Abstract
The response regulator VirR and its cognate sensor histidine kinase, VirS, are responsible for toxin gene regulation in the human pathogen Clostridium perfringens. The C-terminal domain of VirR (VirRc) contains the functional FxRxHrS motif, which is involved in DNA binding and is conserved in many regulatory proteins. VirRc was cloned, purified, and shown by in vivo and in vitro studies to comprise an independent DNA binding domain. Random and site-directed mutagenesis was used to identify further amino acids that were required for the functional integrity of the protein. Random mutagenesis identified a unique residue, Met-172, that was required for biological function. Site-directed mutagenesis of the SKHR motif (amino acids 216 to 219) revealed that these residues were also required for biological activity. Analysis of the mutated proteins indicated that they were unable to bind to the DNA target with the same efficiency as the wild-type protein.
The human pathogen Clostridium perfringens is the major causative agent of gas gangrene. The pathogenesis of this disease is characterized by the involvement of potent extracellular toxins that are responsible for the extensive tissue destruction and necrosis seen in classic gangrene cases (10, 12). The alpha-toxin (phospholipase C) is the major extracellular enzyme produced by C. perfringens and has been shown to be an essential virulence factor (2). The other toxin shown to be involved in the pathogenesis of this disease is perfringolysin O, a cholesterol-dependent cytolysin that has the ability to lyse red blood cells (13). In C. perfringens, extracellular toxin production and virulence is controlled by the VirSR two-component signal transduction system. In this system, VirS is the sensor histidine kinase and VirR is its cognate response regulator (4, 7, 11).
The 236-amino-acid VirR protein has a two-domain structure. The N-terminal receiver domain has strong sequence similarity to the classical phosphoacceptor domain of response regulators (7). The C-terminal domain (VirRc) has been identified as the DNA binding domain (8) and belongs to the newly described LytTR family of transcriptional regulators (9). The LytTR domain is a novel DNA binding domain identified in silico by analysis of bacterial genome sequences (8, 9). Members of this family of regulators can be identified by the presence of a highly conserved FxRxHrS motif within the C-terminal domain (8). In VirR, the FxRxHrS motif was shown to have a functional role in the ability of the protein to bind to its target VirR boxes (8).
Comparative sequence analysis of the LytTR family of proteins identified further regions that show charge conservation, in particular a cluster of Lys and Arg residues closer to the C terminus (8, 9). We have designated this region the SKHR motif, and in this paper we show that it is also essential for the in vivo function of the VirR protein.
VirRc is an independent DNA binding domain.
To test the ability of VirRc to function as an independent DNA binding protein, a deletion derivative was constructed using PCR. The resultant gene, virRΔ2-125, was cloned into pJIR2233 (Table 1), behind the native virR promoter. This plasmid, pJIR2259 (Table 1), was used to transform the virR mutant TS133 (11) to express chloramphenicol resistance. The ability of the truncated gene to complement the chromosomal virR mutation was assessed by a hemolysin assay that measured the level of perfringolysin O activity in the culture supernatant. A shuttle plasmid carrying the wild-type virR gene is capable of restoring perfringolysin O activity to wild-type levels (Table 2). Analysis of the truncated construct in vivo indicated that VirRc was also capable of activating perfringolysin O expression to wild-type levels (Table 2).
TABLE 1.
Relevant strains and plasmids
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| C. perfringens strains | ||
| JIR325 | Strain 13 Nalr Rifr | 7 |
| TS133 | JIR325 virR::Tcr | 11 |
| Plasmids | ||
| pET-22b(+) | Expression vector; C-terminal six-His tag; Apr; 5.5 kb | Novagen |
| pJIR750 | E. coli-C. perfringens shuttle vector; Cmr | 3 |
| pJIR1821 | pJIR1546 with CCA→TAG mutation in VirR boxes 1 and 2 | 5 |
| pJIR1877 | pJIR1859 ΩvirR+(BamHI) (SDM template) | 8 |
| pJIR1897 | pJIR750 ΩvirR+(BamHI) (random mutagenesis template) | 8 |
| pJIR2233 | pJIR750 Ω(BamHI/HindIII; pJIR1877, 0.214 kb, contains virR/S operon promoter) | Recombinant |
| pJIR2253 | pJIR1897 virR M172V | Random mutagenesis |
| pJIR2254 | pJIR1897 virR F184S | Random mutagenesis |
| pJIR2259 | pJIR2233 Ω(BamHI/EcoRI; pJIR2235, 0.3 kb, contains virRΔ2-125 gene) | Recombinant |
| pJIR2491 | pJIR1877 virR S216C | Site-directed mutagenesis |
| pJIR2492 | pJIR1877 virR K217E | Site-directed mutagenesis |
| pJIR2493 | pJIR1877 virR H218F | Site-directed mutagenesis |
| pJIR2494 | pJIR1877 virR R219E | Site-directed mutagenesis |
SDM, site-directed mutagenesis.
TABLE 2.
Effects of virR mutations on perfringolysin O production
| Strain or plasmid | Description or base change(s)a | Amino acid changeb | PfoA titerc |
|---|---|---|---|
| Strains | |||
| JIR325 | Wild type | 8.4 ± 0.9 | |
| TS133 | Strain with chromosomal virR mutation | <1.0 | |
| TS133(pJIR750) | Negative control | <1.0 | |
| TS133(pJIR1897) | Positive control | 8.6 ± 0.9 | |
| Plasmids | |||
| pJIR2259 | virRΔ2-125 | 7.5 ± 0.2 | |
| pJIR2253 | ATG to gTG | M172V | 2.5 ± 0.1d |
| pJIR2254 | TTT to TcT | F184S | <1.0 |
| pJIR2491 | GAA to Gct | S216C | <1.0 |
| pJIR2492 | CTT to gcT | K217E | <1.0 |
| pJIR2493 | CTT to aTT | H218F | <1.0 |
| pJIR2494 | CTT to gTa | R219E | <1.0 |
Lowercase letters in nucleotide sequences represent substituted bases.
All mutant plasmids were obtained by site-directed mutagenesis of pJIR1877 (Table 1) with the exception of pJIR2253 and pJIR2254, which were obtained by random mutagenesis.
PfoA titer, log2 (perfringolysin O titer) obtained when culture supernatants were assayed for hemolytic activity. All values are the means of activity levels obtained in duplicate assays with at least three independent cultures.
P is <0.01 by Student's t test compared to positive control, TS133(pJIR1897).
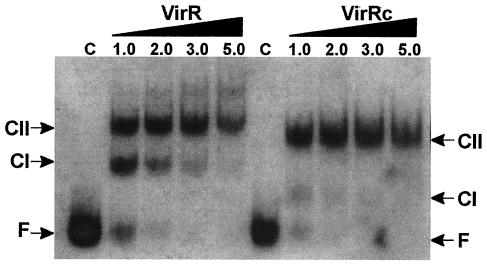
To confirm that VirRc can act as an independent DNA binding protein in vitro, the truncated gene was cloned into the expression vector pET-22b(+). Overexpression and subsequent purification of the 14-kDa protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). In gel mobility shift assays, the wild-type VirR protein binds to a 183-bp DNA fragment that encompasses the two VirR boxes upstream of the pfoA gene (5). After electrophoresis, multiple VirR-DNA complexes are observed. Previous work has shown that the first complex (CI) is formed when either DNA box is bound by VirR and that the second complex (CII) is formed when both VirR boxes are bound (5). As the concentration of protein is increased, higher-order complexes are also observed. The functional protein-DNA conformer is CII (J. K. Cheung and J. I. Rood, unpublished data). Comparative gel mobility shift analysis of VirR and VirRc revealed similar banding patterns; however, little CI formation was observed in the VirRc reactions (Fig. 1). It appears that VirRc may have a higher affinity for the target DNA than does the wild-type protein, with CII being formed at lower concentrations. Interestingly, there is no evidence of the formation of higher-order complexes in the VirRc reactions (Fig. 1). Gel shift reactions performed using a DNA target that has mutations in both VirR boxes (pJIR1821) (Table 1), which had previously been shown to be altered in its ability to bind to the wild-type VirR protein (5), confirmed that the binding observed in the VirRc reactions was specific (data not shown).
FIG. 1.
Comparative gel mobility shift analysis of VirR and VirRc. The target DNA, which carried wild-type VirR boxes (5), was digoxigenin labeled and incubated with purified wild-type VirR or VirRc as indicated. The control reaction mixture (C) was incubated with 2.0 μg of bovine serum albumin. The amount (in picomoles) of purified protein added to each reaction mixture is shown at the top of the panel. Arrows indicate the positions of CI, CII, and free or unbound DNA (F).
Random mutagenesis reveals that Met-172 is required for biological function.
A random mutagenesis approach was used to identify other residues within VirR that were of structural or functional importance. The plasmid pJIR1897, which carries the virR gene on an E. coli-C. perfringens shuttle vector, was passaged through the XL-1 Red mutator strain (Stratagene), and nonhemolytic mutants were detected by their inability to complement the virR mutant TS133. Two independently derived mutants with single point mutations within the virR gene were isolated. The virR F184S mutant was nonfunctional (Table 2), which was in agreement with findings of previous studies (8) that revealed a functional requirement at residue 184 for an amino acid with an aromatic side chain. The other mutation was in a residue that was upstream of the FxRxHrS motif (amino acids 184 to 190). TS133 derivatives carrying the virR M172V gene could restore perfringolysin O activity only to a titer of 2.5 (Table 2) in comparison to the titer of 8.6 in the wild type, indicating that the mutant protein had limited biological activity.
To validate these results, we confirmed that the TS133 strains carrying the mutated plasmids still produced stable VirR protein. Western blots carried out using polyclonal VirR antisera showed that both strains produced wild-type amounts of an immunoreactive protein of the correct size (data not shown). When plasmids carrying the mutated virR genes were introduced into the parent strain, JIR325, the hemolytic ability of the resultant strain was significantly reduced (data not shown), indicating that the mutant proteins were still capable of the protein-protein interactions that are assumed to be essential for VirR function.
The SKHR motif is essential for the function of VirR.
Previous PSI-BLAST (1) searches identified a small, charged, highly conserved region at the C terminus of VirR (8). This pocket of basic amino acids was designated the SKHR motif. To determine the importance of these residues for VirR function, site-directed mutagenesis was performed by the unique site elimination method (6) as described previously (8) using pJIR1877 (Table 1) as the mutagenesis template. In an attempt to introduce minimal secondary structure perturbations, the mutations were designed to alter the charge of particular residues by replacing them with amino acids with side chains of similar sizes and shapes. Accordingly, we constructed mutants that encoded S216C, K217E, H218F, and R219E derivatives of VirR (Table 2). The mutant genes were subcloned into the E. coli-C. perfringens shuttle vector pJIR750 and used to transform TS133 to express chloramphenicol resistance. Determination of the perfringolysin O activity of the resultant derivatives showed that each of the individual residues of the SKHR motif was required for in vivo function. There was no detectable perfringolysin O activity in any of the strains carrying the mutated virR genes (Table 2). Note that extraction and resequencing of the plasmids confirmed that no other changes had been introduced. As before, all of the strains assayed produced an immunoreactive protein of the correct size (data not shown) and no significant differences in levels of VirR production were observed. The negative dominant properties described for the random mutants were also observed with the SKHR mutants (data not shown).
The M172V and SKHR mutant proteins are altered in their ability to bind DNA.
To analyze why the M172V and SKHR mutant proteins were unable to activate pfoA transcription, the mutated genes were PCR amplified and cloned into pET-22b(+). The wild-type VirR protein and mutant VirR proteins M172V, S216C, K217E, H218F, and R219E were successfully purified. The gene encoding VirR F184S was also overexpressed, but the protein was not successfully purified due to the formation of inclusion bodies. The purified proteins all had the expected molecular size of 29 kDa, as observed by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and on a Superdex S200 column eluted as single peaks corresponding to an apparent molecular size of 29 kDa, indicating that the proteins were monomers in solution (data not shown). To confirm their structural integrity, we compared the circular dichroism spectra of the wild-type and mutant proteins. With each peak fraction collected in the gel filtration study as the spectral sample, analysis of far UV spectra from 200 to 250 nm showed that all of the proteins were folded (data not shown).
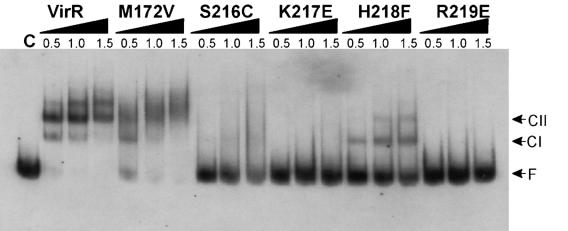
The DNA binding ability of the purified proteins was determined by gel mobility shift analysis. With wild-type VirR protein, CII was the predominant form, with shifts in the levels of CI in favor of those of CII as the VirR concentration increased (Fig. 2). The random mutant M172V had a similar ability to shift the DNA target but did not appear to form the same sharply defined major complexes as the wild-type protein. The smeared bands resulting from the M172V reactions suggested that the complexes were probably formed, with the protein binding to its target, but that the binding interactions were less stable and that the complexes were dissociating during electrophoresis. This result supported the earlier in vivo results that showed that the strain harboring virR M172V produced a low perfringolysin O titer (Table 2). We conclude that this reduced activity is likely to be the result of an unstable or improperly formed protein-DNA conformer.
FIG. 2.
Gel mobility shift analysis of mutant VirR proteins. The target DNA, which carried wild-type VirR boxes (5), was digoxigenin labeled and incubated with purified wild-type VirR or the mutant proteins as indicated. The control reaction mixture (C) was incubated with 2.0 μg of bovine serum albumin. The amount (in micrograms) of purified protein added to each reaction mixture is shown at the top of the panel. Arrows indicate the positions of CI, CII, and free or unbound DNA (F).
The SKHR motif mutants were unable to bind the target DNA with the same affinity as the wild-type protein (Fig. 2). The S216C protein may be able to form small amounts of CI, but even when 1.5 μg of protein was added to the reaction mixture, approximately 90% of the DNA remained unbound. There was no evidence of the formation of CII or higher-order complexes in any of the S216C reactions. From these data, it appears that Ser-216 has a role in the integrity of the DNA binding domain. The two charged residues, Lys-217 and Arg-219, also appear to be required for DNA binding. The mutated proteins K217E and R219E were both incapable of binding the target DNA (Fig. 2). Finally, the results showed that the H218F protein was still capable of forming CI and limited amounts of CII. This result suggested that VirR mutant H218F should be capable of activating pfoA transcription, albeit at a reduced level compared to the wild type. However, in vivo, this VirR derivative behaved in the same manner as the other SKHR mutants, conferring no detectable perfringolysin O activity (Table 2).
Role of M172 and the SKHR motif in VirRc.
Structural comparisons have yielded useful information about the C-terminal domain of the LytTR or FxRxHrS family of transcriptional regulators (8, 9). However, none of these proteins have been crystallized, and structural models of the C-terminal domains show no significant similarity to those of any subfamily of characterized response regulators. Previous fold recognition studies identified the Saccharomyces cerevisiae transcriptional regulator Mbp1 as the top-scoring fold from a prediction server, and the highly conserved FxRxHrS motif aligned with the DNA binding residues KxKR on the recognition helix of Mbp1 (8).
Alignment of the FxRxHrS or LytTR proteins (8, 9) revealed that the M172 residue of VirR is generally a conserved leucine residue in the other members of this family and appears to be located at the beginning of the first predicted α-helix (9). It appears from the gel shift analysis of the VirR M172V protein that this residue has a role in the stabilization of the protein-DNA complex. This conclusion lends support to the hypothesis that the first two α-helices of the LytTR domain form a variant helix-turn-helix, with the M172 residue located at the beginning of the first of these helices, the stabilization helix. Determination of the precise role of M172 must await the elucidation of the structure of VirR.
This study has highlighted the functional importance of a second conserved sequence motif, S[RK][RK][RKHY], in the LytTR or FxRxHrS family. We have shown that the four residues of the SKHR motif are each required for the biological function of VirR. At Ser-216, the replacement of the hydroxyl side chain with a closely related sulfhydryl group completely abolished biological activity and significantly reduced the ability of the S216C protein to bind DNA (Fig. 2). Likewise, the replacement of the positively charged side chains of Lys-217 and Arg-219 with a slightly smaller molecule of opposite charge also rendered the proteins nonfunctional in vivo and in vitro. Finally, the introduction of a hydrophobic aromatic side chain in place of the His-218 imidazole ring abolished in vivo activity in the resultant H218F protein. However, this mutated protein could still bind DNA, although at a reduced efficiency. The H218F mutant is the only mutant isolated in this study and in the previous study (8) that is completely nonfunctional in C. perfringens but that can form at least some of the active protein-DNA conformer, CII. The level of CII was significantly reduced, with approximately 50% of the DNA remaining in its unbound form at maximal protein levels. This reduced level of binding is the likely cause of the lack of biological function in this mutant.
The specific role of the SKHR motif in DNA-protein interaction remains unclear. Structural studies on the VirRc protein and the VirR-DNA complex are required to identify the precise role of all the residues implicated in the function of this domain. Solution of this structure would provide significant insight into the varied biological roles of this important family of proteins.
Acknowledgments
This research was supported by grants from the Australian National Health and Medical Research Council. S.M. is the recipient of an Australian Postgraduate Award.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awad, M. M., A. E. Bryant, D. L. Stevens, and J. I. Rood. 1995. Virulence studies on chromosomal α-toxin and θ-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of α-toxin in Clostridium perfringens-mediated gas gangrene. Mol. Microbiol. 15:191-202. [DOI] [PubMed] [Google Scholar]
- 3.Bannam, T. L., and J. I. Rood. 1993. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid 29:223-235. [DOI] [PubMed] [Google Scholar]
- 4.Ba-Thein, W., M. Lyristis, K. Ohtani, I. T. Nisbet, H. Hayashi, J. I. Rood, and T. Shimizu. 1996. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J. Bacteriol. 178:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, J. K., and J. I. Rood. 2000. The VirR response regulator from Clostridium perfringens binds independently to two imperfect direct repeats located upstream of the pfoA promoter. J. Bacteriol. 182:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng, W. P., and J. A. Nickoloff. 1992. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem. 200:81-88. [DOI] [PubMed] [Google Scholar]
- 7.Lyristis, M., A. E. Bryant, J. Sloan, M. M. Awad, I. T. Nisbet, D. L. Stevens, and J. I. Rood. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12:761-777. [DOI] [PubMed] [Google Scholar]
- 8.McGowan, S., I. S. Lucet, J. K. Cheung, M. M. Awad, J. C. Whisstock, and J. I. Rood. 2002. The FxRxHrS motif: a conserved region essential for DNA binding of the VirR response regulator from Clostridium perfringens. J. Mol. Biol. 322:997-1011. [DOI] [PubMed] [Google Scholar]
- 9.Nikolskaya, A. N., and M. Y. Galperin. 2002. A novel type of conserved DNA-binding in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 30:2453-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rood, J. I. 1998. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 52:333-360. [DOI] [PubMed] [Google Scholar]
- 11.Shimizu, T., W. Ba-Thein, M. Tamaki, and H. Hayashi. 1994. The virR gene, a member of a class of two-component response regulators, regulates the production of perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J. Bacteriol. 176:1616-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevens, D. L., R. K. Tweten, M. M. Awad, J. I. Rood, and A. E. Bryant. 1997. Clostridial gas gangrene: evidence that α and θ toxins differentially modulate the immune response and induce acute tissue necrosis. J. Infect. Dis. 176:189-195. [DOI] [PubMed] [Google Scholar]
- 13.Tweten, R. 1997. Thio-activated clostridial toxins, p. 211-221. In R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press Limited, London, United Kingdom.