Abstract
Early screens for conditional lethal mutations that affected rRNA expression in Escherichia coli identified temperature-sensitive fda mutants (fda encodes the glycolytic enzyme fructose-1,6-diphosphate aldolase). It was shown that these fda(Ts) mutants were severely impaired in rRNA synthesis upon shift to the restrictive temperature, although the mechanism of inhibition was never determined. Here, we bring resolution to this long-standing question by showing that changes in the concentrations of guanosine 5′-diphosphate 3′-diphosphate and initiating nucleoside triphosphates can account for the previously observed effects of fda mutations on rRNA transcription.
Alleles of the gene for fructose-1,6-diphosphate (FDP) aldolase (fda) were isolated in two independent screens for temperature-sensitive mutants affecting macromolecule biosynthesis in Escherichia coli (3, 9). Both fda alleles (h8 [3] and ts8 [9]) caused severe decreases in the synthesis rate of rRNA at the restrictive temperature in cultures grown in the presence of glucose (or other hexoses). Mutations in fda leading to similar phenotypes have also been identified in Bacillus subtilis (11, 22). Originally, the ts8 allele was mapped to another locus (9), but subsequent work definitively mapped the mutation to fda and showed that the temperature-sensitive phenotype resulted from a T-to-G mutation at nucleotide position 901 of the fda coding sequence, resulting in a Val-to-Gly substitution at amino acid 301 (20). Singer and coworkers showed that both ts8 and h8 specifically decreased transcription initiation at rrn P1 promoters (21). The concentration of FDP, the substrate for aldolase, increased sharply in the mutant after temperature shift (3), but it was not reported that FDP inhibited rRNA promoter function. Furthermore, in agreement with the original report (9), Singer and coworkers concluded that effects of the ts8 mutation on guanosine 5′-diphosphate 3′-diphosphate (ppGpp), a direct negative regulator of rrn promoters (1), were not sufficient to account for the effects of the fda mutations on rRNA transcription (1, 4, 21). Therefore, the mechanism of inhibition has remained unclear.
rRNA promoter strength derives in large part from binding of the transcription factor Fis and the α subunit of RNA polymerase to the region upstream of the −35 element in rrn P1 promoters (6). However, neither the binding sites for Fis nor the full α binding site were required for the effects of the fda mutation on rrnB P1 promoter activity (21), implicating the rrn P1 core promoter as the regulatory target of fda. Since rrn P1 core promoters are regulated directly by the concentration of their initiating nucleoside triphosphate (iNTP) as well as by ppGpp (5, 18), here we tested the hypothesis that changes in the concentrations of ppGpp and the iNTP together account for the effects of fda mutations on rRNA transcription.
The ts8 fda mutation was moved to an otherwise wild-type background (strain VH1000 [5]) by transduction with phage P1 using a linked Tn10 (zdg-210::Tn10 [20]) and selecting for tetracycline resistance. Previous reports had suggested that ts8 strains were not competent hosts for growth of P1vir (20); however, we were able to make P1vir lysates on a C600 ts8 strain (a gift from M. Singer, University of California—Davis) using standard protocols (10). We constructed fda and wild-type strains carrying a λ prophage containing an rrn P1 promoter synthesizing a short-lived lacZ mRNA for use as a reporter of rRNA transcription (16). These strains were then transformed with a multicopy plasmid expressing λ cI repressor (pFL122, a gift from M. Filutowicz and J. Wild, University of Wisconsin—Madison) in order to avoid induction of the temperature-sensitive lysogens (cI857) following a temperature shift.
We measured RNA synthesis rates from two promoters, rrnB P1 and rrnB P1(dis), by primer extension (13). rrnB P1 is a well-characterized promoter that forms a short-lived open complex and consequently is regulated negatively by increases in the ppGpp concentration and positively by increases in the ATP concentration (1, 5, 18). rrnB P1(dis), a variant of rrnB P1 with a 3-bp change in the discriminator region (the sequence between the −10 hexamer and the transcription start site), served as a control. rrnB P1(dis) makes exactly the same transcript as rrnB P1, but it forms a much longer-lived open complex, resulting in relative insensitivity to changes in both the ppGpp and ATP concentrations (1, 2, 8, 18). We also measured the concentrations of ppGpp and ATP by formic acid extraction and thin-layer chromatography (7).
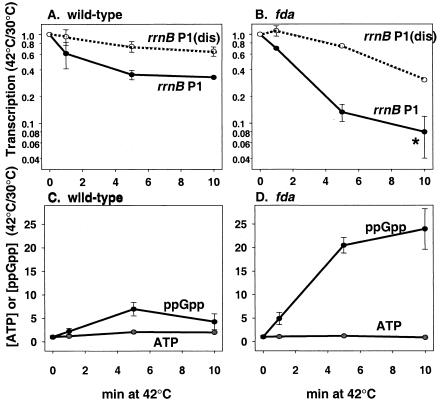
In the wild-type strain, the activities of both the rrnB P1 and rrnB P1(dis) promoters decreased slightly after a shift from 30 to 42°C (approximately three- and approximately twofold, respectively) (Fig. 1A), consistent with previous reports describing the effects of high temperature on rRNA transcription (17). In contrast, rrnB P1 promoter activity dropped sharply (>12-fold) in the fda strain after a shift to the restrictive temperature, much more than the decrease in rrnB P1(dis) promoter activity (Fig. 1B). The RNA synthesized by rrnB P1 was essentially undetectable by 10 min after the shift (Fig. 1B, asterisk); therefore, the ∼12-fold decrease may underestimate the effect of the fda mutation on rrnB P1 activity.
FIG. 1.
Effects of the fda(Ts) mutation on transcription and on ppGpp and iNTP concentrations. rrnB P1 and rrnB P1(dis) promoter activities were measured by primer extension (13) in wild-type (A) and ts8 (B) strains before and as a function of time after a shift from 30 to 42°C. The promoters contained rrnB P1 DNA sequences from −66 to + 9 with respect to the transcription start site (+1) and synthesized an unstable RNA (18). Cultures (rrnB P1, RLG6256; rrnB P1(dis), RLG6257; rrnB P1 fda, RLG6258; rrnB P1(dis) fda, RLG6259) were grown in morpholinepropanesulfonic acid (MOPS) minimal medium (15) supplemented with 0.4% glucose, 0.4% Casamino Acids, 40 μg of Trp/ml, 5 μg of thiamine/ml, 1 mM KH2PO4, and 100 μg of ampicillin/ml. The asterisk in panel B indicates that promoter activity at this time point was at or below background. ATP and iNTP concentrations were extracted with formic acid and measured by thin-layer chromatography as described previously (18) from wild-type (C) and ts8 (D) strains grown in parallel with the cultures in panels A and B, except 20 μCi of KH232PO4/ml was added to the medium. Values were normalized to those at time zero (just before the temperature shift). Each point represents the average of at least two measurements from independent cultures. Error is indicated.
Concurrent with the drop in rrnB P1 promoter activity in the fda mutant, there was a large (∼25-fold) increase in the concentration of ppGpp at the restrictive temperature, much larger than the increase in ppGpp in the wild-type strain (Fig. 1C and D). We did not detect a significant change in ATP concentration in either the mutant or wild-type strain. Thus, contrary to expectations from previous investigations (9, 21), inactivation of aldolase led to induction of ppGpp, accounting for the large decrease in rrnB P1 activity. The mechanisms responsible for induction of ppGpp after inactivation of aldolase, glucose starvation (4), or inhibition of glucose uptake by α-methyl-glucoside (4, 14) remain to be determined.
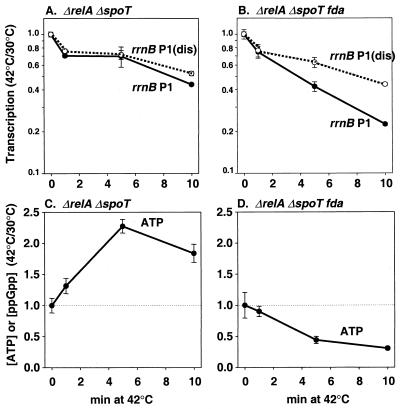
Previous reports indicated that rrnB P1 activity was still inhibited by the fda mutation in cells lacking ppGpp (ΔrelA ΔspoT) (21). Since rrn P1 promoters can also be regulated by the iNTP concentration (14, 18), we asked whether changes in ATP concentration could account for the ppGpp-independent effect of fda. After a shift to 42°C, the activities of the rrnB P1 and rrnB P1(dis) promoters decreased slightly and to the same extent in a strain that cannot make ppGpp (Fig. 2A). In a ΔrelA ΔspoT fda strain, however, the activity of rrnB P1 decreased approximately twofold more than that of the control promoter (Fig. 2B), in agreement with the conclusion reached by Singer and coworkers (21). Notably, the ATP concentration dropped approximately threefold in the ΔrelA ΔspoT fda strain (Fig. 2D), whereas the ATP concentration increased approximately twofold at 42°C in the ΔrelA ΔspoT strain (Fig. 2C). The simplest interpretation of these data is that ppGpp inhibits rRNA synthesis at the restrictive temperature in an fda strain, but in a strain without ppGpp (ΔrelA ΔspoT fda), a decrease in the ATP concentration accounts for the residual inhibition of rRNA transcription. Thus, we suggest that changes in the concentrations of ppGpp and iNTPs are sufficient to account for the observed effects of the fda mutation on rRNA transcription (although our data do not rule out the participation of an additional mechanism[s]). These data reinforce the connections between central metabolism and the concentrations of iNTPs and ppGpp (14), although the mechanisms responsible for some of these connections remain to be determined.
FIG. 2.
Effects of fda(Ts) on promoter activity and ATP concentrations in ΔrelA ΔspoT strains. rrnB P1 and rrnB P1(dis) activities were measured by primer extension in ΔrelA ΔspoT (A) and ΔrelA ΔspoT (B) fda strains as detailed in the legend to Fig. 1. Strains described in the legend to Fig. 1 were sequentially transduced to ΔrelA ΔspoT as described previously (18) and were grown as indicated in the legend, except that the medium contained 1.0% Casamino Acids. ATP concentrations were measured from ΔrelA ΔspoT (C) and ΔrelA ΔspoT fda (D) strains grown as indicated in the legend for panels A and B except 20 μCi of KH232PO4/ml was included in the medium. Each point represents the average of at least two measurements from independent cultures. Error is indicated.
A recent report from Morita and coworkers demonstrated that there is an RNase E-dependent decrease in mRNA stability when certain glycolytic enzymes (including fructose-1,6-diphosphate aldolase) (12) are disrupted. This decrease in message stability might contribute to the nonspecific decrease in promoter activity [i.e., that observed on rrnB P1(dis)] in the fda strain at 42°C.
Although ppGpp and iNTP concentrations have specific, nonredundant roles in the control of rRNA transcription (14), the results reported here illustrate how a decrease in the iNTP concentration can compensate for impaired ability to induce ppGpp. We have observed several other examples where changes in the concentrations of both of these small-molecule regulators compensate for defects in rRNA synthesis: after disruption of the rrn antitermination machinery by mutations in nusB, after inhibition of UP element function by mutations in rpoA, and after deletion of the transcriptional activator fis (19). Taken together, our findings support the model that ppGpp and iNTPs form two separate but complementary homeostatic control loops, fine-tuning rRNA synthesis to the growth conditions.
Acknowledgments
We thank Wilma Ross, Tamas Gaal, Karen Wassarman, and Pierre Rouviere for helpful advice and Mitchell Singer and Jadwiga Wild for strains.
This work was supported by a grant from the National Institutes of Health (GM37048 to R.L.G.) and by predoctoral training grants from the National Science Foundation, National Institutes of Health, and Wisconsin Alumni Research Foundation (to D.A.S.).
REFERENCES
- 1.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. [DOI] [PubMed] [Google Scholar]
- 2.Barker, M. M., and R. L. Gourse. 2001. Regulation of rRNA transcription correlates with nucleoside triphosphate sensing. J. Bacteriol. 183:6315-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bock, A., and F. C. Neidhardt. 1966. Properties of a mutant Escherichia coli with a temperature-sensitive fructose-1,6-diphosphate aldolase activity. J. Bacteriol. 92:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cashel, M., D. R. Gentry, V. H. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 5.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278:2092-2097. [DOI] [PubMed] [Google Scholar]
- 6.Hirvonen, C. A., W. Ross, C. E. Wozniak, E. Marasco, J. R. Anthony, S. E. Aiyar, V. Newburn, and R. L. Gourse. 2001. Contributions of UP elements and the transcription factor FIS to expression from the seven rrn P1 promoters in Escherichia coli. J. Bacteriol. 183:6305-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen, K. F., U. Houlberg, and P. Nygaard. 1979. Thin-layer chromatographic methods to isolate 32P-labeled 5-phosphoribosyl-alpha-1-pyrophosphate (PRPP): determination of cellular PRPP pools and assay of PRPP synthetase activity. Anal. Biochem. 98:254-263. [DOI] [PubMed] [Google Scholar]
- 8.Josaitis, C. A., T. Gaal, and R. L. Gourse. 1995. Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc. Natl. Acad. Sci. USA 92:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liebke, H. H., and J. F. Speyer. 1983. A new gene in E. coli RNA synthesis. Mol. Gen. Genet. 189:314-320. [DOI] [PubMed] [Google Scholar]
- 10.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Mitchell, C., P. W. Morris, L. Lum, G. Spiegelman, and J. C. Vary. 1992. The amino acid sequence of a Bacillus subtilis phosphoprotein that matches an orfY-tsr coding sequence. Mol. Microbiol. 6:1345-1349. [DOI] [PubMed] [Google Scholar]
- 12.Morita, T., W. El-Kazzaz, Y. Tanaka, T. Inada, and H. Aiba. 2003. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J. Biol. Chem. 278:15608-15614. [DOI] [PubMed] [Google Scholar]
- 13.Murray, H. D., J. A. Appleman, and R. L. Gourse. 2003. Regulation of the Escherichia coli rrnB P2 promoter. J. Bacteriol. 185:28-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray, H. D., D. A. Schneider, and R. L. Gourse. 2003. Control of rRNA expression by small molecules is dynamic and non-redundant. Mol. Cell 12:125-134. [DOI] [PubMed] [Google Scholar]
- 15.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao, L., W. Ross, J. A. Appleman, T. Gaal, S. Leirmo, P. J. Schlax, M. T. Record, Jr., and R. L. Gourse. 1994. Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J. Mol. Biol. 235:1421-1435. [DOI] [PubMed] [Google Scholar]
- 17.Ryals, J., R. Little, and H. Bremer. 1982. Control of RNA synthesis in Escherichia coli after a shift to higher temperature. J. Bacteriol. 151:1425-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider, D. A., T. Gaal, and R. L. Gourse. 2002. NTP-sensing by rRNA promoters in Escherichia coli is direct. Proc. Natl. Acad. Sci. USA 99:8602-8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider, D. A., and R. L. Gourse. 2003. Changes in Escherichia coli rRNA promoter activity correlate with changes in initiating nucleoside triphosphate and guanosine 5′-diphosphate 3′-diphosphate concentrations after induction of feedback control of ribosome synthesis. J. Bacteriol. 185:6185-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singer, M., P. Rossmiessl, B. M. Cali, H. Liebke, and C. A. Gross. 1991. The Escherichia coli ts8 mutation is an allele of fda, the gene encoding fructose-1,6-diphosphate aldolase. J. Bacteriol. 173:6242-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singer, M., W. A. Walter, B. M. Cali, P. Rouviere, H. H. Liebke, R. L. Gourse, and C. A. Gross. 1991. Physiological effects of the fructose-1,6-diphosphate aldolase ts8 mutation on stable RNA synthesis in Escherichia coli. J. Bacteriol. 173:6249-6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trach, K., J. W. Chapman, P. Piggot, D. LeCoq, and J. A. Hoch. 1988. Complete sequence and transcriptional analysis of the spo0F region of the Bacillus subtilis chromosome. J. Bacteriol. 170:4194-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]