Abstract
We have identified the mutation responsible for the autosomal recessive wasted (wst) mutation of the mouse. Wasted mice are characterized by wasting and neurological and immunological abnormalities starting at 21 days after birth; they die by 28 days. A deletion of 15.8 kb in wasted mice abolishes expression of a gene called Eef1a2, encoding a protein that is 92% identical at the amino acid level to the translation elongation factor EF1α (locus Eef1a). We have found no evidence for the involvement of another gene in this deletion. Expression of Eef1a2 is reciprocal with that of Eef1a. Expression of Eef1a2 takes over from Eef1a in heart and muscle at precisely the time at which the wasted phenotype becomes manifest. These data suggest that there are tissue-specific forms of the translation elongation apparatus essential for postnatal survival in the mouse.
The autosomal recessive wasted (wst) mutation arose spontaneously in the inbred mouse colony HRS/J at the Jackson laboratory in 1972 (1). The phenotype of homozygous wst/wst mice is intriguing; the mice have neurological defects, waste away, and have immune system abnormalities, which include a defective response to DNA damage in lymphoid cells (1).
Homozygous wst/wst mice appear completely normal until weaning age; at ≈21 days, they develop tremors and ataxia. Histological examination reveals extensive neuronal vacuolar degeneration of anterior horn cells of the spinal cord with less severe abnormalities of motor nuclei in the brain stem (2). No abnormalities are seen in the rest of the central nervous system, including Purkinje cells, and myelin appears to be unaffected (2). The mice then lose weight (presumably through muscle wasting), develop progressive paralysis, and die by ≈28 days (1). The time course of this disease progression varies very little, regardless of genetic background, weaning time, or environmental factors such as specific pathogen free conditions (ref. 3 and J.P., unpublished observations). During this postweaning period, wst/wst mice also develop progressive atrophy of the spleen and thymus and a concomitant decrease in the levels of circulating lymphocytes (1). Cells derived from lymphoid tissues of wasted mice from 26 days onwards show a defective response to radiation-induced DNA damage, which is reminiscent of a checkpoint defect; however, this defect is not seen in fibroblasts nor is it seen in lymphoid-derived cells of mice of 22 days (4–6).
The wst gene was mapped initially to distal chromosome 2, within 1 centiMorgan of the gene for ragged (Ra), a semi-dominant visible mutation, which affects coat quality (7). To map the wasted gene precisely with respect to molecular markers, we set up an interspecific backcross and showed wst to lie within the most distal group of markers on chromosome 2 (3).
Because there are no obvious biochemical clues to the basis of the wasted mutation, we have used a positional cloning/positional candidate approach to isolate the wst gene. Because those genes that map close to wst on Chr 2 and have been mapped in humans reside without exception on distal human Chr 20q (8), genes which map to human Chromosome 20q13 have been considered as a source of new markers and/or candidate genes. One such gene encodes a protein that displays a high level of homology (92% identity and 98% similarity at the amino acid level) to the translation elongation factor EF1α (human locus EEF1A; mouse locus Eef1a) and is called EF1α2 or S1 (human locus EEF1A2; mouse locus Eef1a2; ref. 9 and 10). EF1α promotes the GTP-dependent binding of aminoacyl-tRNA to the A site of the ribosome (11). EF1α is distributed widely, although expression is very low or undetectable in skeletal and cardiac muscle postnatally (12, 13), and expression in the brain declines between embryonic life and adulthood in rats (12). EF1α2, on the other hand, is found exclusively in terminally differentiated cells of skeletal muscle, heart, and certain areas of the brain (12–14). In those tissues where both genes are expressed, Eef1a2 is consistently more highly expressed than Eef1a (15). In rat muscle and heart, EF1α levels decline by 95% within the first month of postnatal life; EF1α2 levels on the other hand increase during this period but are constant through adult life (15). There appears, therefore, to be some degree of reciprocal expression of the two genes. Although fibroblasts normally do not express Eef1a2, the mouse 3T3 fibroblast cell line does, and this result was used to study regulation of Eef1a2 at different stages of the cell cycle (9). It was found that Eef1a2 was highly expressed when cells were serum-starved, in Go phase, but expression declined dramatically when cells were serum stimulated and reentered the cell cycle. Eef1a, on the other hand, was expressed at all stages of the cell cycle in this cell line (9). Whereas EEF1A2 appears to be single copy, numerous pseudogenes for EEF1A exist in the human genome (10).
We now show that the wasted mutation is a deletion spanning 15.8 kilobases (kb) that removes the promoter region and first noncoding exon of the mouse Eef1a2 gene, abolishing transcription of the gene. No other gene has been detected within this deleted region.
MATERIALS AND METHODS
Mouse Strains.
All of the mouse strains used in this study were maintained at the Medical Research Council Mammalian Genetics Unit. The maintenance of the ragged/wasted stock and the setting up of the wasted backcross was as described (3). Control animals were either littermates of wst/wst mice (of the genotype +/wst) or age-matched animals from the C3H/HeH strain, which represents the major component of the genetic background of the wasted stock. DNA from HRS/J control mice was purchased from The Jackson Laboratory. The animal studies described in this paper were carried out under the guidance issued by the Medical Research Council in Responsibility in the Use of Animals for Medical Research (1993) and Home Office Project License no. 30/00875.
PCR of Genomic DNA.
PCR primers for analysis of the deletion are given in the legend of Fig. 2. The primers used for mapping Eef1a2 in the wasted backcross were 5′-GAGAGCTTCTCACAGTACCC and 5′-ATGTCTCGCACGGCGAAGC. Other primers used for mapping were as described (3). Typically, 50 ng of genomic DNA was amplified in 30-μl reaction volumes with 200 μM each dNTP and 1 unit of Taq polymerase. Reactions were carried out in buffer comprising 20 mM (NH4)2SO4, 75 mM Tris⋅HCl (pH 9.0), 0.01% Tween 20, 1.5 mM MgCl2, with the addition of dimethyl sulfoxide to 10% for all Eef1a2 primers. Cycling conditions were 95°C for 3 min, followed by 30 cycles of 94°C for 40 sec, 55°C for 1 min, and 72°C for 1 min. Reaction products were visualized on 2% agarose gels. PCR across the wasted deletion was achieved using the Expand Long template PCR system (Boehringer Mannheim), according to the manufacturers’ conditions, using primers 1r and 4f.
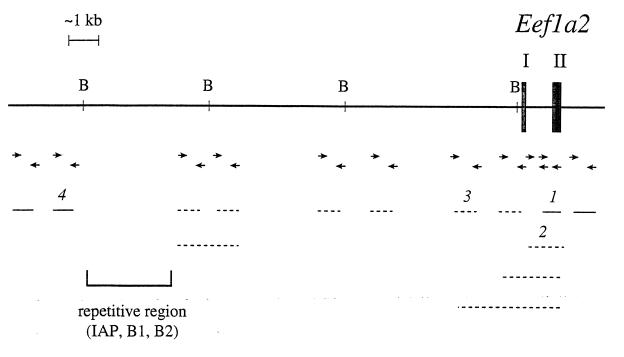
Figure 2.
Analysis of the region upstream of the Eef1a2 gene. BamHI restriction sites are marked “B.” Exons I and II of Eef1a2 are indicated as filled boxes. The lines represent the PCR product obtained with the primers listed below (according to the number above the line). Solid lines represent products that were obtained from both HRS/J and wst/wst DNA and dotted lines represent those that were produced only from HRS/J DNA and gave no product from wst/wst DNA. The extent of the deletion is indicated by arrows. PCR primers were as follows: product 1, 1f: 5′-TGTGGCTGAATGGGGTTAGG, 1r: 5′-CACTGTGGGGGCTCTGGTTT (product size 226 bp); product 2, 2f: 5′-CAGAGCTTCACTCAGTCTG,1r: as above (product size 379 bp); product 3, 3f: 5′-TAGTGGCTCCTTGGAACAG, 3r: 5′-CTACTCTCCCTGAATGCCTT (product size 456 bp); and product 4, 4f: 5′-TGACTATCCCATTGCCAGG, 4r: 5′-CTGCAACTCAACAGCCCATT (product size 191 bp).
Bacterial Artificial Chromosome (BAC) Library Screening.
PCR pools from a mouse ES cell BAC library were purchased from Research Genetics, and screened for Eef1a2 according to the supplier’s instructions by using the primers described above.
Sequencing.
Sequencing of PCR products, BAC, and plasmid DNA was carried out using the ThermoSequenase kit (Amersham) according to the manufacturer’s instructions.
RNA Preparations.
RNA was prepared from mouse tissues flash-frozen in liquid nitrogen using the Total RNA Isolation Reagent from Advanced Biotechnologies, based on the method of Chomczynski and Sacchi (25).
Reverse Transcription (RT)-PCR.
First strand cDNA synthesis was carried out on 2 μg of total RNA (which had previously been DNase I treated) by using the First-Strand cDNA synthesis kit from Pharmacia Biotech and random hexamer primers. 5 μl aliquots of a given RT product were then used as the template for PCR reactions under the conditions described above, again for 30 cycles. Three reactions were carried out for each product corresponding to β-actin (primers 5′-TGAAGTACCCCATTGAACACG and 5′-GTGCTAGGAGCCAGGGCAGT; product size 770 bp), elongation factor 1α (primers 5′-ATATTACCCCTAACACCTGG and 5′-CTGTGACAGATTTTTGGTCAAG; product size 246 bp), and elongation factor 1α2 (primers 5′-CCTCAGGAGGCTGCCCAG and 5′-ATGTCTCGCACGGCGAAGC; product size 286 bp). RT-negative controls were carried out for each tissue sample, with each pair of primers. Primers for Eef1a and Eef1a2 were designed so as to maximize sequence differences between the two genes, to prevent crossreaction, and also to span introns. Products were analyzed on 2% agarose gels.
Computing.
blast, grail, and geneid programs were accessed through the computing facilities provided by the Medical Research Council Human Genome Mapping Project Resource Centre, UK.
RESULTS
Mapping of Eef1a2.
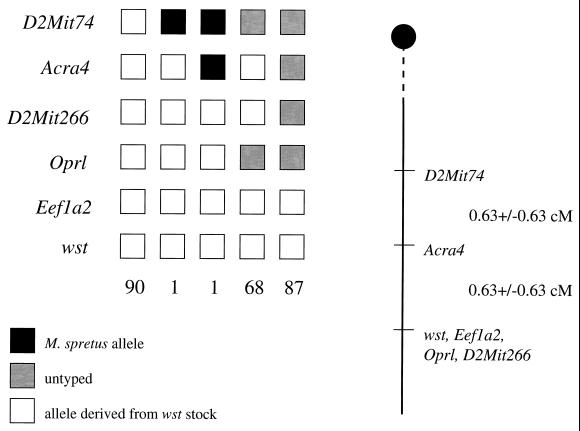
The wasted backcross described in ref. 3 has now yielded 255 N2 offspring. Before the mapping of the EEF1A2 gene to human chromosome 20q13, the closest markers to wst were the microsatellite D2Mit266 and the gene Oprl, which encodes an orphan opioid receptor. Both of these are nonrecombinant with wst, but Oprl has been excluded as a candidate gene because there is no difference in expression level of the gene between wasted and wild-type mice, and sequencing of the whole Oprl coding region in wasted and wild-type control mice has revealed no changes (unpublished data). We have now mapped the mouse homolog of EEF1A2, Eef1a2, by PCR. Primers were designed using published mouse cDNA sequence to amplify intron IV, which in the rat contains a microsatellite (9). The PCR product obtained from mouse DNA showed size variation between the wasted stock and Mus spretus, and this product was used to follow the segregation pattern of Eef1a2 relative to wst (Fig. 1). Because Eef1a2 did not recombine with wst in 247 animals tested, putting the genes within 1.21 centiMorgans of each other at the 95% confidence limit, we investigated the gene further as a candidate for wst.
Figure 1.
Pedigree analysis of N2 offspring in the wasted backcross. Briefly, Ra+/+wst female mice were crossed to M. spretus males. Wild-type female offspring from this cross, presumptive +/wst heterozygotes, then were backcrossed to Ra+/+wst male mice. Only those offspring identified as wst/wst homozygotes were selected for further analysis. Two of the recombinants between wst and D2Mit266 listed in Abbott et al. (3) now have been shown to have been misclassified as wst/wst and have been eliminated from further analysis. The loci are listed to the left of the boxes, and the numbers below the boxes indicate the number of N2 offspring carrying that haplotype. The chromosomes shown were inherited from the F1 parent. To the right of the figure is the map derived from the haplotype analysis, giving the distances between loci as centiMorgans +/− standard error. The figure given for the distance between D2Mit74 and Acra4 is based on a total number of 166 animals scored because all of these mice were typed for the proximal marker D2Mit73 also (3).
Mutation Analysis of Eef1a2.
Published mouse cDNA and rat genomic sequences for Eef1a2 (16, 9) were used to design PCR primers with which to amplify each exon of the gene in wst/wst and control mice (HRS/J, the strain on which wst first arose). Exons III to VIII were amplified and sequenced in this way; no sequence differences were found between HRS/J and wst/wst (data not shown). However, all attempts, using multiple pairs of primers, to amplify exons I and II from wst/wst DNA failed, although PCR products were obtained easily from wild-type DNA. The relative positions of the various primers used and their pattern of failure to amplify DNA from wasted mice was strongly suggestive of a deletion in wst/wst mice (Fig. 2). We sought therefore to define this region further. We screened an ES cell-derived mouse BAC library and isolated two overlapping clones containing the entire Eef1a2 gene, each ≈65 kb. After sequencing the ends of the BACs, we designed PCR primers corresponding to each end of each BAC; these amplified both wild-type and wst/wst DNA, putting the upper size limit of the putative deletion at 55 kb (because most of the Eef1a2 gene, including all of the coding sequence, is intact in wasted mice). Subcloning of the BACs, contig assembly of the subclones, sequence analysis, and PCR amplification of segments across the region enabled us to narrow down the deletion interval to between 18 and 11.5 kb (Fig. 2). Again, the positioning of the primers was consistent with the presence of a deletion in wasted mice and not a simple insertion or inversion. PCR products were sequenced to confirm their identity (data not shown).
By database searching (using blast; ref. 17) within the apparently deleted region, a truncated intracisternal A particle (IAP) element (present at ≈1,000 copies in the mouse genome) and various highly repetitive elements were found (Fig. 2). These repeats complicated the analysis because they prevented both the design of unique sequence PCR primers with which to narrow down the possible extent of the deletion and prevented Southern blot analysis.
Eventually, however, we were able to amplify across the boundaries of the deletion in wst/wst DNA by using primers 1r and 4f (see Fig. 2 legend), which gave a product of just under 3 kb (no product was obtained from PCR of HRS/J DNA because the distance between the primers in wild-type DNA is predicted to be >18 kb). Sequence analysis of this PCR product showed that one end of the deletion lies 206 bp upstream of exon II of Eef1a2. The other end of the deletion is in the gag gene of the IAP element shown in Fig. 2. Sequence analysis of the deleted region is shown in Fig. 3. The site of the deletion appears to be a clean break with no sequences gained or lost. The size of the wasted deletion is thus precisely defined as 15.8 kb. There is no significant region of homology between the regions immediately flanking the deletion.
Figure 3.
Sequence analysis across the deletion breakpoint region in wasted and wild-type (HRS/J) mice. Underlined sequence represents the gag gene of the IAP element and nonunderlined sequence is intron 1 of Eef1a2. Sequence in bold is exon II of Eef1a2. Dotted lines indicate the 15.8 kb deleted in wst/wst mice.
This 15.8-kb region was sequenced completely from wild-type DNA. No significant homology was detected to any known gene or EST by searching the EMBL and GenBank databases using the blast and tblastx programs (17). No CpG islands were found within the deletion, other than that associated with Eef1a2; there were a number of sites for rare-cutters, but these sites all fell within the IAP element. We used a number of gene identification programs [geneid (18), grail 1a, and grail 2 (19)] to identify putative intron/exon boundaries, and ORFs, but no significant regions were identified consistently by more than one program. Although we cannot rule out the presence of another gene, we have found no evidence for one.
Expression of Eef1a2.
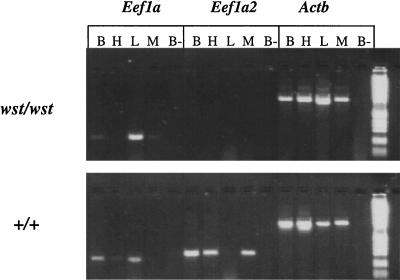
Because the deletion present in wasted mice starts in the first intron of the Eef1a2 gene, we examined the level of expression of Eef1a2 in wasted and control mice. We carried out RT-PCR using tissues isolated from 21-day-old wst/wst mice, their normal littermates (genotype +/wst), and age-matched controls from strain C3H/HeH, which represents the genetic background that predominates in the wasted stock maintained at Harwell (it is not possible to obtain mice from the same litters with the genotype +wst/+ wst, as Ra is maintained in repulsion to wst in this stock, so that +wst/+wst animals are also Ra/Ra, and thus usually die during early postnatal life.) Fig. 4 shows the results obtained. It can be seen clearly that although Eef1a maintains a normal pattern of expression in wasted mice, with expression of the gene in brain and liver and at a low level in muscle, expression of Eef1a2 is abolished completely, as expected for a deletion that removes the first noncoding exon and promoter of the gene.
Figure 4.
RT-PCR of RNA from age-matched 21-day-old homozygous wst/wst and wild-type (+/+) mice. Expression of Eef1a, Eef1a2, and a β-actin control are shown. The tissues assayed are brain (B), heart (H), liver (L), and skeletal muscle (M). The RT-negative control is shown for brain only (B-); similar controls were done for all tissues, and all were completely negative. The molecular weight marker is the 1-kb ladder (GIBCO/BRL).
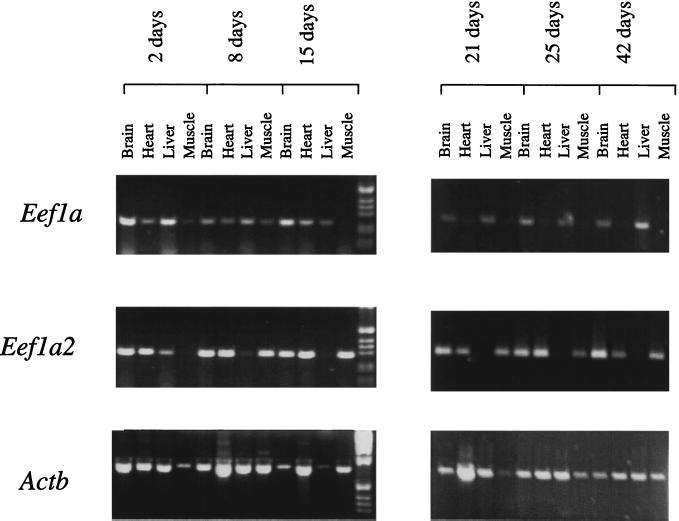
Because the developmental expression profile of Eef1a2 has been determined for rat (15) but not for mouse, we also sought to examine the relative expression levels of Eef1a2 and Eef1a in a number of mouse tissues in wild-type mice between 2 and 42 days of postnatal development, again using RT-PCR. The results obtained are shown in Fig. 5. In skeletal muscle and heart from mice of 21 days onwards, there is a dramatic reciprocity of expression between Eef1a and Eef1a2. No expression of Eef1a can be detected in skeletal muscle or heart of mice from 25 days onwards, and only trace amounts are seen at 21 days. Similarly, no Eef1a2 expression can be seen in liver at these ages. However, when mice of 2 to 15 days are examined, the clear-cut relationship between Eef1a and Eef1a2 breaks down. In skeletal muscle and heart from mice of 2 days, Eef1a is clearly expressed; in fact, Eef1a2 is not detectable in skeletal muscle at this stage, although it is strongly expressed in heart at all ages tested. At 8 days, both genes are clearly expressed in muscle, but by 15 days Eef1a is only just detectable. These results confirm and extend the findings in rat (15). Between 21 and 25 days, Eef1a2 expression in skeletal muscle has replaced expression of Eef1a. Thus Eef1a2 appears to be predominant over Eef1a in skeletal muscle, which is profoundly affected by the wasted mutation.
Figure 5.
RT-PCR of RNA from +/+ mice of different ages, from 2 to 42 days after birth. Expression of Eef1a, Eef1a2, and a β-actin control are shown. No-RT controls were negative in all cases. The molecular weight marker is the 1-kb ladder (GIBCO/BRL).
The situation in brain is less clear-cut, consistent with findings in the rat, in that both Eef1a2 and Eef1a are expressed in RNA prepared from total brain at all ages tested. Expression analysis in the rat, however, using in situ hybridization and primary culture of cells from rat neonatal cerebral cortex, has demonstrated that expression of Eef1a2 is restricted to neurons, whereas Eef1a is expressed in nonneuronal cell types in addition to neurons (14).
DISCUSSION
We have demonstrated that the primary lesion in wasted mice is a deletion that abolishes expression of a translation elongation factor-related gene, Eef1a2. The expression pattern of Eef1a2, and its putative function based on homology to Eef1a, offer an immediate explanation for several aspects of the wasted phenotype. Eef1a2 is switched on in skeletal muscle and heart postnatally; expression of Eef1a gradually declines in these tissues such that by 25 days only Eef1a2 is detectable. The implications for the development of the wasted phenotype are clear. Wasted mice have no observable defect until they are 21 days of age; they deteriorate markedly over the next few days and are almost invariably dead by 28 days. Along with tremors and ataxia, the most obvious defect is the weight loss of the mice. By 27 days, the average body weight of a homozygous wst/wst mouse is just over 7 g, compared with 16.5 g for normal littermate controls (6). Because fat stores are very small at this age, the majority of the weight loss must result from loss of muscle bulk. If, as the expression data of Eef1a2 and Eef1a in wild-type mice would suggest, Eef1a2 is essential for translation elongation in skeletal muscle and heart after 25 days, this weight loss can be accounted for by loss of Eef1a2 and the resultant failure of translation. No findings have been reported on the pathology of heart in wasted mice.
Because both Eef1a and Eef1a2 are expressed in brain, it is less easy at first sight to account for the neurological abnormalities. Eef1a2 expression in rat brain is confined to neurons, but even in these cells, there is some expression of Eef1a. The RT-PCR results suggest that both genes are expressed from early in postnatal life, whereas the tremors and ataxia typical of wst/wst mice are not seen until the mice reach 21 days. In rat, in which quantitative studies have been carried out, it has been shown that expression of Eef1a2 in brain increases gradually throughout postnatal life, peaking at around 30 days. After 10 days, Eef1a expression starts to decline (15). The pattern of expression of Eef1a2 coincides with the pattern of synaptogenesis in the rat, with the number of synaptic junctions peaking between 26 and 35 days (20). It is possible, then, that Eef1a2 only becomes necessary for normal function once synaptogenesis has been completed, and this is why the onset of neurological defects in the wasted mice occurs postweaning. Alternatively, Eef1a2 might be involved directly in some aspect of synaptogenesis. It will be of great interest to determine the pattern of expression of the EEF1A2 protein within neurons.
It is less clear at this stage whether the loss of Eef1a2 could be responsible for the immune system defects seen in wasted mice, namely atrophy of spleen and thymus and the defective response to DNA damage seen in lymphoid derived cells of wasted mice from 26 days onwards. It has been reported (12) that no expression of Eef1a2 is detectable in rat spleen by either Northern blotting or RNase protection analysis. We too were unable to detect expression of Eef1a2 in mouse spleen by RT-PCR at 21, 24, or 42 days (data not shown). Although we can still not rule out the possibility that the gene is expressed in a very small subset of cells within the lymphoid system, there is as yet no evidence for this. It seems, then, if Eef1a2 is the only gene affected by the deletion in wasted mice, that the immune system abnormalities are in some way secondary to the loss of Eef1a2 expression. It is conceivable that Eef1a2 is required for the translation of a protein, which is required for the maintenance of lymphoid precursor cell populations. A specific knockout of Eef1a2 will be required to test this hypothesis because transgenic correction of the wasted phenotype would still not categorically rule out the possibility of the involvement of a second gene.
Sequence analysis of the deleted region has failed to reveal significant homology to any known gene or EST other than Eef1a2, but clearly it will be necessary to carry out more extensive analysis of the region adjacent to the deletion end point upstream of Eef1a2 before we can establish whether or not a second gene is involved. It is worth noting that, on the side of the deletion upstream of Eef1a2, a further 2–3 kb of IAP element and other repeats remains intact; this may act to insulate any possible effect of the deletion on an adjacent gene. Again, the only functional test of whether deletion of Eef1a2 is responsible for all aspects of the wasted phenotype will be to knock out Eef1a2 by homologous recombination and compare the phenotype of the resulting Eef1a2−/Eef1a2− mice to those with a wst/wst genotype.
Although it remains to be proved whether EF1α2 does actually take part in translation elongation, the circumstantial evidence for its involvement (high level of homology with EF1α and reciprocal expression of the two genes) is strong. Why would tissue-specific isoforms of such a basic component of the translational apparatus exist? Possibly, the requirements of tissues such as skeletal muscle, heart, and certain terminally differentiated cells within the brain for translation may be different from those of actively growing cells. It is of note that a tissue-specific isoform of elongation factor 1β, which interacts with EF1α, also has been isolated (21). The expression of this isoform, like EF1α2, is confined to brain and skeletal muscle, again suggesting that there is a form of EF1 specific to these cells. It has been suggested that because increased expression of EF1α increases translational fidelity in yeast (22), EF1α2 may have a similar role in mammalian muscle and brain and that terminally differentiated cells have a requirement for enhanced translational fidelity (12). However, the more striking aspect that these cell types have in common is that they are required to survive throughout the lifespan of the organism. There are several lines of evidence to connect a decrease in translational fidelity, and down-regulation of EF1α in particular, to aging (23). Overexpression of EF1α in Drosophila has been shown to increase lifespan (24). It seems possible, therefore, that the function of tissue-specific translation elongations factors may be in some way to increase translational fidelity in cell types that are not being turned over in mature animals.
It remains to be seen whether Eef1a2 really does have a role in translation elongation, and if so whether synthesis of all proteins in affected tissues, or just a subset, is affected. However, the finding that wasted mice have a complete deficiency of Eef1a2 should assist enormously in establishing the precise role of Eef1a2 in mammals. We have now demonstrated that this putative tissue-specific variant of the translation elongation apparatus is essential for postnatal viability of the postweaning mouse.
Acknowledgments
We are grateful to A. Southwell, S. Ball, and V. Clarke for their excellent technical assistance. We thank A. Pilz and S. Malas for their contributions to the earlier mapping analysis of the wst gene. We thank A. Bird, D. Bonthron, N. Hastie, and I. Jackson for helpful discussions and critical reading of the manuscript.
ABBREVIATIONS
- EF1α
translation elongation factor 1 alpha
- EF1α2
translation elongation factor 1 alpha 2
- BAC
bacterial artificial chromosome
- RT
reverse transcription
- kb
kilobases
- IAP
intracisternal A particle
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ 223837).
References
- 1.Shultz L D, Sweet H O, Davisson M T, Coman D R. Nature (London) 1984;297:402–404. doi: 10.1038/297402a0. [DOI] [PubMed] [Google Scholar]
- 2.Lutsep H, Rodriguez M. J Neuropathol Exp Neurol. 1989;48:519–533. doi: 10.1097/00005072-198909000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Abbott C, Malas S, Pilz A, Pate L, Ali R, Peters J. Genomics. 1994;20:94–98. doi: 10.1006/geno.1994.1131. [DOI] [PubMed] [Google Scholar]
- 4.Inoue T, Aikawa K, Tezuka H, Kada T, Shultz L D. Cancer Res. 1986;46:3979–3982. [PubMed] [Google Scholar]
- 5.Nordeen S, Schaefer V, Edgell M, Hutchison C, III, Shultz L, Swift M. Mutat Res. 1984;140:219–222. doi: 10.1016/0165-7992(84)90081-2. [DOI] [PubMed] [Google Scholar]
- 6.Tezuka H, Inoue T, Noguti T, Kada T, Shultz L D. Mutat Res. 1986;161:83–90. doi: 10.1016/0027-5107(86)90102-8. [DOI] [PubMed] [Google Scholar]
- 7.Sweet H. Mouse News Lett. 1984;71:31. [Google Scholar]
- 8.Siracusa L D, Abbott C M, Morgan J L, Zuberi A R, Pomp D, Peters J. Mamm Genome. 1997;7:S28–S44. doi: 10.1007/s003359900314. [DOI] [PubMed] [Google Scholar]
- 9.Ann D, Moutsatsos I, Nakamura T, Lin H, Mao P, Lee M, Chin S, Liem R, Wang E. J Biol Chem. 1991;266:10429–10437. [PubMed] [Google Scholar]
- 10.Lund A, Knudsen S, Vissing H, Clark B, Tommerup N. Genomics. 1996;36:359–361. doi: 10.1006/geno.1996.0475. [DOI] [PubMed] [Google Scholar]
- 11.Hershey J W B. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, Francoeur A-M, Liu S, Wang E. J Biol Chem. 1992;267:24062–24068. [PubMed] [Google Scholar]
- 13.Knudsen S, Frydenberg J, Clark B, Leffers H. Eur J Biochem. 1993;215:549–554. doi: 10.1111/j.1432-1033.1993.tb18064.x. [DOI] [PubMed] [Google Scholar]
- 14.Lee S, LeBlanc A, Duttaroy A, Wang E. Exp Cell Res. 1995;219:589–597. doi: 10.1006/excr.1995.1268. [DOI] [PubMed] [Google Scholar]
- 15.Lee S, Wolfraim L, Wang E. J Biol Chem. 1993;268:24453–24459. [PubMed] [Google Scholar]
- 16.Lee S, Ann D, Wang E. Biochem Biophys Res Commun. 1994;3:1371–1377. doi: 10.1006/bbrc.1994.2336. [DOI] [PubMed] [Google Scholar]
- 17.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Guigo R, Knudsen S, Drake N, Smith T. J Mol Biol. 1992;226:141–157. doi: 10.1016/0022-2836(92)90130-c. [DOI] [PubMed] [Google Scholar]
- 19.Uberbacher E C, Mural R J. Proc Natl Acad Sci USA. 1991;88:11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aghajanian G, Bloom F. Brain Res. 1967;6:716–727. doi: 10.1016/0006-8993(67)90128-x. [DOI] [PubMed] [Google Scholar]
- 21.Pizzuti A, Gennarelli M, Novelli G, Colosimo A, Lo Cicero S, Caskey C T, Dallapiccola B. Biochem Biophys Res Commun. 1993;197:154–162. doi: 10.1006/bbrc.1993.2454. [DOI] [PubMed] [Google Scholar]
- 22.Song J M, Picologlou S, Grant C M, Firoozan M, Tuite M F, Liebman S. Mol Cell Biol. 1989;9:4571–4575. doi: 10.1128/mcb.9.10.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rattan S. Mutat Res. 1991;256:115–125. doi: 10.1016/0921-8734(91)90005-v. [DOI] [PubMed] [Google Scholar]
- 24.Shepherd J, Walldorf U, Hug P, Gehring W. Proc Natl Acad Sci USA. 1989;86:7520–7521. doi: 10.1073/pnas.86.19.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]