex with wild-type MexR protein.
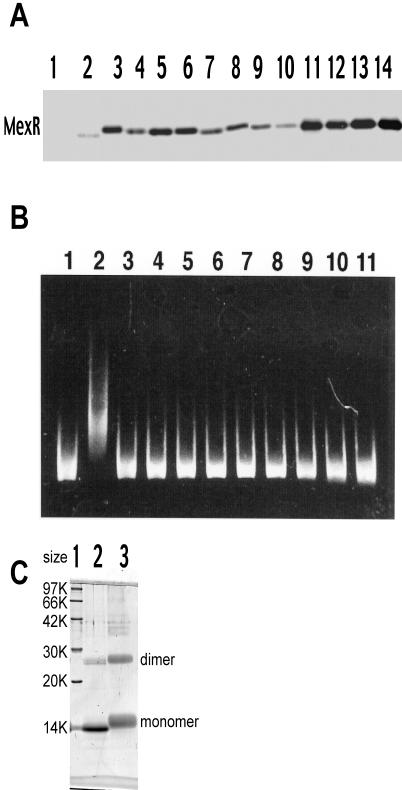
The level of the MexR-His6 proteins in cell extracts was analyzed by the Western blot method using rabbit antibody raised against MexR. Wild-type MexR-His6 showed a low level of expression (Fig. 1A, lane 2) that is most likely due to its strong repression by plasmid-encoded MexR. In fact, the MIC of aztreonam for the cells expressing plasmid-borne MexR appeared to be 32 times lower than that for the cells without plasmid-borne MexR, suggesting that transcription of mexAB-oprM was also strongly repressed by plasmid-borne MexR (Table 1). Cells harboring the plasmid with the mexR mutants produced an elevated level of MexR, probably because the transcription was freed from self-repression. Considerable variations in the level of MexR were observed. Mutant MexR-His6 was purified by His · Bind-Resin (Novagen) chromatography except for Leu45Pro, Ile46Asn, and Leu134Pro (data not shown). The gel retardation assay showed that a 0.2 μM concentration of the wild-type MexR-His6 formed a complex with the 10 nM mexOP DNA as expected (Fig. 1B, lane 2) (12). An attempt to stain MexR with Coomassie blue was unsuccessful, probably due to a limit of assay sensitivity. Specificity of MexR for mexOP DNA was confirmed in a previous study (12). On the other hand, all nine of the MexR-His6 mutants tested failed to bind to the probe DNA (Fig. 1B, lanes 3 through 11), suggesting that the mutations are likely to affect the DNA-binding domain. The possibility that the mutant MexR may have lost its dimerizing capability was ruled out by a cross-linking experiment (Fig. 1C).
FIG. 1.
Properties of the mutant MexR-His6 protein. (A) Expression of the MexR protein. PAO4290 cells harboring pMMB67HE with or without mexR-his6 were grown in the presence of 1 mM IPTG. Cell lysate was prepared by disintegrating cells in a solution of 10 mM Tris-2% Triton X-100-1 mM MgCl2 (pH 7.9) with ultrasonic oscillation and then centrifuging at 100,000 × g for 60 min. About 10 μg of protein was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12). Preparation of the mexOP probe DNA and the in vitro MexR DNA binding assay were done as described elsewhere (12). Lanes: 1, pMMB67HE without mexR; 2, pMMB67HE encoding wild-type MexR-His6; 3, Leu45Pro; 4, Ile46Asn; 5, Leu57Pro; 6, Leu57Arg; 7, Thr69Ile; 8, Ile72Asn; 9, Leu75Pro; 10, Leu75Arg; 11, Arg83Cys; 12, Arg91Cys; 13, Arg91His; 14, purified MexR-His. (B) Gel retardation assays by MexR-His6. The mexOP probe DNA (final concentration, 10 nM) was incubated with homogeneously purified MexR-His6 derived from E. coli DH5α (final concentration, 0.2 μM) in a solution of 4 mM Tris-4 mM HEPES-25 mM KCl-1 mM EDTA-25% glycerol (pH 7.9), and the 7.5-μl mixture was subjected to electrophoresis in 4% polyacrylamide gels (in 0.25× Tris-borate-EDTA). The gel was soaked in 10,000-fold-diluted CYBR Green I (BioWhittaker Molecular Applications), and DNA was visualized with UV light at 254 nm. Lanes: 1, DNA probe without MexR; 2, wild-type MexR-His; 3, Leu57Pro; 4, Leu57Arg; 5, Thr69Ile; 6, Ile72Asn; 7, Leu75Pro; 8, Leu75Arg; 9, Arg83Cys; 10, Arg91Cys; 11, Arg91His. (C) Cross-linking experiment with mutant MexR. A representative mutant MexR (Arg91His) protein, 6 μg/20 μl, was mixed with 10 μg of disuccinimydyl suberate/μl and incubated at 4°C for 30 min. To the mixture was added 10 μl of 500 mM Tris-HCl (pH 8.0) and 30 μl of solubilizer, and the mixture was heated at 100°C for 5 min. A sample containing 2 μg of MexR was applied to a 14% polyacrylamide gel. Lane 1, size markers; lane 2,Arg91His without disuccinimydyl suberate; lane 3, Arg91His with disuccinimydyl suberate. Wild-type MexR and other mutant MexR proteins showed essentially the same gel profile (data not shown). A significant amount of protein bands at the position corresponding to the dimer in the sample without disuccinimydyl suberate also appeared with wild-type MexR (not shown). Retarded mobility of disuccinimydyl suberate-treated MexR is due to bound disuccinimydyl suberates.
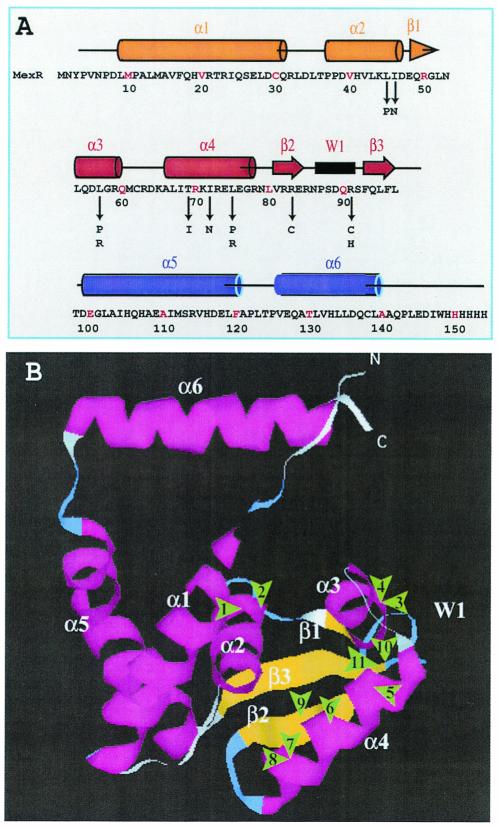
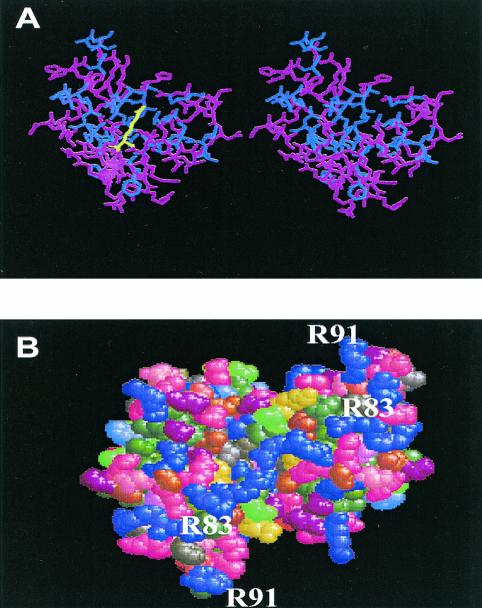
The repressor proteins, which regulate the transcription of the multidrug efflux pump, have been assigned for the MarR family (1, 11). Recently the three-dimensional structures of MexR and MarR have been solved by X-ray crystallography (2, 6). We mapped the mutations on the proposed MexR structure. Among 25 mutations, 16 were localized to three specific sites, in the α4-helix (residues 66 to 79) of MexR, suggesting the importance of the α4-helix structure for DNA binding (Fig. 2). In fact, these sites were highly conserved in the DNA-binding region. These sites faced opposite to the charged residues at the α4-helix, which might be important for maintaining the structural integrity of the DNA-binding site. Mutations may disturb the hydrophobic interaction as depicted in Fig. 3A. Changes in the α3-helix (residues 54 to 59) occurred as Leu57Arg and Leu57Pro. Amino acids in the hydrophobic region of the α3-helix, such as Leu54 and Leu57, may interact with the hydrophobic surface of the α4-helix to stabilize the DNA-binding region (Fig. 2B). Arg83 and Arg91 are located at the bottom of the MexR structure and form a line of positive charges that forms the DNA-binding site (Fig. 3B).
FIG. 2.
Mapping of mutations on the MexR structure. (A) Amino acid sequence, secondary structures, and mutation sites. Matching of amino acid sequence and the secondary structure was based on X-ray crystallographic data. Numbers indicate amino acid residues. Arrows mark mutation sites reported in this study. (B) Mutations were localized on the MexR structural model based on X-ray crystallography. Arrowheads indicate the approximate locations of the mutations. Distances are arbitrary. The numbers refer to the mutations as follows: 1, Leu45Pro; 2, Ile46Asn; 3, Leu57Pro; 4, Leu57Arg; 5, Thr69Ile; 6, Ile72Asn; 7, Leu75Pro; 8, Leu75Arg; 9, Arg83Cys; 10, Arg91His; 11, Arg91Cys.
FIG. 3.
Computer-aided visualization of mutation. (A) Stereo view molecular model of the Leu75Arg mutation as a representative of mutation in the hydrophobic core region. The mutation was inserted on the crystallographic structure of MexR by Program O for Windows NT, version 7.0. Figures were drawn by using Program Ras Win Molecular Graphics Windows, version 2.6-ucb. Hydrophobic amino acid residues and other residues are shown by blue and red, respectively. Mutation sites are given in yellow. (B) Surface potential of the bottom part of the MexR dimer. Arg residues are emphasized by a purple color in the space-fill model. Distribution of the electrostatic potential was drawn by the Swiss-pdb Viewer, version 3.7. The locations of Arg83 and Arg91 are shown in the region containing the α4, β2, W1, and β3 structures.
In this study, we obtained several classes of MexR proteins with impaired DNA-binding capability by selecting for the dominant-negative phenotype. Mutations were mapped on a limited region of MexR, which might be important in forming the DNA-binding domain.
Acknowledgments
We thank D. Lim and N. C. J. Strynadka of the University of British Columbia for providing raw X-ray crystallographic data of MexR.
This study was supported by grants from the Ministry of Education, Culture, Sport, Science and Technology, the Japan Society for Promotion of Science, the Ministry of Health, Labor and Welfare, Tokai University (Project Research Grant), and Tokai University School of Medicine (Research Project Grant).
REFERENCES
- 1.Alekshun, M. N., Y. S. Kim, and S. B. Levy. 2000. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 35:1394-1404. [DOI] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 3.Evans, K., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 5.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim, D., K. Poole, and N. C. J. Strynadka. 2002. Crystal structure of the mexR repressor of the mexRAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J. Biol. Chem. 277:29253-29259. [DOI] [PubMed] [Google Scholar]
- 7.Moreshed, S. R. M., Y. Lei, H. Yoneyama, and T. Nakae. 1995. Expression of genes associated with antibiotic extrusion in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 210:356-362. [DOI] [PubMed] [Google Scholar]
- 8.Perez-Rueda, E., and J. Collado-Vides. 2001. Common history at the origin of the position-function correlation in transcriptional regulators in archaea and bacteria. J. Mol. Evol. 53:172-179. [DOI] [PubMed] [Google Scholar]
- 9.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. E. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rella, M., and D. Haas. 1982. Resistance of Pseudomonas aeruginosa PAO to nalidixic acid and low levels of beta-lactam antibiotics: mapping of chromosomal genes. Antimicrob. Agents Chemother. 22:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saito, K., S. Eda, H. Maseda, and T. Nakae. 2001. Molecular mechanism of MexR-mediated regulation of MexAB-OprM efflux pump expression in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 195:23-28. [DOI] [PubMed] [Google Scholar]
- 13.Saito, K., H. Yoneyama, and T. Nakae. 1999. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 179:67-72. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y..
- 15.Sanchez, P., F. Rojo, and J. L. Martinez. 2002. Transcriptional regulation of mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 207:63-68. [DOI] [PubMed] [Google Scholar]
- 16.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoneyama, H., A. Ocaktan, M. Tsuda, and T. Nakae. 1997. The role of mex-gene products in antibiotic extrusion in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 233:611-618. [DOI] [PubMed] [Google Scholar]
- 18.Ziha-Zarifi, L., C. Llanes, T. Koehler, J.-C. Pechere, and B. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:283-291. [DOI] [PMC free article] [PubMed] [Google Scholar]