Abstract
Unique, small sequences (sequence tag sites) have been identified at the 3′ ends of most human genes that serve as landmarks in genome mapping. We investigated whether a single copy gene could be isolated directly from total human DNA by transformation-associated recombination (TAR) cloning in yeast using a short, 3′ unique target. A TAR cloning vector was constructed that, when linearized, contained a small amount (381 bp) of 3′ hypoxanthine phosphoribosyltransferase (HPRT) sequence at one end and an 189-bp Alu repeat at the other end. Transformation with this vector along with human DNA led to selective isolations of the entire HPRT gene as yeast artificial chromosomes (YACs) that extended from the 3′ end sequence to various Alu positions as much as 600 kb upstream. These YACs were retrofitted with a NeoR and a bacterial artificial chromosome (BAC) sequence to transfer the YACs to bacteria and subsequently the BACs to mouse cells by using a Neo selection. Most of the HPRT isolates were functional, demonstrating that TAR cloning retains the functional integrity of the isolated material. Thus, this modified version of TAR cloning, which we refer to as radial TAR cloning, can be used to isolate large segments of the human genome accurately and directly with only a small amount of sequence information.
Keywords: gene isolation, yeast artificial chromosome
Isolation of a single copy gene from a genome is a laborious process because it relies on characterization of random clones in libraries comprised of yeast or bacterial artificial chromosomes (YACs or BACs, respectively). Even if a YAC or BAC contains an entire gene, the specific recovery of the gene is an arduous process. Often a gene is available as a contig set of fragments that must be pieced together.
Recently we demonstrated through our direct cloning of the breast cancer genes BRCA2 and BRCA1 (ref. 1, and unpublished results) that transformation-associated recombination (TAR) in the yeast Saccharomyces cerevisiae can be used to selectively isolate single copy genes from total human DNA as large circular YACs. Independently from us the idea to use recombination in yeast for mammalian gene isolation has been proposed by Ketner et al. (2). TAR cloning is based on copenetration into yeast spheroplasts of gently isolated genomic DNA along with vector DNA that contains 5′ and 3′ sequences (hooks) specific for a gene of interest, followed by recombination between the vector and the human DNA to establish a YAC. Propagation of the YAC can occur if the human DNA contains sequences that can function as an origin of replication (ARS) in yeast. These sequences are common in human DNA with approximately one ARS-like sequence per 20–40 kb (ref. 3 and see references in ref. 4), suggesting that many human genes can be isolated by TAR cloning using vectors with two specific hooks.
The use of vectors with two specific targeting sequences has limitations for TAR cloning. First, some chromosomal regions may be unclonable because they lack yeast ARS-like sequences. Second, often there is only limited sequence information available, such as a 3′ end expressed sequence. In view of these limitations, we decided to determine whether a single copy gene could be isolated by TAR cloning directly from total human DNA when one of the two hooks was a single specific gene sequence, while the other was a commonly repeated sequence such as an Alu. Use of a common repeat as a second targeting sequence provides an additional opportunity to isolate various size regions extending from the specific 3′ hook to different upstream Alu positions so that there is a higher likelihood to obtain clones containing an ARS-like sequence (see Fig. 1). Because one of the ends is fixed, we refer to this approach as radial TAR cloning. For this study, we chose the human hypoxanthine phosphoribosyltransferase (HPRT) gene. It provided the opportunity to determine TAR cloning fidelity, because HPRT function can be assessed after transfection of the resulting clone into mouse cells (5–7).
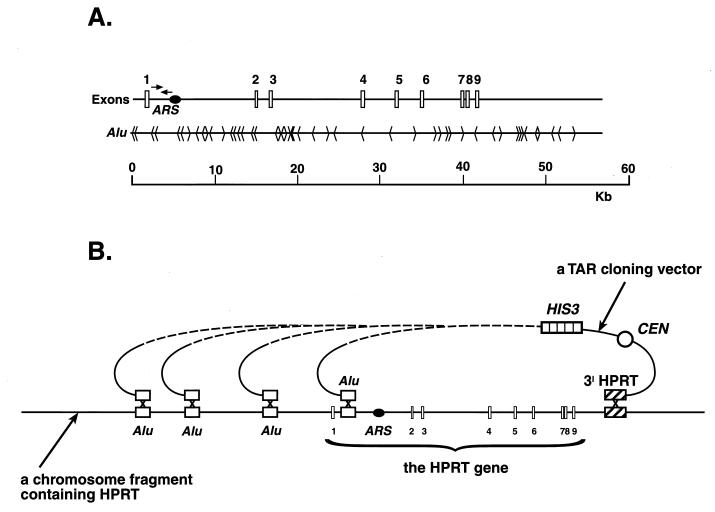
Figure 1.
Isolation of the human HPRT gene by radial TAR cloning. (A) The structure of the human HPRT gene (5). The gene contains nine exons. The position and orientation of 49 Alu sequences within the locus are shown in the context of the known ARS in intron 1. Arrows indicate the positions of primers used for initial identification of clones containing the HPRT gene. (B) A scheme of isolation of HPRT as a series of circular YACs using a TAR cloning vector containing a 3′ HPRT sequence and an Alu repeat. Yeast spheroplasts are transformed with genomic human DNA along with a TAR cloning vector containing the 3′ sequence (dotted box) and the Alu (see Materials and Methods) at the ends of the linearized plasmid. Recombination between the sequences in the vector and genomic DNA containing HPRT leads to the establishment of circular YACs that extend from the 3′ sequence to various Alu positions. Because the vector lacks an ARS, the only YACs that will be stably maintained are those that include human DNA fragments containing a yeast ARS-like sequence. In the present scheme, only YACs that include a fragment extending upstream from intron 1 can be propagated because that is the position of the first ARS-like sequence in the HPRT region extending from the 3′ hook (15). CEN corresponds to the yeast chromosome VI centromere, and HIS3 is a selectable marker.
MATERIALS AND METHODS
Yeast Strain and Transformation.
The highly transformable Saccharomyces cerevisiae strain VL6–48 (MATa, his3-Δ200, trp1-Δ1, ura3–52, lys2, ade2–101, met14) (8), which has HIS3 deleted was used for transformations. Spheroplasts that enable efficient transformation were generated by using a previously described protocol (1). Agarose plugs (100 μl) containing approximately 5 μg of high molecular weight human DNA were prepared from normal human fibroblasts MRC-5 (American Type Culture Collection) and used for yeast transformation (1, 8). Linearized TAR cloning vector (1 μg) was added to the DNA-containing plugs before treating with agarase and presented to spheroplasts. Yeast transformants were selected on synthetic complete medium plates lacking histidine.
Construction of TAR Cloning Vector.
The TAR circularizing vector pVC-HP1 containing a 3′ sequence of the human HPRT gene and an Alu sequence was constructed from the vector pVC-cdc27hs (CDC27-CEN6-HIS3-TEL) that was previously used for selective TAR cloning of human ribosomal DNA (9). The human CDC27 and the TEL sequences in pVC-cdc27hs were replaced by a 381-bp EcoRI–BamHI fragment corresponding to a 3′ sequence of HPRT (positions 53, 462–53,842 in the genomic sequence, accession number G184369) and a 189-bp XhoI–EcoRI fragment containing the Alu BLUR13 sequence (10). The 3′ HPRT hook lacked any human repeat elements or yeast ARS-like sequence based on sequence analysis. The 3′ targeting sequence is approximately 12 kb downstream of the 3′ end of mature mRNA of HPRT. The 3′ HPRT sequence was PCR-amplified from genomic DNA by using a pair of primers, HP3 (5′-CCGGAATTCCTCAGGTTAACGATATATTGTCAG-3′) and HP4 (5′-CGCGGATCCGTGTCAACCTTCCCAGCTCTTGG-3′). The HPRT hook was cloned into the TAR vector in the orientation shown in Fig. 1B. The vector pVC-HP1 was cut with EcoRI (the site is located between the hooks) before transformation to yield a linear molecule bounded by the HPRT and Alu hooks.
PCR Analysis.
Two pairs of primers were used for PCR characterization of YAC pools: IN1R and IN1L specific for intron 1 sequence and 46R and 47L specific for exon 2 of HPRT. IN1L (5′- CCCCATCAGCCTCTGGTATCTTAGC-3′) and IN1R (5′-AGCCAGCACCTCAGATATACA-3′) amplify a 516-bp sequence of intron 1 (11); 46L (5′-TGCTGGGATTACACGTGTGAACC-3′) and 47R (5′-GACTCTGGCTAGAGTTCCTTCTTCC-3′) amplify a 575-bp sequence of exon 2 along with flanking introns (12). Both PCR products are diagnostic for recombination between the TAR vector and 3′ region of the genomic HPRT. The presence of HPRT coding region in YAC clones was examined by PCR using nine pairs of the previously described primers for exons 1–9 (12). Yeast genomic DNA isolated from the transformants was amplified by using primers under the following standard PCR conditions: 50 mM KCl, 10 mM Tris⋅HCl, pH 9.0, 3.0 mM MgCl2, 0.2 mM dTTP, dCTP, dGTP and dATP in a final volume of 50 μl. Thermocycling conditions consisted of 35 cycles of 1 min at 94°C, 45 s at 55°C, and 5 min at 68°C, followed by one cycle of 10-min extension at 72°C in a 9600 Thermocycler (Perkin–Elmer).
Characterization of YAC Clones.
Chromosomal size DNAs from yeast transformants were separated by transverse alternating field electrophoresis (TAFE), blotted and hybridized with human DNA as previously described (8). To estimate the size of circular YACs, agarose DNA plugs were exposed to 5 krad from a Cs137 irradiator (8, 16) before TAFE analysis. At this dose, because many of the molecules receive only a single break, the size of the lagging band reveals the size of the circular DNA. To identify sequences upstream of HPRT, 1 μg of total DNA isolated from yeast transformants was digested to completion with SrfI in combination with different endonucleases. Samples were run by gel electrophoresis, transferred to a nylon membrane, and hybridized with a 516-bp intron 1 probe generated by PCR using the primers IN1L and IN1R (see above).
Retrofitting of YACs for Propagation in Bacterial and Mammalian Cells.
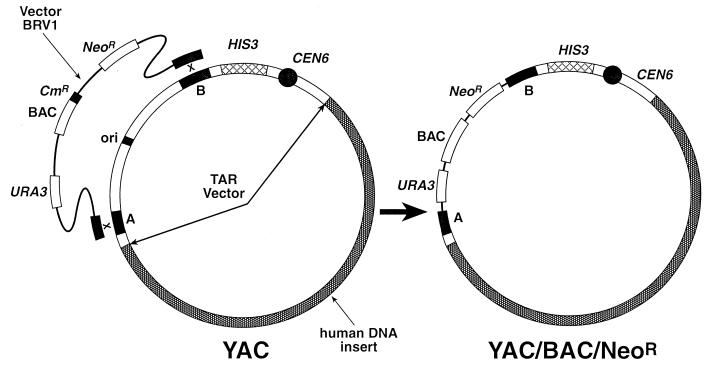
A new yeast-bacteria-mammalian cell shuttle vector, BRV1, was used for retrofitting the large circular YACs for propagation as BACs and subsequent transfection into mammalian cells using the selectable marker NeoR (1). The vector contains two short (approximately 300 bp each) targeting sequences, A and B, flanking the ColE1 origin of replication in the pRS303-based TAR cloning vectors (13). These targeting sequences are separated by an unique BamHI site. Recombination of the BamHI-linearized BRV1 vector with a YAC in yeast leads to replacement of the ColE1 origin of replication in the TAR cloning vector by a cassette containing the F factor origin of replication, the chloramphenicol acetyltransferase (CmR) gene, the NeoR gene, and the URA3 yeast selectable marker (Fig. 2). A standard lithium acetate transformation procedure was used for retrofitting of HPRT YACs. YAC retrofitting was highly efficient: more than 95% of Ura+ His+ transformants obtained with BRV1 contained retrofitted YACs. These constructs were moved to Escherichia coli by electroporation as described previously (1, 8). In brief, yeast chromosome-size DNAs were prepared in agarose plugs and, after melting and agarase treatment, the DNAs were electroporated into DH10B competent cells (GIBCO/BRL) by using a Bio-Rad Gene Pulser.
Figure 2.
Retrofitting of a circular YAC into a BAC containing the NeoR mammalian selectable marker. The retrofitting vector BRV1 contains two targeting sequences, A and B, flanking the ColE1 origin of replication in a TAR cloning vector used for a gene isolation. Recombination between the BamHI-linearized BRV1 vector and a YAC during yeast transformation leads to replacement of the ColE1 origin of replication in the TAR cloning vector by a cassette containing the F factor origin of replication (BAC), the chloramphenicol acetyltransferase (CmR) gene, the NeoR gene, and the URA3 yeast selectable marker.
Transfer of HPRT Containing BAC/YACs into Mouse Cells.
The NeoR/BAC/YAC DNAs were isolated from bacterial cells by using a standard alkaline lysis procedure, purified on Qiagen columns, and used for transfection into HPRT-deficient A9cl18 mouse cells using the NeoR gene as a selectable marker. Three methods of delivering BAC/YAC DNA into mammalian cells (electroporation, lipofection, and calcium phosphate precipitation) were used in this study. A9 cells (2 × 106) were electroporated at 300 volts and 960 μF capacitance with 10- 15 μg of purified DNA. Lipofection was performed on A9 cells by using the Life Technologies standard protocol with Lipofectamine. Conditions for calcium phosphate transfection were similar to those previously described (14) except only 1 μg of DNA was used and the precipitate was added to 5 × 105 cells for 6 hr. HPRT-positive clones were identified among neomycin-resistant clones (800 μg/ml G418, Life Technologies) by selection on DMEM medium supplemented with 10% fetal bovine serum, 100 μM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine (HAT selection).
RESULTS
Strategy for Radial TAR Cloning.
The radial TAR cloning scheme for isolation of a single copy gene from total genomic DNA (in this case the HPRT gene) is described in Fig. 1. A mixture of human DNA and a linearized TAR cloning vector is presented to yeast spheroplasts. The vector contains a yeast centromere (CEN6), a yeast selectable marker (HIS3), and a small 3′ fragment of HPRT as a specific targeting sequence at one end and an Alu repeat at the other end. A double recombination event involving (i) the gene-specific hook and a DNA fragment containing HPRT, and (ii) the Alu hook and one of the several Alus that are present in a large fragment (in the correct orientation and 5′ proximal) will result in a circular YAC (see Fig. 1B). The minimum size of YAC that can be cloned with this scheme is dictated by the position of the first ARS-like sequence proximal to the 3′ hook. YAC propagation depends on the presence of such a sequence, because the TAR vector lacks an ARS. For the HPRT gene the first ARS-like sequence is located in intron 1 (Fig. 1A). This scheme predicts a variety of YAC sizes with inserts that extend from the 3′ sequence to various Alu positions that are upstream to the first ARS-like sequence.
Radial TAR Cloning of the Human HPRT Gene.
A centromere-based yeast TAR vector, pVC-HP1 (hprt- HIS3-CEN6-Alu), was created in such a way that when linearized, one end contained 381 bp of the 3′ HPRT sequence approximately 12 kb downstream of the 3′ end of mature mRNA of HPRT whereas the other end contained 189 bp of an Alu sequence (see Materials and Methods). Six transformation experiments were carried out with freshly prepared yeast spheroplasts as previously described for TAR cloning of the human BRCA2 gene (1). Approximately 1,400 His+ colonies were obtained. (Using 5 μg of human DNA, 1 μg of vector and 2 × 109 spheroplasts, there were approximately 100–300 transformants per experiment.) To identify transformants containing the HPRT gene, 1,200 of the transformants were combined into 40 pools and examined by PCR. A pair of primers was used that identifies a sequence of intron 1 that is about 52 kb upstream of the 3′ HPRT hook (see Fig. 1A). All presumptive HPRT clones should contain this intron because it is the first yeast ARS-like sequence upstream from the hook (15). Seven pools were identified that yielded PCR products specific to intron 1. Individual clones containing the HPRT sequence were isolated from each pool, clones 1–7. Among these seven clones, at least six were independent, because they arose from different experiments.
Physical Analysis of YAC Clones Containing the HPRT Gene.
Several approaches were taken to establish the integrity and stability of the cloned material in the seven isolates. The presence of exons 1–9 of HPRT (12) was determined by PCR using primers for each exon. The PCR products of all exons in isolates 1–4 were identical to those obtained with total genomic DNA (Table 1). Isolates 5 and 6 lacked only exon 1. Based on the size of these YACs (see below), they probably arose through recombination between the Alu hook and one of the two Alus within the intron 1 sequence that is proximal to the ARS sequence (see Fig. 1A). The isolate 7 contained exons 2, 6, 7, 8, and 9 but lacked exons 1, 3, 4 and 5 sequences, suggesting that rearrangements occurred within the cloned fragment during its establishment.
Table 1.
Characterization of HPRT-positive YAC clones by PCR
| YAC isolate | Size, kb | Exons present
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7+8* | 9 | ||
| 1 | 80 | + | + | + | + | + | + | + | + |
| 2 | 100 | + | + | + | + | + | + | + | + |
| 3 | 120, 300† | + | + | + | + | + | + | + | + |
| 4 | 140, 250, 600† | + | + | + | + | + | + | + | + |
| 5 | 55 | − | + | + | + | + | + | + | + |
| 6 | 55 | − | + | + | + | + | + | + | + |
| 7 | ND | − | + | − | − | − | + | + | + |
ND, not determined.
The primers amplify exon 7 and exon 8 sequences along with short sequences of flanking introns (12).
The colony consists of a mixture of cells with different size YACs. All YACs contain the entire HPRT gene.
Genomic DNAs were isolated from the original transformants and analyzed by TAFE. As expected for a circular YAC (8), the HPRT hybridizing material was retained in the starting wells of the gel (data not shown). To estimate the size of the cloned material the agarose plugs were irradiated with a low dose of ionizing radiation (15). Based on TAFE analysis of the irradiated DNAs, isolates 2–4 contained circular YACs from 100 kb to 600 kb. Isolate 1 contained an 80-kb YAC. Isolates 5 and 6, which lacked exon 1, contained YACs that were 55 kb (Table 1). Because the size of the entire HPRT gene is about 60 kb (5), we concluded that at least four among seven isolates contain YACs that are bigger than the predicted HPRT gene.
To establish the stability of the cloned HPRT-containing fragments, original colonies of transformants were streaked, and 10 subclones of each isolate were characterized in terms of YAC size and presence of exons. For isolates 1, 2, 5, and 6, the YACs in each of 10 subclones were identical to the original isolates. Isolates 3 and 4 contained different size circular YACs (Table 2). Based on PCR analysis, the different size YACs contained an entire HPRT gene (data not shown). All YACs were mitotically stable and did not exhibit rearrangements during subsequent cell divisions. Because broken yeast chromosomes can pass several cell divisions (17), the presence of different size YACs in the original transformants may be explained by circularization of the targeted human chromosomal fragment after the first division of the transformed cell. Alternatively, small YACs may result from deletions of large YACs during their establishment.
Table 2.
Transfection of the NeoR retrofitted HPRT containing BAC/YACs into A9 mouse cells
| Size of BAC/YAC | Method of transfection | Frequency of NeoR transfections × 10−6 per μg BAC/YAC DNA | Number of NeoR transfectants expressing HPRT* |
|---|---|---|---|
| 80 kb | Lipofection | 38.0 | 10/10 |
| Electroporation | 0.2 | 0/2 | |
| Calcium phosphate | 0.4 | ND | |
| 100 kb | Lipofection | 36.0 | 10/10 |
| Electroporation | 0.01 | ND | |
| 120 kb | Lipofection | 50.0 | 10/10 |
| Electroporation | 1.0 | 5/5 | |
| Calcium phosphate | 16.0 | 5/5 | |
| 140 kb | Lipofection | 75.0 | 10/10 |
| Electroporation | 0.8 | 5/5 | |
| 110 kb (BRCA2)† | Lipofection | 35.0 | 0/10 |
ND, not determined.
*Capable to grow on hypoxanthine/aminopterin/thymidine medium.
Human BRCA2 YAC (1) was retrofitted into BAC/NeoR and used as a control in transfection experiments.
The YAC clones containing the entire HPRT gene were analyzed for the presence of the expected common sequences upstream of HPRT. The DNAs from isolates 1 (with an 80-kb YAC), 2 (with a 100-kb YAC), 3 (with 120- and 300-kb YACs) and 4 (with 140- and 250-kb YACs) were digested by SrfI in combination with different restriction endonucleases (NheI, SalI, and EcoRI), separated on TAFE gels and blot-hybridized with an intron 1 probe. Because the endonuclease SrfI cuts the HPRT gene within the intron 1 distal to the sequence used for probing, the visualized bands consist of the 5′ HPRT sequence plus a sequence upstream of HPRT (up to the first endonuclease recognition site). Identical fragments were identified for each endonuclease digestion for all these YACs (data not shown). Because the bands were identical to those obtained with genomic DNA and all exons were present, we concluded that the radial TAR cloning enabled the isolation of chromosomal fragments that extended from the 3′ end HPRT sequence to various Alu positions as much as several hundred kbs upstream of the HPRT gene.
A TAR-cloned human HPRT gene is functional. The four different size YAC isolates containing the entire HPRT sequence were examined for the presence of functional copies of HPRT. To do this, they were first retrofitted by recombination with the vector BRV1 that contained a NeoR marker and sequences that would enable subsequent propagation as a BAC (see Fig. 2). These BAC/YACs then were transferred to E. coli by electroporation, as described in Materials and Methods. Based on inter Alu PCR patterns and the presence of exons (data not shown), the HPRT genes of the original isolates remained unchanged in the BAC/YAC/NeoR derivatives. The BAC/YACs were isolated and transfected into HPRT-deficient mouse cells using the NeoR gene as a selectable marker and then subsequently were tested for HPRT function.
Three methods were examined for the introduction of the BAC/YACs into mouse cells: electroporation, lipofection, and calcium phosphate precipitation. As shown in Table 2, lipofection was the most efficient, yielding ≈5 × 10−5 neomycin-resistant transfectants per μg of BAC/YAC DNA. The efficiency was much lower when using the other procedures. Among 62 of the NeoR transfectants obtained with the various methods, nearly all were able to grow in hypoxanthine/aminopterin/thymidine-containing medium. Thus, the radial TAR cloning approach in yeast resulted in the direct isolation of the complete human gene whose structural and function integrity was retained.
DISCUSSION
We have described the isolation of the entire single copy gene HPRT from total human DNA by TAR cloning using a 3′ end-specific targeting sequence along with a commonly occurring repeat. This radial TAR cloning method is highly selective and results in nearly 1% of the yeast transformants containing the HPRT sequence. The enrichment was comparable to that previously observed for TAR cloning with vectors containing two gene-specific sequences (ref. 1 and unpublished results). This finding at first may seem surprising; however, the actual mechanism(s) of interaction between a TAR vector and potentially cloned regions has not yet been determined. For example, as suggested earlier, recombination may be preferred between the vector and the ends of fragments (4), in which case TAR cloning would be efficient only for those fragments with a 3′ HPRT sequence near one end.
Although in this study we used an Alu sequence as a nonspecific hook, other types of repetitive sequences also can be used for radial TAR cloning. Use of less frequent repeats, such as LINEs, can increase likelihood of cloning of larger regions.
The modified TAR cloning procedure for gene isolation has many advantages compared with the previously described TAR cloning that was based on the use of two specific targeting sequences. Use of a common repeat as a second targeting sequence excludes the requirement for the presence of a yeast ARS-like sequence in the specific region to be isolated. A gene that lacks an ARS sequence can be isolated because radial TAR cloning allows adjacent sequences that contain an ARS to be included in the isolated fragment. Because yeast ARS-like sequences are quite frequent in the human genome (3), and fragments up to 600 kb can be cloned by TAR as circular molecules (8), most genes are accessible by this method. We note that replacement of an Alu repeat by a B1 repeat in the TAR vector allows us to apply the radial TAR cloning for a single copy gene isolation from mouse genome (unpublished data).
Recently we have demonstrated that the size of the HPRT-specific hook can be reduced up to 200 bp at least without affecting on efficiency of the gene isolation by radial TAR cloning (unpublished data). Thus the method also could be applied for isolation of chromosomal regions by using sequence tag sites (STS) information. Because approximately one STS has been identified per 100 kb of the human genome, radial TAR cloning could simplify the mapping and sequencing of the human genome. We emphasize the radial nature of this cloning scheme, because by simply changing the arrangement of the targeting hook, it is possible to clone large regions in both directions from the STS. We also propose that radial TAR cloning can be used for direct chromosome walking. For example, once a region is isolated, sequence that is distal to an STS could be used in subsequent TAR cloning to get the next adjacent region in the chromosome.
Our results demonstrate that TAR cloning provides an efficient means for the accurate isolation of genome material as YACs, unlike previous methods for developing YACs where chimeras and other artifacts are a common problem (18, 19). In this report we have shown that radial TAR cloning provides for the isolation of a complete gene that exhibits both structural and functional integrity. Previously, we have demonstrated that the seven BRCA2 and five BRCA1 clones isolated by TAR cloning were complete based on physical analysis (ref. 1 and unpublished results). Thus, for most genes complete copies can be isolated directly because of the opportunity to isolate rapidly many putative copies of the gene. The ease of isolation of many copies of a gene by TAR cloning is particularly valuable, because it may be difficult to obtain a complete gene in a library. Before the TAR cloning approach, no complete BRCA1 or BRCA2 genes had been identified in any YAC or BAC library. Many features may contribute to accurate recovery of genomic material by TAR cloning. For example, there are no restriction or ligation steps and handling of human DNA is greatly reduced. It is also possible that circular TAR cloning provides greater stability to the cloned DNA because there is no need to establish telomeres on the cloned material.
We have demonstrated that after retrofitting, circular YACs containing a NeoR marker can be efficiently and accurately transferred into mouse cells. Nearly all NeoR transfectants contained a functional HPRT. Thus TAR cloning provides the opportunity to study expression of isolated genes and to characterize control sequences even when they are fairly far from the coding region of the gene. As discussed earlier (9), TAR cloning also provides the opportunity to isolate families of genes and genes with diverged sequences, because recombination during transformation in yeast is efficient even between highly diverged sequences (20, 21).
We note that the strategy of gene isolation described above could be applied to many organisms, because yeast ARS-like sequences are frequent in eukaryotic genomes (3). Because the entire isolation procedure, including PCR characterization, requires only 2–3 weeks, TAR cloning provides a powerful tool for genomes studies.
Acknowledgments
We gratefully acknowledge the critical reading of the manuscript with S. Bennett. We appreciate the technical assistance of Eric Jenkins in some experiments. Support was provided in part by an interagency grant (1-YO2-HG-60021-01) from the National Institutes of Health Center for Human Genome Research and in part by a Cooperative Research and Development Agreement (CRADA) with Life Technologies, Inc.
ABBREVIATIONS
- TAR
transformation-associated recombination
- YAC
yeast artificial chromosome
- HPRT
hypoxanthine phosphoribosyltransferase
- BAC
bacterial artificial chromosome
- TAFE
transverse alternating field electrophoresis
References
- 1.Larionov V, Kouprina N, Solomon G, Barrett J C, Resnick M A. Proc Natl Acad Sci USA. 1997;94:7384–7387. doi: 10.1073/pnas.94.14.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ketner G, Spencer F, Tugendreich S, Connelly C, Hieter P. Proc Natl Acad Sci USA. 1994;91:6186–6190. doi: 10.1073/pnas.91.13.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stinchomb D T, Thomas M, Kelly I, Selker E, Davis R W. Proc Natl Acad Sci USA. 1980;77:4559–4563. doi: 10.1073/pnas.77.8.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larionov V, Kouprina N, Graves J, Chen X-N, Korenberg J R, Resnick M A. Proc Natl Acad Sci USA. 1996;93:491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards A, Voss H, Rice P, Civitello A, Stegemann J, Schwager C, Zimmermann J, Erfle H, Caskey C T, Ansorge W. Genomics. 1990;6:593–608. doi: 10.1016/0888-7543(90)90493-e. [DOI] [PubMed] [Google Scholar]
- 6.Huxley C, Hagino Y, Schlessinger D, Olson M V. Genomics. 1991;9:742–750. doi: 10.1016/0888-7543(91)90369-p. [DOI] [PubMed] [Google Scholar]
- 7.Wada M, Ihara Y, Tatsuka M, Mitsui H, Kohno K, Kuwano M, Schlessinger D. Biochem Biophys Res Commun. 1994;200:1693–1700. doi: 10.1006/bbrc.1994.1647. [DOI] [PubMed] [Google Scholar]
- 8.Larionov V, Kouprina N, Graves J, Resnick M A. Proc Natl Acad Sci USA. 1996;93:13925–13930. doi: 10.1073/pnas.93.24.13925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kouprina N, Graves J, Cancilla M, Resnick M A, Larionov V. Gene. 1997;197:269–276. doi: 10.1016/s0378-1119(97)00271-0. [DOI] [PubMed] [Google Scholar]
- 10.Pavan W J, Hieter P, Reeves R H. Mol Cell Biol. 1990;10:4163–4169. doi: 10.1128/mcb.10.8.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sternlicht M, Mirell C, Safarians S, Barsky S H. Biochem Biophys Res Commun. 1994;199:511–518. doi: 10.1006/bbrc.1994.1258. [DOI] [PubMed] [Google Scholar]
- 12.Steingrimsdottir H, Rowley G, Dorado G, Cole J, Lehmann A R. Nucleic Acids Res. 1992;20:1201–1208. doi: 10.1093/nar/20.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomassen D G, Gilmer T M, Annab L A, Barrett J C. Cancer Res. 1985;45:726–732. [PubMed] [Google Scholar]
- 15.Sykes R C, Lin D, Hwang S J, Framson P E, Chinault A C. Mol Gen Genet. 1988;212:301–309. doi: 10.1007/BF00334700. [DOI] [PubMed] [Google Scholar]
- 16.Game J C, Sitney K C, Cook V E, Mortimer R K. Genetics. 1989;123:695–713. doi: 10.1093/genetics/123.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandell L L, Zakian V. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 18.Green E D, Riethman H C, Dutchik J E, Olson M V. Genomics. 1991;11:658–669. doi: 10.1016/0888-7543(91)90073-n. [DOI] [PubMed] [Google Scholar]
- 19.Neil D L, Villasante A, Fisher R B, Vetrie D, Cox B, Tyler-Smith C. Nucleic Acids Res. 1990;18:1421–1428. doi: 10.1093/nar/18.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mezard C, Pompon D, Nicolas A. Cell. 1992;70:659–670. doi: 10.1016/0092-8674(92)90434-e. [DOI] [PubMed] [Google Scholar]
- 21.Larionov V, Kouprina N, Eldarov M, Perkins E, Porter G, Resnick M A. Yeast. 1994;10:93–104. doi: 10.1002/yea.320100109. [DOI] [PubMed] [Google Scholar]