Abstract
The period (per) gene in Drosophila melanogaster provides an integral component of biological rhythmicity and encodes a protein that includes a repetitive threonine-glycine (Thr-Gly) tract. Similar repeats are found in the frq and wc2 clock genes of Neurospora crassa and in the mammalian per homologues, but their circadian functions are unknown. In Drosophilids, the length of the Thr-Gly repeat varies widely between species, and sequence comparisons have suggested that the repeat length coevolves with the immediately flanking amino acids. A functional test of the coevolution hypothesis was performed by generating several hybrid per transgenes between Drosophila pseudoobscura and D. melanogaster, whose repetitive regions differ in length by about 150 amino acids. The positions of the chimeric junctions were slightly altered in each transgene. Transformants carrying per constructs in which the repeat of one species was juxtaposed next to the flanking region of the other were almost arrhythmic or showed a striking temperature sensitivity of the circadian period. In contrast, transgenes in which the repeat and flanking regions were conspecific gave wild-type levels of circadian rescue. These results support the coevolutionary interpretation of the interspecific sequence changes in this region of the PER molecule and reveal a functional dimension to this process related to the clock’s temperature compensation.
Keywords: per, coevolution, Thr-Gly, repeat, circadian
Genes that are essential circadian clock components have been identified at the molecular level in Drosophila (per and tim), Neurospora (frq, wc-1, and wc-2), and the mouse (Clock) (reviewed in ref. 1). per, frq, and wc-2 encode regions with repetitive sequences, which include runs of Thr-Gly or Ser-Gly dipeptides (2–4). Putative mammalian homologues of the fly per gene also encode similar repeats (5–8). The function of these repeats is unknown, but structural studies of (Thr-Gly)n dipeptides reveal that they generate β-turn conformations that are extremely flexible and dynamic when challenged with different temperatures and polarities (9). In addition, the Thr-Gly repeats in Drosophila melanogaster are polymorphic in length and are distributed as a significant latitudinal cline in Europe, with high levels of the shorter length variants distributed predominantly in the south (10). This suggests that natural selection might be predisposing the length alleles to different types of environments. Statistical analyses of the Thr-Gly and surrounding DNA sequences with a variety of models designed to reveal the signature of natural selection have suggested that in both D. melanogaster (11) and Drosophila simulans (12) the frequencies observed of different natural Thr-Gly haplotypes are not consistent with the expectations of neutrality or drift. In addition, interspecific sequence analyses of the repetitive regions of over a dozen Drosophila species have suggested that the immediate flanking regions of the repeat coevolve with repeat length (13, 14). The evolutionary dynamics being played out in this region of the PER molecule suggest that the repeat “domain” may play a functional role.
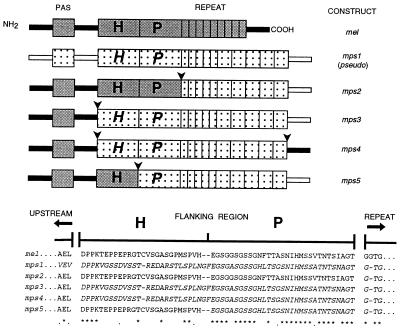
To perform an experimental test for the proposed coevolution, we have generated a number of hybrid per genes between two species that have very different repeat lengths. We juxtaposed the repeat of one species next to the flanking region of the other, thereby breaking up the suspected coevolutionary interaction between the two regions. It would be predicted that this might have some effect on circadian behavior. A potential problem is that selection can act on fitness increments that are as small as the inverse of the effective population size (15), about 106 in D. melanogaster (16). Therefore, any disruption of the proposed coevolution between the two regions may be phenotypically undetectable given the limitations of laboratory experiments. Nevertheless, we selected two per genes, one from Drosophila pseudoobscura (17) and the other from D. melanogaster (2); the former has the longest repetitive region of all per genes so far identified. It consists of about 10 copies of a degenerate Thr-Gly motif to which is added a pentapeptide cassette, which has been derived by slippage from the dipeptide Thr-Gly repeat (14). There are 30–35 copies of this pentapeptide in different D. pseudoobscura strains (18), giving the repetitive region a length in excess of 200 amino acids (13, 14, 17, 18). In comparison, the D. melanogaster repetitive region is composed of about 20 pairs of Thr-Gly, although different strains also have different repeat copy number (18). Fig. 1 illustrates the Thr-Gly repeat and its proposed coevolving 5′ flanking sequences (13, 14), which are labeled as block P and consist of amino acids 665–695 in the D. melanogaster sequence (2). Block H (amino acids 639–664) cannot be aligned between Drosophila species (13, 14, 17) but may nevertheless represent additional sequences that could be involved in the putative interaction between the repeat and the immediately adjacent region. The 40-amino acid region immediately upstream of block H (amino acids 598–638) does not appear to coevolve with interspecific repeat length (14).
Figure 1.
(Top) per transgenes of D. melanogaster N- and C-terminal sequences (filled bars) D. pseudoobscura (open bars). The approximate positions of the N-terminal PAS domain (39) are shown as filled or speckled boxes for the two species. D. melanogaster sequences in blocks H (amino acids 639–664, ref. 2 for numbering) and P (amino acids 665–695) are in gray, and corresponding sequences in D. pseudoobscura are speckled and italicized. Thr-Gly repeats of D. melanogaster (amino acids 696–737) and D. pseudoobscura are shown as narrow repetitive units, 9 for D. melanogaster, representing the 20 pairs of Thr-Gly dipeptides, and 5 for D. pseudoobscura, representing the 10 imperfect dipeptide repeats in this species (13, 14). The 5 broader repeat units in D. pseudoobscura represent the 30 copies of the pentapeptide encoding sequence (see refs. 13 and 14). Arrows show positions of chimeric junctions. (Bottom) Coevolving amino acid blocks (marked H and P, see text). A few amino acids N-terminal to block H and the first few residues of the Thr-Gly repeat (C-terminal to block P) are shown. The length of this repeat is 66 amino acids in D. melanogaster and 209 amino acids in D. pseudoobscura (13). Asterisks indicate identical amino acids, and dots indicate conservative substitutions. D. melanogaster amino acids are shown in roman type, and D. pseudoobscura sequences are shown in italic type.
Four chimeric per transgenes were created (mps2–5; see Fig. 1) by using sequences from D. melanogaster and D. pseudoobscura. All hybrid per genes except mps4 encode the 5′ half of D. melanogaster and the 3′ half of D. pseudoobscura per (Fig. 1). The position of the chimeric junction between the coding regions of the two species was manipulated (Fig. 1). The repeat and its immediate flanking regions H and P were made conspecific for D. pseudoobscura (chimeric constructs mps3 and mps4), or the repeat was heterospecific compared with D. melanogaster flanking regions H and P (construct mps2), or the repeat and region P were conspecific for D. pseudoobscura but heterospecific with regard to D. melanogaster region H and all upstream per sequences (mps5). Our results reveal striking phenotypic differences between the transformants, which supports the idea of an intragenic coevolution between the repeat and the flanking regions.
MATERIALS AND METHODS
Generation of Chimeric per Genes.
The mel transgene consists of the 13.2-kb per transcription unit from D. melanogaster inserted into the cp20.1 transformation vector marked with rosy+ and has been described previously (2). This fragment was also inserted into the pW8 transformation vector marked with white+ (19), and two further independently transformed mel lines were generated. mps1 has also been described previously (20). It contains the complete coding sequence of the D. pseudoobscura per gene fused to the 5′ upstream region of D. melanogaster at a position close to the 3′ end of the large first intron. These sequences are also carried within the cp20.1 transformation vector. To generate the novel mps2–5 chimeric transgenes, PCR strategies were used. The D. melanogaster primers are represented with nucleotide numbering as in ref. 2 and D. pseudoobscura primers with numbers from the amino acid sequence from ref. 17. Transgenes mps3–5 were inserted into pW8 and were generated in Leicester, whereas mps2 was inserted into cp20.1 and made at Brandeis.
mps2.
A 3′ junction primer, 5′-CGTGACCGTACCAGTGCCAGTGCCGGCAATGCTCG-3′, with the D. pseudoobscura sequence (amino acids 659–664) and D. melanogaster sequence (amino acids 5090–5107) was used with an upstream D. melanogaster 5′ primer, 5′-AAAGAGCTCGATCCGCCCAAAACG-3′ (SstI site underlined). Included were three extra As that were digested before ligation. The D. pseudoobscura 3′ primer, 5′-AAATCTAGAGTTATCGGCTCG-3′, also had three extra As, which were digested before ligation. The chimeric SstI-XbaI fragment obtained (21) was sequenced for errors and then ligated to the XbaI-EcoRI 3′ D. pseudoobscura fragment. This fragment was then inserted in front of the relevant SstI site of the cp20.1 vector carrying the 5′ D. melanogaster BamHI-SstI fragment to give the final mps2 clone. The mps2 construct was confirmed with diagnostic restriction analysis and sequencing of the chimeric junction.
mps3.
A 5′ D. pseudoobscura primer, 5′-CACCCGTGGAGCTCGACCCG-3′ (amino acids 619–638, SstI site underlined, mismatch bold), was used with a 3′ D. pseudoobscura primer 5′-TTCTCCATCTCGTCGTTGTG-3′ (amino acids 878–884) to generate a 0.6-kb fragment, which was sequenced for errors and cut with SstI and XbaI. This was ligated to a D. pseudoobscura 2.4-kb XbaI-EcoRI fragment reconstituting the 3′ D. pseudoobscura sequences. A BamHI-SstI D. melanogaster per fragment representing the 5′ part of the construct was ligated to the D. pseudoobscura SstI-EcoRI fragment generating the mps3 transgene in pW8. Diagnostic restriction analysis and DNA sequencing of the chimeric junction confirmed the correct construction of the chimeric gene.
mps4.
The mps3 0.6-kb D. pseudoobscura fragment above was coamplified with a D. melanogaster fragment with a 5′ primer, 5′-AAGCACAACGACGAGATGGA-3′ (amino acids 5333–5352), and a 3′ primer, 5′-GCTACGCCTGTTCCGGATCC-3′ (amino acids 5627–5646, BamHI site underlined). The D. pseudoobscura 3′ primer and the D. melanogaster 5′ primer are complementary. Further amplification of the initial products in the presence of the external 5′ and 3′ primers generates a chimeric fragment, which was restricted and used to replace the corresponding D. melanogaster SstI-BamHI fragment in pW8.
mps5.
A D. pseudoobscura fragment was generated with a 5′ primer, 5′-GAGGGCAGTGGCGCCAGTGG-3′ (amino acids 628–634), and 3′ primer, 5′-TTCTCCATCTCGTCGTTGTG-3′ (amino acids 878–884). A D. melanogaster product was amplified with 5′ primer, 5′-AACTATAACGAGAACCTGCT-3′ (4874–4893), and 3′ primer, 5′-CCACTGGCGCCACTGCCCTCGTGGACGGGACT-3′ (italics, complementary to amino acids 628–634, 5002–5013). The two fragments, the 5′ D. melanogaster and 3′ D. pseudoobscura primers, amplify a chimeric fragment, which was sequenced for errors and digested with SstI and XbaI to give a 0.6-kb fragment, which replaces the corresponding fragment in mps3.
P-Element Transformation and Behavioral Analysis.
The four hybrid genes and two further per constructs carrying the cloned parental D. melanogaster (mel) and D. pseudoobscura (mps1) coding sequences were transformed into the appropriate hosts by using standard methods (22). per01 males carrying an autosomal copy of the transgene were examined with respect to their circadian locomotor activity under free running conditions (DD, constant darkness) at 18, 25, and 29°C. The activity of each fly was analyzed by using autocorrelation (23) and a high resolution spectral analysis (24, 25). Flies were monitored for 7 days in constant darkness (DD) after a previous entrainment period in a light/dark 12:12 cycle (LD 12:12). Data were collected in 30-min bins in an automated event recorder similar to ones described previously (26). Transformants were reared at 25°C in an LD 12:12 cycle, and monitoring was carried out at 18, 25, or 29°C after 2 days of acclimatization in LD conditions. Data collection began 18 h after the last light to dark transition. The data were fed into the autocorrelation procedure of the Statistical Programs for the Social Sciences (spss, Chicago) statistical package. Significant rhythmicity in an autocorrelogram was one where the correlation coefficient (r) itself showed cycling, and the peak was equal or greater than the 95% confidence limits (2/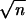 ). The spectral analysis (24), which gave a more accurate estimate of the period as well as confirming or rejecting the significance of the autocorrelation, was also employed. Monte-Carlo simulations were used to generate approximate 95 and 99% confidence limits by randomizing the data for each fly 100 times and performing spectral analyses on these data (27). Autocorrelograms and spectrograms were given a numerical code and judged as being rhythmic or arrhythmic on the basis of a blind assessment of their correlogram by three independent assessors (A.A.P., J.M.H., and C.P.K.). If a record gave a significant period with autocorrelation, but not spectral analysis, or vice versa, then the record was judged “arrhythmic.”
). The spectral analysis (24), which gave a more accurate estimate of the period as well as confirming or rejecting the significance of the autocorrelation, was also employed. Monte-Carlo simulations were used to generate approximate 95 and 99% confidence limits by randomizing the data for each fly 100 times and performing spectral analyses on these data (27). Autocorrelograms and spectrograms were given a numerical code and judged as being rhythmic or arrhythmic on the basis of a blind assessment of their correlogram by three independent assessors (A.A.P., J.M.H., and C.P.K.). If a record gave a significant period with autocorrelation, but not spectral analysis, or vice versa, then the record was judged “arrhythmic.”
RESULTS AND DISCUSSION
All four lines of the D. melanogaster per+ transgene mel rescue rhythms in a high percentage of per01 hosts with periods in the circadian 24–25-h range at all three temperatures (Table 1). The D. pseudoobscura per coding region (mps1) is not as effective in rescuing per01 behavior, yet 50–75% of the transformants show significant rhythmicity at 18 and 25°C (see Fig. 2 and Table 1) with periods of ca. 28 h. These periods are stable, even at the highest temperature, when only 15% of the transformants are rhythmic. Previous results with the same D. pseudoobscura (mps1) construct (20) found fewer rhythmic individuals at 25°C, but this was because of the use of a less sensitive statistical measure of rhythmicity (20, 23, 24). The mps2 construct (three lines) generated a small percentage of weakly rhythmic flies, barely above the level found for per01 mutants (Table 1; Figs. 2 and 3). Thus in mps2 transformants, where the repeat of D. pseudoobscura lies directly adjacent to the heterospecific 5′ flanking material of D. melanogaster (Fig. 1), per gene function is severely disrupted. In contrast, the mps3 gene (two lines) produced excellent rescue with 75–95% of transformants showing statistically significant rhythms and with mean periods in the 23–25-h range at all temperatures (Figs. 2 and 3). In mps3 the repeat of D. pseudoobscura lies next to its conspecific 5′ flanking sequences (blocks H and P in Fig. 1).
Table 1.
Free running circadian locomotor activity periods of various per transformants at different temperatures based on spectral analysis (24)
| Line | 18°C
|
25°C
|
29°C
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| N | n | Period, h | N | n | Period, h | N | n | Period, h | |
| mel | |||||||||
| 2a | 61 | 41 | 24.6 ± 0.1 | 25 | 23 | 25.0 ± 0.1 | 53 | 45 | 24.8 ± 0.1 |
| 17a | 59 | 31 | 24.1 ± 0.2 | 24 | 20 | 24.3 ± 0.7 | 46 | 26 | 24.2 ± 0.2 |
| 34a | 51 | 30 | 24.2 ± 0.2 | 47 | 39 | 25.0 ± 0.1 | 32 | 32 | 24.9 ± 0.2 |
| 116a | 50 | 41 | 23.5 ± 0.1 | 33 | 30 | 25.5 ± 0.1 | 33 | 31 | 25.4 ± 0.2 |
| Pooled | 221 | 143 | 24.1 ± 0.1 | 129 | 112 | 25.0 ± 0.1 | 164 | 134 | 24.9 ± 0.1 |
| mps1 | |||||||||
| I20 | 26 | 17 | 29.4 ± 0.8 | 31 | 18 | 28.3 ± 1.2 | 61 | 13 | 29.0 ± 1.2 |
| I26 | 25 | 21 | 27.4 ± 0.6 | 37 | 17 | 28.9 ± 0.8 | 82 | 8 | 29.5 ± 1.0 |
| Pooled | 51 | 38 | 28.3 ± 0.5 | 68 | 35 | 28.6 ± 0.6 | 143 | 21 | 29.2 ± 0.8 |
| mps2 | |||||||||
| 6 | 31 | 4 | 22.7 ± 4.1 | 61 | 5 | 33.0 ± 1.6 | 65 | 5 | 29.8 ± 1.6 |
| 9 | 28 | 6 | 24.6 ± 2.1 | 29 | 8 | 28.6 ± 2.0 | 39 | 7 | 28.6 ± 2.7 |
| 22 | 28 | 11 | 27.5 ± 2.2 | 68 | 8 | 28.1 ± 2.7 | 34 | 5 | 28.8 ± 2.4 |
| Pooled | 87 | 21 | 25.8 ± 1.5 | 158 | 21 | 29.5 ± 1.3 | 138 | 17 | 29.0 ± 1.3 |
| mps3 | |||||||||
| 65c | 19 | 15 | 24.1 ± 0.1 | 44 | 42 | 25.7 ± 0.1 | 15 | 13 | 25.6 ± 0.2 |
| 67a | 22 | 15 | 23.3 ± 0.2 | 32 | 32 | 24.4 ± 0.1 | 12 | 12 | 25.0 ± 0.2 |
| Pooled | 41 | 30 | 23.7 ± 0.1 | 76 | 74 | 25.1 ± 0.1 | 27 | 25 | 25.3 ± 0.2 |
| mps4 | |||||||||
| 6f | 17 | 15 | 24.3 ± 0.1 | 23 | 30 | 24.8 ± 0.1 | 66 | 62 | 24.8 ± 0.1 |
| 16b | 27 | 16 | 23.9 ± 0.1 | 20 | 17 | 25.1 ± 0.2 | 24 | 21 | 24.7 ± 0.1 |
| Pooled | 44 | 31 | 24.1 ± 0.1 | 43 | 37 | 25.0 ± 0.1 | 90 | 83 | 24.8 ± 0.1 |
| mps5 | |||||||||
| 23 | 25 | 20 | 24.0 ± 0.1 | 68 | 65 | 27.2 ± 0.1 | 51 | 48 | 28.3 ± 0.2 |
| 25 | 67 | 47 | 24.9 ± 0.3 | 73 | 48 | 30.9 ± 0.7 | 97 | 74 | 38.9 ± 1.0 |
| Pooled | 92 | 67 | 24.6 ± 0.2 | 141 | 113 | 28.8 ± 0.4 | 148 | 122 | 34.7 ± 0.8 |
| per01 | 31 | 5 | 22.6 ± 1.1 | 32 | 3 | 19.3 ± 1.9 | 57 | 4 | 21.5 ± 2.1 |
Results based on autocorrelation-derived periods are almost identical (data not shown, but see Fig. 2). Two-way ANOVA omitting the largely arrhythmic mps2 transformants revealed significant effects of temperature (T), genotype (G), and T × G interaction (P < 0.001 in all cases). The Newman–Keuls a posteriori procedure revealed significant changes in period over temperature for both lines of the mps5 transformants (P < 0.002 for both lines). N, number of males examined; n, number of significant rhythmic males. Results are mean ± SEM.
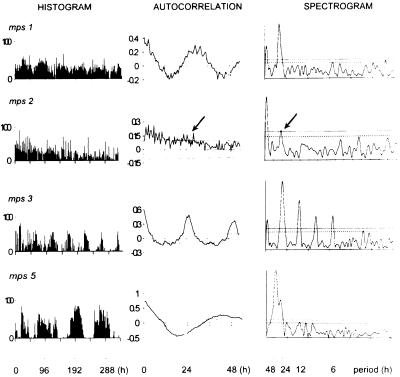
Figure 2.
Free running locomotor activity histograms, autocorrelation, and spectral analysis for four male per01 transformants monitored at 29°C carrying the mps1, mps2, mps3, and mps5 transgenes. Histograms give raw activity events in 30-min bins. The correlograms show r and 95% confidence limits either side of r = 0. Spectrograms show 95% (dashed, Lower) and 99% (dotted, Upper) confidence limits (27). From top to bottom are shown: mps1, 28-h cycle; mps2, one of few males rhythmic by our criteria (see Materials and Methods), and barely so, period ca. 26 h (arrow); mps3, typical of mel and mps4 transformants (not shown), period ca. 24.5 h; mps5, >45-h period.
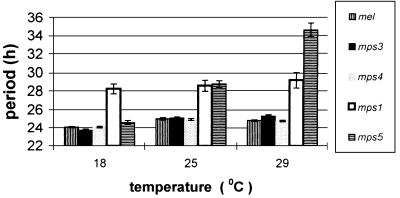
Figure 3.
Pooled mean (±SEM) free running locomotor activity periods for the different transformants at 18, 25, and 29°C. Data for the individual lines are in Table 1. mps5 gives a dramatic increase in period at higher temperatures. mps2 results are not shown as most transformants were arrhythmic, and those that were rhythmic showed a wide range of periodicities (see Table 1).
The dramatic phenotypic differences observed between the mps2 and mps3 transformants strongly support the idea of an intragenic coevolution between the length of the repeat and the immediate flanking upstream ≈60 amino acids in blocks H and P (13, 14). In addition, the mps5 transgene generated a phenotype intermediate between mps2 and mps3, in that both lines showed robust rhythms, but the periods were extremely temperature-dependent, remarkably so in line 25 (Table 1; Fig. 2). The phenotype displayed by these mps5 transgenic flies is reminiscent of the classic perL1 mutation, which also gives a temperature-sensitive lengthening of period (26, 28). However, even mps5 transformant line 23, which gives the least dramatic phenotype of the two (Table 1), shows a larger change in period than that reported in perL1 between 17 and 25°C (28). These mps5 results confirm that the unalignable amino acids in block H must also play a role in the coevolutionary interaction between the flanking region and the repeat, and suggest that these residues are under positive natural selection rather than drift. The mps4 transgene produced essentially wild-type rhythms in terms of strength of rescue, period length, and period stability over temperature, revealing that a D. pseudoobscura repeat plus its flanking sequences represents a “coevolved” module, which can function perfectly well within an otherwise D. melanogaster PER protein.
Within the per genes of Diptera, the Thr-Gly encoding hexamer ACNGGN has mutated to give both perfect and imperfect repetitive motifs of various lengths (13, 14, 17, 18). D. pseudoobscura represents the species with the longest repetitive region that has been identified to date and contains both the Thr-Gly dipeptide and a related pentapeptide (13, 14, 17, 18). Secondary structure predictions of the repetitive regions in a large number of Drosophila species suggest that, irrespective of sequence composition, the amino acids in these regions generate a series of flexible turns (13, 18). Conformational analysis of poly(Thr-Gly) peptides, the major component of the D. melanogaster repeat, reveals that Thr-Gly repeats generate type II or type III β-turns (9). Consequently, it would appear that it is the length of the repeat that coevolves with the flanking region rather than the species-specific amino acid composition of the arrays.
Overall, these results suggest that the coevolutionary interaction between the repeat and its flanking sequences may serve a function related to the clock’s cardinal property of temperature compensation (29). The improved rescue of mps3 over mps1 shows that the D. melanogaster N-terminal half of PER is also required for robust behavioral rhythms in addition to a coevolved Thr-Gly region. This may reflect the fact that the PAS dimerization domain of PER (1) encoded in mps3 is conspecific with the Timeless (TIM) partner molecule of the host (30). Furthermore, it is important to note that in spite of the mps1 transgene’s relatively poor rescue, its temperature compensation is maintained, even at the highest temperature. Therefore it does not follow that a poorly rescuing per gene will inevitably also suffer from defects in temperature compensation.
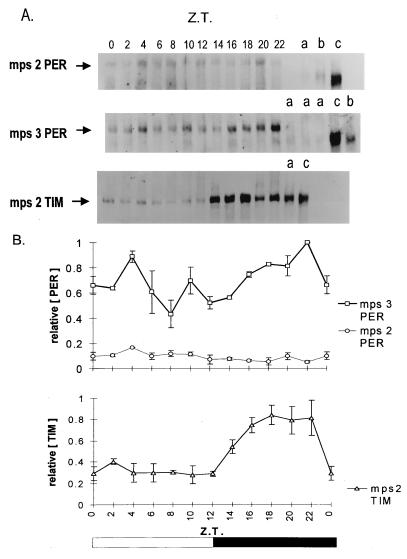
The sequences flanking the repeat in blocks H and P (Fig. 1) contain 27 amino acid substitutions between the two parental species, most of which are distributed in block H (Fig. 1). Comparing this flanking region to sequences in the databases did not reveal any significant similarities to motifs that might illuminate the function of this region. To learn more about the mps products, Western blots were performed with head extracts from mps2 and mps3 transformants over the circadian cycle in an LD 12:12 regime at 25°C, with a polyclonal anti-PER antibody (Fig. 4). Serial dilutions of total protein were used to quantify the amount of mps2PER signal in relation to wild-type and mps3PER (data not shown). Whereas mps3 and wild-type PER produced similar levels of signal [compare mps3PER at Zeitgeber time (ZT 22) with wild-type at ZT 0 in lane c], mps2PER detected was 5–7 times less abundant (Fig. 4A, see top panel) and similar to the levels of wild-type PER found in tim0 mutants (31). The mps3 transformants show significant differences in PER levels over the circadian cycle, with all three individual mps3PER blots showing a peak in intensity just before dawn at ZT 22 (Fig. 4). This temporal profile is similar to that observed with wild-type PER (e.g., ref. 32, and data not shown). Wild-type D. melanogaster PER also shows a change in mobility and a “broadening” of the PER band over the circadian cycle because of phosphorylation (32). This was present in our wild-type control blots (data not shown) and can also be appreciated by inspecting the broader wild-type control bands that can be seen in lanes c of Fig. 4, which represents PER mobility at ZT 0. However, the changes in mobility because of phosphorylation appeared to be considerably reduced with mps3PER (Fig. 4), even though these transformants gave robust behavioral rhythms.
Figure 4.
(A) Western blots of mps2 and mps3 transformant heads collected at different Zeitgeber times (ZT 0, lights on; ZT 12, lights off) in LD 12:12 at 25°C. Lanes on the right show per01 (a), tim0 (b), and per+ Canton-S (c) controls at ZT 0, except for the TIM blot where the controls were taken at ZT 18. From top to bottom are shown: mps2 blotted with anti-PER; mps3 blotted with anti-PER; mps2 blotted with anti-TIM. The predicted primary Mr of the mps2–5PER product is 136,000, heavier than either D. melanogaster (128,000) or D. pseudoobscura (132,000) proteins. However, D. melanogaster PER runs at about 180,000 (32), and the chimeric MPS proteins run a little higher. (B) Densitometry analysis for three separate anti-PER and anti-TIM blots. The highest intensity PER or TIM band within each blot was given a value of unity. Means ± SEM are shown. Control experiments in which serial amounts of total protein were loaded for mps3PER, wild-type PER, and mps2PER at ZT 0, followed by Western blotting with anti-PER, revealed an approximate 5- to 7-fold reduction in mps2PER intensity for the same amount of protein (data not shown). No characteristic wild-type PER cycling (32) was ever observed in mps2 blots, although all three blots showed the highest protein levels at ZT 4. Western blotting was performed as in ref. 32 with minor modifications. Rabbit anti-PER antibody (gift of J. Hall and R. Stanewsky) was used at a concentration of 1:10,000, and rat anti-TIM antibody (gift of M. Myers) was used at 1:1,000. On each Western blot, equal amounts of proteins were used for each time point and for controls, but the amounts loaded varied between 50 and 200 μg, depending on the genotype.
In sharp contrast, the mps2PER product appears to be expressed both in the light and dark phases, with little evidence for any robust circadian cycling or for any mobility shifts indicative of posttranslational modifications (Fig. 4). The apparent failure of mps2PER to cycle suggests either altered stability of the chimeric protein or less effective synthesis due perhaps to low activity of mps2PER, which in turn could lead to a defective cycle, low mRNA levels, and low synthesis of mps2PER. Because the levels of mps2PER are so low, we cannot state definitively that cycling was not present. On the other hand, mps2 transformant head extracts, blotted with an anti-TIM antibody (29), revealed a low amplitude (3× peak-to-trough) TIM cycling (Fig. 4), similar to that of per01 mutants under LD 12:12 conditions (33).
We favor an altered stability of mps2PER, based on recent work with PER–reporter fusions, which suggests that a region of PER encoded by a ≈700-bp fragment that contains the D. melanogaster Thr-Gly region may be a target for degradation (34, 35). Changes in PER stability would be expected to affect the negative feedback loop of per by altering the period (36). Perhaps then, the proximal cause for the dramatic lack of phenotypic temperature compensation in mps5 flies lies in a subtle temperature-sensitive change in PER degradation. The arrhythmicity of mps2 transformants might therefore reflect a more serious instability in the mps2PER molecule. A dysfunction in the mRNA, although possible, is unlikely given the statistical analysis of DNA and protein sequences that gave rise to these experiments (13, 14). These revealed a significant correlation between amino acid changes in the flanking regions, but not between synonymous nucleotide positions, and repeat length differences between pairs of Drosophila species. As the substitution rate in the synonymous position largely reflects the molecular clock, then the correlation between flanking region amino acid changes and repeat length differences was not simply because of the evolutionary time elapsed because any two species shared a common ancestor (13, 14). If RNA structure was involved in this coevolutionary process, we might expect that any two species with different repeat lengths would also have more changes in their flanking RNA sequences and so generate a positive correlation between the silent nucleotide position and repeat length.
This coevolution could thus represent the signature of natural selection as it attempts to maintain the appropriate circadian degradation kinetics for PER in the presence of a relatively high mutation rate in the repetitive region (11). It is not clear whether mutations in the flanking region may have imposed selection pressure on the length of the repeat or vice versa. However, because mutations in the repeat and flanking region are tightly linked, they conform to models where compensatory neutral mutations can become fixed even when the individual mutations are deleterious (37). We can envisage a scenario where a small change in the length of the repeat, for example by a gain or loss of a single repeat unit, may be slightly deleterious under certain conditions (see ref. 38). A mutation in the flanking region may restore the status quo, even though this compensating mutation may be slightly deleterious in the absence of the initial change. After many rounds of mutation, heterospecific combinations of flanking regions and repeat arrays could become incompatible with function, as in the case of the two species studied here. This process would be enhanced in a region associated with a high mutation rate such as the Thr-Gly repeat (11) and if the domain determined an adaptive character, which circadian temperature compensation is likely to represent.
Like the mps5 transformant, the perL1 mutant also shows significant increases in the period with higher temperatures (26, 28). The mutation maps to the PAS region of the PER protein, which is involved in protein-protein interactions (39, 40) and which may mediate the negative feedback mechanism believed to be at the heart of per function (41). PERL1/TIM protein–protein interactions are temperature-sensitive in a yeast assay (41), so perhaps the mps2 and mps5 transgenes produce defective PER–TIM dimerization (but see ref. 42). In addition, it may be more than coincidence that the frq3 and frq7 mutations in Neurospora crassa, which disrupt the temperature compensation of the circadian period, map to the immediate flanking regions of the fungal Thr-Gly/Ser-Gly motif (43). Perhaps these repetitive regions within the two clock genes serve a common function related to temperature compensation. In Neurospora this could be in addition to the different forms of FRQ product that are differentially translated at high and low temperatures (44).
Coevolution within molecules is rarely studied, although a notable example concerns the Drosophila Adh gene, where compensatory mutations appear to maintain long range interactions between the 5′ and 3′ regions of the transcript (45). The interspecific compensatory changes in the flanking regions that track the length changes in the Thr-Gly repeat seem to be driven, at least in part, by selection aimed at maintaining the temperature compensation of the circadian oscillator in the face of the instability provided by the rapidly evolving repetitive region. However, this might form part of a bigger overall picture, which has the Thr-Gly domain as a key feature of the degradation kinetics of the PER protein. Thus the Thr-Gly and associated regions may not provide part of the mechanism for the temperature compensation of the clock per se but rather represent an evolutionarily dynamic motif, which must maintain a particular conformation to mediate the thermal stability of the circadian phenotype.
Recent studies of the natural Thr-Gly length polymorphism within D. melanogaster have also revealed a relationship between Thr-Gly length and temperature compensation (38). Within D. melanogaster, the natural variation in repeat length of different Thr-Gly alleles (from 14 to 24 Thr-Gly pairs) is not accompanied by any significant flanking amino acid alterations, and small changes in temperature compensation can be detected between the different variants generating further support for the coevolution hypothesis (38). Thus not only do the statistical analyses of Thr-Gly sequences suggest the action of natural selection on this part of the PER molecule (11–14), but the phenotypic analyses of chimeric transgenes reported in this study and the work of Sawyer et al. (38) identify the temperature compensation of the clock as a possible target for this evolutionary pressure.
In conclusion, the most remarkable feature of the results reported here is that a functional interaction between subregions of the Thr-Gly “domain” was predicted by relatively simple statistical analyses of the relevant DNA sequences from a number of Dipteran species (13, 14). Without this evolutionary framework, there would have been no reason to create the appropriate transgenes, and their effects on temperature compensation would have remained undiscovered. Our experiments have defined a functional module of the PER protein whose subregions can maintain function if they diverge together. The results highlight the dangers inherent in studying chimeric genes between species without having an evolutionary perspective of the sequences to be joined. As can be seen, a very slight difference in the position of the chimeric junction can produce major phenotypic effects.
Acknowledgments
We thank the Science and Engineering Research Council, Biotechnology and Biological Sciences Research Council, and Human Frontiers Science Program for grants (to C.P.K.) and the Commission of the European Communities for a grant (to C.P.K. and R.C.). We acknowledge a Ministerio dell’Universitá e della Ricerca Scientifica e Technologia-British Council award, a Biotechnology and Biological Sciences Research Council studentship (to I.T.), and the Brazilian Government for a Conselho Nacional de Pesquisas scholarship (to A.A.P.). M.R. and G.H. were supported by National Institutes of Health Grant GM33205.
ABBREVIATIONS
- LD
light/dark
- ZT
Zeitgeber time
References
- 1.Rosato E, Piccin A, Kyriacou C P. BioEssays. 1997;19:1075–1082. doi: 10.1002/bies.950191206. [DOI] [PubMed] [Google Scholar]
- 2.Citri Y, Colot H V, Jacquier A C, Yu Q, Hall J C, Baltimore D, Rosbash M. Nature (London) 1987;326:42–47. doi: 10.1038/326042a0. [DOI] [PubMed] [Google Scholar]
- 3.McClung C R, Fox B A, Dunlap J C. Nature (London) 1989;339:558–562. doi: 10.1038/339558a0. [DOI] [PubMed] [Google Scholar]
- 4.Linden H, Macino G. EMBO J. 1997;16:98–109. doi: 10.1093/emboj/16.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Z S, Ulbrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C C. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 6.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Nature (London) 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 7.Albecht U, Sun Z S, Eichele G, Lee C C. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 8.Shearman L O, Zylka M J, Weaver D R, Kolakowski L F, Jr, Reppert S M. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 9.Castiglione-Morelli M A, Guantieri V, Villani V, Kyriacou C P, Costa R, Tamburro A M. Proc R Soc London Ser B. 1995;260:155–163. doi: 10.1098/rspb.1995.0073. [DOI] [PubMed] [Google Scholar]
- 10.Costa R, Peixoto A A, Barbujani G, Kyriacou C P. Proc R Soc London Ser B. 1992;258:43–49. doi: 10.1098/rspb.1992.0128. [DOI] [PubMed] [Google Scholar]
- 11.Rosato E, Peixoto A A, Costa R, Kyriacou C P. Genet Res. 1997;69:89–99. doi: 10.1017/s001667239700267x. [DOI] [PubMed] [Google Scholar]
- 12.Rosato E, Peixoto A A, Barbujani G, Costa R, Kyriacou C P. Genetics. 1994;138:693–707. doi: 10.1093/genetics/138.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peixoto A A, Campesan S, Costa R, Kyriacou C P. Mol Biol Evol. 1993;10:127–139. doi: 10.1093/oxfordjournals.molbev.a039993. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen J, Peixoto A A, Piccin A, Costa R, Kyriacou C P, Chalmers D. Mol Biol Evol. 1994;11:839–853. doi: 10.1093/oxfordjournals.molbev.a040167. [DOI] [PubMed] [Google Scholar]
- 15.Kimura M. Genetics. 1962;47:713–719. doi: 10.1093/genetics/47.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aquadro C F. Trends Genet. 1992;8:355–362. doi: 10.1016/0168-9525(92)90281-8. [DOI] [PubMed] [Google Scholar]
- 17.Colot H, Hall J C, Rosbash M. EMBO J. 1988;7:3929–3937. doi: 10.1002/j.1460-2075.1988.tb03279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa R, Peixoto A A, Thackeray J R, Dalgleish R, Kyriacou C P. J Mol Evol. 1991;32:238–246. doi: 10.1007/BF02342746. [DOI] [PubMed] [Google Scholar]
- 19.Klemenz R, Weber U, Gehring W J. Nucleic Acids Res. 1987;15:3947–3959. doi: 10.1093/nar/15.10.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersen G, Hall J C, Rosbash M. EMBO J. 1988;7:3939–3947. doi: 10.1002/j.1460-2075.1988.tb03280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yon J, Fried M. Nucleic Acids Res. 1989;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubin G M, Spradling A C. Science. 1982;218:348–352. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 23.Dowse H B, Ringo J M. J Theor Biol. 1989;139:487–515. doi: 10.1016/s0022-5193(05)80468-0. [DOI] [PubMed] [Google Scholar]
- 24.Roberts D H, Lehar J, Dreher J W. Astron J. 1987;93:968–989. [Google Scholar]
- 25.Kyriacou C P, Hall J C. Anim Behav. 1989;37:850–859. [Google Scholar]
- 26.Ewer J, Hamblen-Coyle M, Rosbash M, Hall J C. J Neurogenet. 1990;7:31–73. doi: 10.3109/01677069009084151. [DOI] [PubMed] [Google Scholar]
- 27.Konopka R J, Kyriacou C P, Hall J C. J Neurogenet. 1996;11:117–140. doi: 10.3109/01677069609107066. [DOI] [PubMed] [Google Scholar]
- 28.Konopka R J, Pittendrigh C, Orr D. J Neurogenet. 1989;6:1–10. doi: 10.3109/01677068909107096. [DOI] [PubMed] [Google Scholar]
- 29.Pittendrigh C S. Proc Natl Acad Sci USA. 1954;40:1018–1029. doi: 10.1073/pnas.40.10.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers M P, Wager-Smith K, Rothenfluh-Hilfiker A, Young M W. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 31.Price J L, Dembinska M A, Young M W, Rosbash M. EMBO J. 1995;14:4044–4049. doi: 10.1002/j.1460-2075.1995.tb00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edery I, Zweibel L J, Dembinska M E, Rosbash M. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng H, Qian Z, Myers M P, Rosbash M. Nature (London) 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 34.Dembinska M E, Stanewsky R, Hall J C, Rosbash M. J Biol Rhythms. 1997;12:157–172. doi: 10.1177/074873049701200207. [DOI] [PubMed] [Google Scholar]
- 35.Stanewsky R, Frisch B, Brandes C, Hamblen-Coyle M J, Rosbash M, Hall J C. J Neurosci. 1996;17:676–696. doi: 10.1523/JNEUROSCI.17-02-00676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardin P E, Hall J C, Rosbash M. Nature (London) 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 37.Kimura M. Proc Natl Acad Sci USA. 1991;88:5969–5971. doi: 10.1073/pnas.88.14.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawyer L, Hennessy J M, Peixoto A A, Rosato E, Parkinson H E, Costa R, Kyriacou C P. Science. 1997;278:2117–2120. doi: 10.1126/science.278.5346.2117. [DOI] [PubMed] [Google Scholar]
- 39.Huang Z J, Edery I, Rosbash M. Nature (London) 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 40.Huang Z J, Curtin K D, Rosbash M. Science. 1995;267:1169–1172. doi: 10.1126/science.7855598. [DOI] [PubMed] [Google Scholar]
- 41.Gekakis N, Saez L, Delahaye-Brown A M, Myers M P, Sehgal A, Young M W, Weitz C J. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 42.Rutila J E, Zeng H, Le M, Curtin K D, Hall J C, Rosbash M. Neuron. 1996;17:921–929. doi: 10.1016/s0896-6273(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 43.Aronson B D, Johnson K A, Dunlap J C. Proc Natl Acad Sci USA. 1994;91:7683–7687. doi: 10.1073/pnas.91.16.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Garceau N Y, Loros J J, Dunlap J C. Cell. 1997;89:477–486. doi: 10.1016/s0092-8674(00)80228-7. [DOI] [PubMed] [Google Scholar]
- 45.Parsch J, Tanda S, Stephan W. Proc Natl Acad Sci USA. 1997;94:928–933. doi: 10.1073/pnas.94.3.928. [DOI] [PMC free article] [PubMed] [Google Scholar]