Abstract
In populations that are small and asexual, mutations with slight negative effects on fitness will drift to fixation more often than in large or sexual populations in which they will be eliminated by selection. If such mutations occur in substantial numbers, the combined effects of long-term asexuality and small population size may result in substantial accumulation of mildly deleterious substitutions. Prokaryotic endosymbionts of animals that are transmitted maternally for very long periods are effectively asexual and experience smaller effective population size than their free-living relatives. The contrast between such endosymbionts and related free-living bacteria allows us to test whether a population structure imposing frequent bottlenecks and asexuality does lead to an accumulation of slightly deleterious substitutions. Here we show that several independently derived insect endosymbionts, each with a long history of maternal transmission, have accumulated destabilizing base substitutions in the highly conserved 16S rRNA. Stabilities of Domain I of this subunit are 15–25% lower in endosymbionts than in closely related free-living bacteria. By mapping destabilizing substitutions onto a reconstructed phylogeny, we show that decreased ribosomal stability has evolved separately in each endosymbiont lineage. Our phylogenetic approach allows us to demonstrate statistical significance for this pattern: becoming endosymbiotic predictably results in decreased stability of rRNA secondary structure.
Intracellular prokaryotic associates, or endosymbionts, are common in the Metazoa (1). Molecular phylogenetic evidence, in combination with fossil-based dating, indicates that maternal transmission has been in place for millions of years in endosymbionts of several insect groups (2), including aphids (3), carpenter ants (4), and tsetse flies (5). As compared with free-living relatives, these endosymbiont lineages exhibit faster rates of substitution in 16S rDNA, and their coding genes show an excess of nucleotide substitutions causing amino acid replacements (6). This finding is consistent with the hypothesis that a substantial proportion of new mutations are slightly deleterious and that more of these mutations drift to fixation in populations that are small and asexual as compared with populations that are very large or sexual (7–10). Maternally transmitted endosymbionts must possess small population sizes compared with those of related free-living bacteria: the observation of faster evolution at nucleotide sites under selection is consistent with reduced effectiveness of purifying selection at those sites, because of drift.
With this hypothesis, the functioning of gene products is predicted to be worse in endosymbionts than in free-living relatives because of the accumulation of deleterious mutations. One test involving direct effects on fitness is an evaluation of the stability of secondary structure of gene products (11). For rRNA molecules, the majority of mutations will lower stabilities of secondary structure. Thus, using the hypothesis that slightly deleterious mutations are fixed more often in endosymbionts, their rRNA structure is predicted to be less stable. To test this prediction, we calculated stabilities of secondary structures for 16S rRNA sequences for endosymbionts and for related free-living bacteria and compared the values.
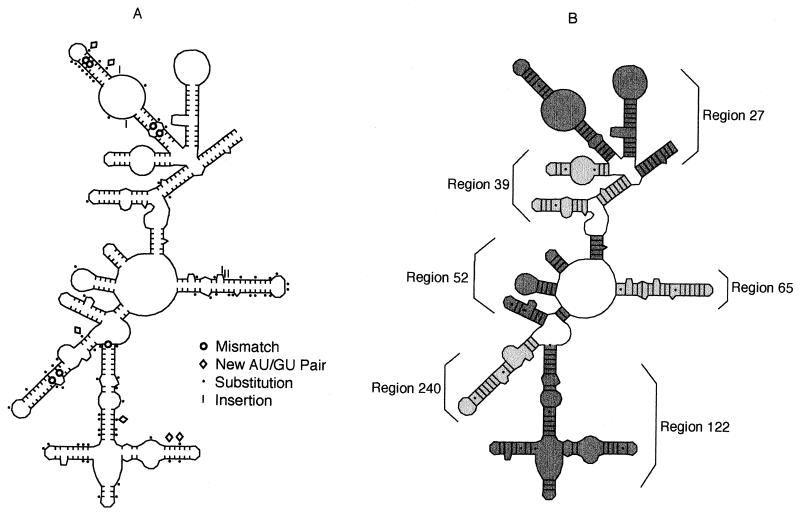
The 16S rRNA is ideally suited for testing effects of nucleotide evolution on the stability of a gene product. Its wide use for bacterial systematics has generated a large dataset (12, 13). Its sequence is conserved among taxa, reflecting functional constraint on translational machinery; constraint is corroborated by slower growth rates in cells with point mutations in this gene (14). The conserved secondary structure of the subunit has been characterized by using a combination of phylogenetic and enzymatic approaches (15, 16). Rules governing the relationship of RNA sequence to helix stability have been determined on the basis of experiments with oligomers of known sequence (17). These values allowed us to estimate stabilities of homologous stem–loops in related organisms. Initial reconstructions of the entire endosymbiont 16S rRNA secondary structure indicated that destabilizing substitutions occurred throughout the gene. For quantitative estimates of stability, we focused on Domain I (Fig. 1) in which regions vary widely in degree of phylogenetic conservatism and, presumably, functional constraint.
Figure 1.
Secondary structure of Domain I of the 16S rRNA (15, 16). (A) Structure for the endosymbiont B. aphidicola of the aphid Schlechtendalia chinensis, showing differences from corresponding positions in Escherichia coli. (B) Stabilities were calculated separately for each of the six labeled regions.
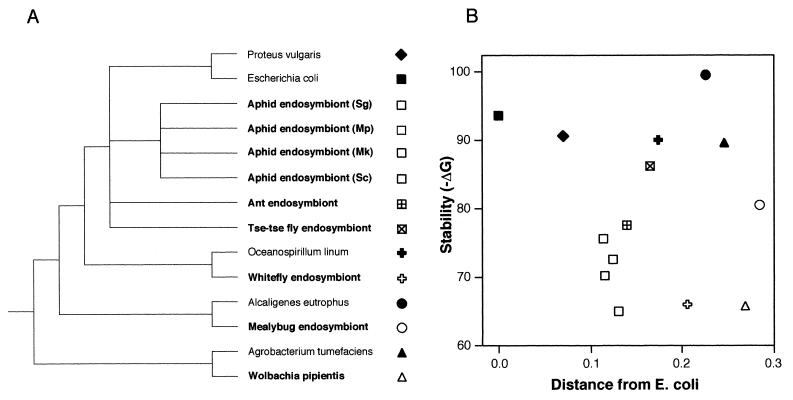
Six endosymbiont lineages were included: the bacteriocyte-associated symbionts of aphids, tsetse flies, mealybugs, whiteflies, and carpenter ants, plus Wolbachia pipientis. The latter is a maternally inherited pathogen that causes reproductive abnormalities in arthropods (18). All included groups are in the Proteobacteria, which includes Escherichia coli and other well-studied bacteria. The six lineages comprise all Proteobacteria for which recent molecular phylogenetic studies support a long history of endosymbiosis (2–5, 18–22). The aphid endosymbionts (Buchnera aphidicola) show the greatest estimated minimum age (2). For this group, we examined several taxa including lineages that diverged early (>108 years ago) and thus that have evolved independently for much of their history as endosymbionts. Free-living taxa were chosen to be as closely related to endosymbionts as possible, excluding other symbiotic or parasitic organisms. Relationships of organisms included in the study are shown in Fig. 2A.
Figure 2.
Stabilities of Domain I in endosymbiotic and free-living bacteria. (A) Relationships of taxa included in this study, based on molecular phylogenetic studies (2–5, 18–22). (B) Stabilities (−ΔG) summed over Domain I for each organism. Related free-living and endosymbiotic organisms are given the same symbol shape, with free-living taxa represented by a solid symbol. The horizontal axis shows distance from E. coli, measured as proportion sequence differences in the 16S rRNA.
METHODS
Secondary structures were deduced from sequence data by aligning sequences to find homologous regions and superimposing sequence onto the most closely related published structure (16). The free energy of folding for these structures was measured by using the program mfold (23) in the Genetics Computer Group package (24). Domain I was divided into six subsets of helices, and free energy (−ΔG) was measured for each. Three subsets (27, 39, and 52, Fig. 1B) were not composed of a continuous RNA strand, because other helix subsets were terminal. Parts of the resulting subsets are artificial; for example, containing a helix with no terminal loop or a terminal loop that is actually a bifurcated loop in the real structure. Likewise, for some real features such as bifurcated loops, lone base pairs, and tertiary contacts, estimates of stability are less reliable than for helices, internal loops, and terminal loops. However, both artificial and unpredictable regions were congruent in all comparisons and do not contribute to differences in folding energy. The resulting stabilities are not estimates of global stability for Domain I, but do allow comparisons of stem–loop stability between taxa.
In free-living taxa, secondary structure of all stems was unambiguously congruent with closely related published structures. Among endosymbionts, 6 (of 153) stems contained helices for which predicted structure was ambiguous, not including several regions where insertions had extended stem–loops. For ambiguous helices, the energy optimization function of mfold was used to predict the most stable configuration for the part of the structure in question. Because all ambiguous regions were in endosymbionts, this method yields a conservative estimate for our hypothesis: the mfold algorithm always picks either the correct structure or one that is measured to be more stable than the correct structure. For taxa with ambiguous helices, removing these helices resulted in destabilization of 16–23% compared with free-living outgroups; including regions predicted by mfold resulted in destabilization of 18.1–24.9%.
Names of organisms (with their GenBank accession numbers) included in this study were: aphid endosymbionts B. aphidicola of Schizaphis graminum (M63246), B. aphidicola of Myzus persicae (M63249), B. aphidicola of Mindarus kinseyi (M63253), B. aphidicola of S. chinensis (Z19056); endosymbiont of the ant Camponotus camponotii strain rufipes (X92552); endosymbiont Wigglesworthia glossinidia of the tsetse fly Glossina tachinoides (L37342); endosymbiont of the mealybug Pseudococcus longispinus (M68889); endosymbiont of the whitefly Trialeurodes vaporariorum (Z11928); Wolbachia pipientis from the mosquito Culex pipiens (X61768); E. coli (J01695); Oceanospirillum linum (M22365); Alcaligenes eutrophus (M32021); Agrobacterium tumefaciens (M11223); and Proteus vulgaris (X07652).
RESULTS
All endosymbionts were found to have less stable rRNAs than their free-living closest relatives (Fig. 2, Table 1). Moreover, the most stable endosymbiont score was less stable than any of the free-living bacterial scores, all of which were very similar. Free-living taxa showed relatively little variation in estimated stabilities; endosymbionts varied more widely. This variation is consistent with the hypothesis that decreased stability reflects the fixation of deleterious mutations by drift, because effective population size and duration of endosymbiotic history must vary among the groups.
Table 1.
Predicted stability of endosymbionts compared with outgroups (ΔG, in kcal/mol)
| Taxon | Regions of Domain I of 16S rRNA
|
Total | |||||
|---|---|---|---|---|---|---|---|
| 122 | 240 | 65 | 52 | 39 | 27 | ||
| P. vulgaris | −13.9 | −8.2 | −5.7 | −17.2 | −14.9 | −30.7 | −90.6 |
| E. coli | −17.0 | −12.0 | −4.2 | −14.3 | −14.9 | −31.2 | −93.6 |
| Aphid endosymbiont (Sg) | −15.0 | −7.2 | −4.5 | −14.3 | −11.9 | −21.6 | −75.6 |
| Aphid endosymbiont (Sc) | −7.8 | −0.9 | −8.0 | −14.3 | −11.9 | −22.1 | −65.0 |
| Aphid endosymbiont (Mk) | −9.6 | −7.4 | −7.6 | −14.3 | −11.9 | −21.8 | −72.6 |
| Aphid endosymbiont (Mp) | −6.8 | −7.2 | −8.4 | −14.3 | −11.9 | −21.6 | −70.2 |
| Ant endosymbiont | −13.3 | −8.9 | −4.8 | −18.3 | −15.4 | −16.8 | −77.5 |
| Tsetse fly endosymbiont | −13.9 | −9.4 | −7.2 | −18.3 | −13.3 | −24.0 | −86.1 |
| O. linum | −12.2 | −9.6 | −7.3 | −15.7 | −16.8 | −28.3 | −89.9 |
| Whitefly endosymbiont | −4.0 | −6.1 | −1.0 | −17.2 | −15.4 | −22.3 | −66.0 |
| A. eutrophus | −17.2 | −9.1 | −7.5 | −21.2 | −15.4 | −29.0 | −99.4 |
| Mealybug endosymbiont | −9.4 | −4.6 | −6.0 | −21.2 | −9.7 | −29.4 | −80.4 |
| A. tumefaciens | −14.3 | −11.3 | −3.1 | −16.4 | −17.9 | −26.5 | −89.5 |
| W. pipientis | −0.8 | −6.9 | −7.9 | −15.5 | −17.9 | −16.7 | −65.7 |
Stabilities calculated separately for six parts of Domain I. Numbers are −ΔG, the change in free energy associated with the structure.
Based on phylogenetic relationships, destabilization evolved four to six separate times (Fig. 2). The uncertainty in number of independent occurrences arises because relationships of the symbiont lineages of aphids, ants, and tsetse flies are not resolved by 16S rDNA analyses. Thus, although observations on bacterial morphology and host association suggest three independent origins (1), these groups potentially represent only one or two origins of endosymbiosis. Each of the four to six independent sister-group comparisons supports the prediction of lowered stability in endosymbionts. Considering a null hypothesis that endosymbionts are equally likely to show increased or decreased rRNA stability relative to free-living relatives, this result is statistically improbable (P = 0.016, P = 0.032, and P = 0.063 depending on whether the three related groups are considered as three origins, two origins, or one origin, respectively).
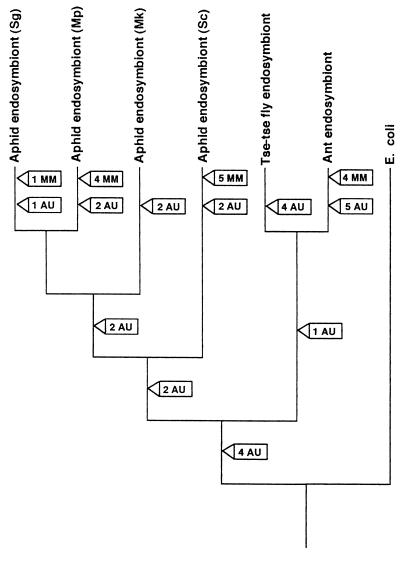
Regardless of their phylogenetic relationships, most substitutions conferring decreased stability evolved independently among the endosymbionts of the four aphid species, the tsetse fly, and the carpenter ant. This can be observed by reconstructing destabilizing events on a phylogenetic tree, assuming that the correct phylogeny and the correct mapping of destabilizing events are those that minimize the number of such events (Fig. 3). Even considering these conservative hypotheses, most destabilizing evolution occurs in separate lineages, near the tips of the tree.
Figure 3.
Positions of destabilizing substitutions of endosymbionts related to E. coli, assuming both the tree topology and the mapping of substitutions that minimize independent events. Destabilizing substitutions are divided into two categories, those causing a stem mismatch (MM) and those causing an AU or GU pair to replace a GC pair in a stem (AU). Where two substitutions are associated with an event in either category (e.g., an AU pair replaces a GC pair), only one destabilization event is mapped onto the tree. Even with these assumptions, which minimize independent events, most destabilizing substitutions are unique to individual lineages, indicating largely independent evolution of destabilization.
Destabilization of endosymbiont sequences is distributed among regions of Domain I (Table 1, Fig. 1A). This dispersion is consistent with our hypothesis: slightly deleterious mutations are expected to be distributed along the gene.
The sequence changes underlying destabilization can be divided into two major types, those causing base pair mismatches in helices and those increasing A/U content. The latter, which occur in a variety of organisms, are generally considered to result from mutational bias rather than selection (25, 26). Using this interpretation, increased A/U content of endosymbionts reflects a greater rate of fixation of new mutations, consistent with the hypothesis of reduced effectiveness of selection. However, if the bias instead reflects selection on genomic base composition, A/U substitutions that decrease stability could be interpreted as reflecting a change in selection rather than the fixation of deleterious mutations through drift. Thus, destabilization because of mismatches provides more clearcut support for the hypothesis that the substitutions observed in endosymbionts are deleterious and fixed through drift. In our comparisons, both sources of destabilization, A/U bias and mismatches, contribute substantially to observed differences in stability between endosymbionts and free-living bacteria. This can be demonstrated by composing a hypothetical sequence, identical to that of a free-living taxon except for the introduction of mismatches that characterize a related endosymbiont. For example, changing the E. coli sequence at six positions to introduce the six mismatches of the endosymbiont of the aphid S. chinensis (Fig. 1A) results in a structure with very similar A/U content but stability that is 15% lower; the real endosymbiont sequence is destabilized 30% relative to E. coli. The converse experiment, repairing the mismatches in the S. chinensis endosymbiont sequence, reduces destabilization to 15% relative to E. coli.
DISCUSSION
Organelles have population structures resembling those of vertically transmitted endosymbionts. Lynch (9, 10) has presented evidence for the accumulation of slightly deleterious mutations in mitochondria and chloroplasts, based on comparisons between the evolution and properties of transfer RNAs of corresponding organellar and nuclear genomes. In animals, mitochondrial tRNAs evolve faster and have less stable secondary structure than nuclear tRNAs (9); in plants, animals, and yeast, effects of purifying selection on sequence evolution of tRNAs is less for mitochondrial and chloroplast genomes than for nuclear genomes (10). Our analysis uses independent comparisons of endosymbionts to related free-living prokaryotes to demonstrate that accumulation of deleterious mutations is a predictable consequence of the endosymbiotic habit; this result allows greater generalization about effects of population structure on deleterious evolution.
An alternative explanation for the unusual pattern of nucleotide substitutions in endosymbionts and mitochondria is that destabilization results from a change in the magnitude of selection coefficients (s) rather than a change in effective population size (Ne). The observation that a greater proportion of new mutations behave as neutral alleles implies that a greater proportion have the value Nes < 1. This may result from either a decrease in Ne, as proposed, or a decrease in s. The latter could occur if endosymbionts experience relaxed selection in the context of the intracellular environment, which might be more constant than environments of free-living prokaryotes. At present, this alternative cannot be excluded. However, some observations weigh against it. First, the pattern of increased rate of evolution at sites under selection occurs at all loci examined (6). A speedup based on a change in selection might be expected to be more idiosyncratic in effects on different loci, whereas an effect because of population structure would affect all loci similarly, as observed. Second, ribosomes and other cellular structures of insect symbionts evolve in the context of a wide variation in temperature, a primary determinant of stability. For example, aphids and their symbionts live at temperatures from below 0°C to greater than 40°C. Third, the observation that Wolbachia shows the same pattern as bacteriocyte associates (Fig. 2B, ref. 6) excludes the explanation that selection on stability is relaxed because of coadaptation of a mutualistic host. Wolbachia does not benefit its hosts (18) and invades a variety of host tissues rather than living only in host cells specialized to house it. Nonetheless, further tests will be required to settle the issue of the relative roles of reduced population size vs. reduced selection as the cause of increased rates of evolution in symbionts.
Selection among host lineages will counter deterioration of symbiont fitness. If each host individual contains an essentially pure culture of symbionts, because of the bottleneck imposed at the time of inoculation of progeny, then selection will act primarily between host maternal lineages. Thus, Ne of the symbiont will be related to Ne of the host (adjusted to include only females). Even for insects, which can have much larger populations than vertebrates, effective population sizes are much smaller than those of bacteria: upper estimates of Ne for insects are about 106–107 (see refs. 27 and 28) and some estimates are much lower, at 104 or less (see refs. 29–31). Lower estimates of Ne for E. coli are about 109 (32). Thus, selection at the host level may slow but not prevent accumulation of deleterious mutations in endosymbiotic bacteria.
The replacement of GC stem pairs with AU pairs implies the occurrence of compensatory substitutions. Initially, a destabilizing mismatch mutation (e.g., a GC pair replaced by a GA pair) is fixed through drift; subsequently, selection drives a compensatory substitution at the paired site (e.g., a GA replaced by UA). The greater effectiveness of selection on the second mutation suggests fluctuation either in the strength of selection or in effective population size (33). In long-term endosymbionts, the continued deterioration and eventual breakdown of gene function may be prevented by compensatory substitutions that are enabled by such fluctuations in the effectiveness of selection.
CONCLUSION
Our study demonstrates repeated evolution of rRNA destabilization in endosymbiotic bacteria. It provides strong support for the proposal that the small effective population size of maternally transmitted endosymbionts results in the increased fixation by drift of slightly deleterious mutations. For mutualistic endosymbionts, selection at the level of the host animals would oppose any decline in fitness but may be ineffective depending on host population size. Over long periods of evolution, deleterious substitutions may accumulate, causing measureable effects on gene products as documented here for the 16S rRNA. The magnitude of these effects on fitness, for either the symbionts or the hosts that depend on them, is not yet clear.
Acknowledgments
We thank M. Nachman, L. Nagy, R. Parker, J. B. Walsh, and E. Waters for valuable comments. This work was partially supported by grants to N.A.M. from the National Science Foundation (DEB-9527635, with Paul Baumann) and the U.S. Department of Agriculture (9601607), and by a National Science Foundation Graduate Research Fellowship (9602246) and a fellowship from the University of Arizona Research Training Grant in the Analysis of Biological Diversification to J.D.L.
References
- 1.Buchner P. Endosymbiosis of Animals with Plant Microorganisms. New York: Wiley Interscience; 1965. [Google Scholar]
- 2.Moran, N. A. & Telang, A. (1998) Bioscience 48, in press.
- 3.Baumann P, Moran N A, Baumann L. Bioscience. 1997;47:12–20. [Google Scholar]
- 4.Schröder D, Deppisch H, Obermeyer M, Krohne G, Stackebrandt E, Hölldobler B, Goebel W, Gross R. Mol Microbiol. 1996;21:479–490. doi: 10.1111/j.1365-2958.1996.tb02557.x. [DOI] [PubMed] [Google Scholar]
- 5.Aksoy S, Pourhosseini A A, Chow A. Insect Mol Biol. 1995;4:15–22. doi: 10.1111/j.1365-2583.1995.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 6.Moran N A. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller H J. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 8.Ohta T. Nature (London) 1974;252:351–354. doi: 10.1038/252351a0. [DOI] [PubMed] [Google Scholar]
- 9.Lynch M. Mol Biol Evol. 1996;13:209–220. doi: 10.1093/oxfordjournals.molbev.a025557. [DOI] [PubMed] [Google Scholar]
- 10.Lynch M. Mol Biol Evol. 1997;14:914–925. doi: 10.1093/oxfordjournals.molbev.a025834. [DOI] [PubMed] [Google Scholar]
- 11.Ohta T. Nature (London) 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- 12.Woese C R. Microbiol Rev. 1987;52:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maidak B L, Olsen G J, Larsen N, Overbeek R, Mccaughey M J, Woese C R. Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Connor M, Brunelli C A, Firpo M A, Gregory S T, Lieberman K R, Lodmell J S, Moine H, Van Ryk D I, Dahlberg A E. Biochem Cell Biol. 1995;73:852–868. doi: 10.1139/o95-093. [DOI] [PubMed] [Google Scholar]
- 15.Gutell R R, Larsen N, Woese C R. Microbiol Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutell R R. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaeger J A, Turner D H, Zuker M. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werren J H. Annu Rev Entomol. 1997;42:587–609. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- 19.Moran N A, Munson M A, Baumann P, Ishikawa H. Proc R Soc London Ser B. 1993;253:167–171. [Google Scholar]
- 20.Clark M A, Baumann L, Munson M A, Baumann P, Campbell B C, Duffus J E, Osborne J S, Moran N A. Curr Microbiol. 1992;25:119–123. [Google Scholar]
- 21.Munson M A, Baumann P, Moran N A. Mol Phylogenet Evol. 1993;1:26–30. doi: 10.1016/1055-7903(92)90032-c. [DOI] [PubMed] [Google Scholar]
- 22.O’Neill S L, Giordano R, Colbert A, Karr T L, Robertson H M. Proc Natl Acad Sci USA. 1992;89:2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genetics Computer Group. Program Manual for the Wisconsin Package. Madison, WI: Univ. of Wisconsin; 1996. , Version 9. [Google Scholar]
- 24.Zuker M. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 25.Sueoka N. J Mol Evol. 1992;34:95–114. doi: 10.1007/BF00182387. [DOI] [PubMed] [Google Scholar]
- 26.Jermiin L S, Graur D, Lowe R M, Crozier R H. J Mol Evol. 1994;39:160–173. doi: 10.1007/BF00163805. [DOI] [PubMed] [Google Scholar]
- 27.Kreitman M. Nature (London) 1983;304:412–417. doi: 10.1038/304412a0. [DOI] [PubMed] [Google Scholar]
- 28.Nei M, Graur D. Evol Biol. 1984;9:73–118. [Google Scholar]
- 29.Britten H B, Brussard P F, Murphy D D, Austin G T. Great Basin Naturalist. 1994;54:97–105. [Google Scholar]
- 30.Owen R E, Mydynski L J, Packer L, McCorquodale D B. Biochem Genet. 1992;30:443–453. doi: 10.1007/BF01037585. [DOI] [PubMed] [Google Scholar]
- 31.Taylor C E, Toure Y T, Coluzzi M, Petrarca V. Med Vet Entomol. 1993;7:351–357. doi: 10.1111/j.1365-2915.1993.tb00704.x. [DOI] [PubMed] [Google Scholar]
- 32.Selander R K, Caugant D A, Whittam T S. In: Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology. Neidhardt F, editor. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 1625–1648. [Google Scholar]
- 33.Ohta T. Nature (London) 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]