Abstract
CREB, the cAMP response element binding protein, is a key transcriptional regulator of a large number of genes containing a CRE consensus sequence in their upstream regulatory regions. Mice with a hypomorphic allele of CREB that leads to a loss of the CREBα and Δ isoforms and to an overexpression of the CREBβ isoform are viable. Herein we report the generation of CREB null mice, which have all functional isoforms (CREBα, β, and Δ) inactivated. In contrast to the CREBαΔ mice, CREB null mice are smaller than their littermates and die immediately after birth from respiratory distress. In brain, a strong reduction in the corpus callosum and the anterior commissures is observed. Furthermore, CREB null mice have an impaired fetal T cell development of the αβ lineage, which is not affected in CREBαΔ mice on embryonic day 18.5. Overall thymic cellularity in CREB null mice is severely reduced affecting all developmental stages of the αβ T cell lineage. In contrast γδ T cell differentiation is normal in CREB mutant mice.
Many of the transcriptional effects of cAMP are mediated by the cAMP response element binding protein CREB, which binds to the CRE element in the upstream region of a variety of genes. CREB is a member of the CREB/ATF family of transcription factors and is phosphorylated by protein kinase A after an intracellular increase in cAMP (1). More recently it has become apparent that CREB is an in vivo substrate for a variety of other kinases including calmodulin kinases II and IV (2) or RSK2 (3), implying that CREB activates transcription in response to cAMP, Ca2+, and growth factor stimulation (4–6).
A targeted mutation of the CREB gene has been reported (7). Unexpectedly, these mice were healthy, fertile, and did not show any developmental abnormalities. However, they showed abnormalities in studies of learning and memory (8) and of behavioral responses to opiate withdrawal (9). Interestingly, a detailed analysis of these mice led to the identification of the isoform CREBβ (10), which was strongly up-regulated in CREB mutant mice. This suggests that its presence in CREB mutant mice, in addition to an up-regulation of the cAMP response element modulator (CREM) (7) could explain the mild phenotype observed in these mice (CREBαΔ mice).
To evaluate the in vivo significance of the newly identified CREBβ isoform, we have generated CREB null mice by homologous recombination in embryonic stem cells. In contrast to CREBαΔ mice, CREB null mice die perinatally. Because CREM is also up-regulated in CREB null mice, CREBβ is responsible for viability of CREBαΔ mice. In this report, we describe the phenotype of the CREB null mice, which gives insights into the developmental role of CREB.
Functionally important CRE elements are found in the regulatory regions of a variety of T cell-specific genes (11, 12), implicating members of the CREB/ATF family in the regulation of gene expression during T cell differentiation and activation. However, the physiological importance of these findings remains to be determined. It has recently been reported that T cell activation leads to phosphorylation of CREB (13), suggesting that CREB is important for T cell function. This is also suggested by the analysis of transgenic mice expressing a dominant negative CREB protein under the control of a T cell-specific CD2 promoter/enhancer (13). Thymocytes and T cells from these mice show a proliferative defect after activation. However, it remains unclear which of the CREB/ATF family members is critically involved, because expression of the dominant negative CREB protein inactivates all family members. Herein, we show the importance of CREB for fetal T cell development. CREBαΔ mice show normal fetal T cell development, underlining the physiological importance of the CREBβ isoform in CREBαΔ mutant mice.
MATERIALS AND METHODS
Generation of CREB −/− Mice.
λ phage clones containing the CREB gene were isolated from a mouse embryonic stem (ES) cell (strain 129) library. A gene targeting vector was constructed by using the promoterless β-galactosidase containing pHM3 vector (14). As a 5′ homology sequence, a 5-kb AvaII/HindIII fragment containing part of exon 10, exon 9, and intronic sequences was used. A 1.8-kb HindIII fragment located 3′ of exon 11 was used as a 3′ homology sequence. Twenty micrograms of the targeting vector was digested to linear fragments with NotI and electroporated into 107 E14-J ES cells. Homologous recombination events were identified by Southern blot analysis using external probes. ES cells from 3 of 10 positive clones were injected into C57BL/6 blastocysts to generate chimeric mice, which were mated with C57BL/6 females. F1 heterozygotes were used to generate −/− mice. Animals from two clones were analyzed separately and were shown to be phenotypically identical. Adult and newborn mice were genotyped by using three PCR primers (LacZ, AAGCGCCATTCGCCATTCAGGC; 5′, GATGTACAAACATACCAGATCCGC; 3′, CACAGAACCTACTGTTAGCAGATG) under the following conditions: 2 mM MgCl2, 35 cycles of 94°C for 1 min, 67°C for 1 min, and 72°C for 1 min. The mutant band was 370 bp; wild-type band was 230 bp.
RNA Analysis.
Total RNA was isolated from tissue or flow-cytometry-sorted thymocytes after homogenization in guanidinium thiocyanate (15). Northern blot analyses were performed as described (16). Briefly, Hybond N filters (Amersham) were hybridized in 50% formamide/5× SSC/50 mM NaH2PO4, pH 6.5/8× Denhardt’s solution/1% SDS/total yeast RNA (0.5 mg/ml) at 65°C with the probes indicated, except for the β-actin probe, which was used at 42°C. Filters were washed at 65°C for one 10-min period with 2× SSC/1% SDS and for two 10-min periods with 0.1× SSC/1% SDS or for the β-actin probe at 42°C with 2× SSC/0.1% SDS for 10 min, followed by two 10-min washes with 2× SSC/1% SDS and 0.1× SSC/1% SDS, respectively. For reverse transcription-coupled PCR (RT–PCR) analyses, RNA was treated for 30 min with RNase-free DNase I (10 units) at 37°C. After phenol/chloroform extraction, cDNA synthesis was performed with Superscript II (GIBCO/BRL) according to the manufacturer’s instructions using random hexamer primers. PCR amplification was performed in a 50-μl reaction using 0.5 μl of the cDNA, 10 μM primers, all four dNTPS (each at 200 μM), and the reaction buffer for the pfu polymerase (Stratagene) provided by the company. The following primers were used simultaneously: 5′ CREB, ACTGGCTTGGCACAACCAGA; 3′ CREB, GGCAGAAGTCTCTTCATGATT; 5′ Hprt, AGCGTCGTGATTAGCGATGAT; 3′ Hprt, GTCAGGGCATATCCAACAACAAC (1 min at 94°C, 1 min at 56°C, 1 min at 72°C; 35 cycles); or 5′ TCRα, CAGAACCCAGAACCTGCTGTG; 3′ TCRα, GGCCCCATTGCTCTTGGAATC; 5′ β-actin, GGACGACATGGAGAAGATCTG; 3′ β-actin, CCGCTCGTTGCCAATAGTGAT (1 min at 94°C, 1 min at 56°C, 1 min at 72°C). For quantification Southern blot analyses of the PCR products were performed and filters were hybridized with polynucleotide kinase 32P-labeled gene-specific oligonucleotides (TCRα, CGGCACATTGATTTGGGAGTC; β-actin, AGCTGTGCTATGTTGCTCTAG) at 37°C in 7% SDS/250 mM Na2HPO4/1 mM EDTA. Filters were washed at 37°C in 1% SDS/25 mM Na2HPO4/1 mM EDTA.
Central Nervous System (CNS) Histology and Immunohistochemistry.
Brains were fixed with 4% (wt/vol) paraformaldehyde for 24 h. Histological evaluation of CNS structures was performed on hematoxylin/eosin- and Nissl-stained coronal paraffin sections (3 μm). Immunocytochemical analysis was performed on serial free-floating 70-μm coronal Vibratome sections. Vibratome sections were incubated in buffer A (10% normal swine serum in PBS/0.2% Triton X-100, pH 7.4) for 30 min and then with the primary antiserum, diluted in buffer A, for 48 h at 4°C. The following primary antibodies were used: rabbit antiserum against CREB protein diluted 1:10,000, rabbit antiserum against CREM protein diluted 1:2,000, antiserum against SCIP/Oct-6 diluted 1:1,000 (a gift from D. Meijer, Erasmus University, Rotterdam, The Netherlands) (17), and rabbit antiserum against CGRP diluted 1:1,000 (Camon, Wiesbaden, Germany). Immunoreactivity was visualized by the avidin–biotin complex method (Vectastain, Vector Laboratories) as described (10). Sections were developed in 0.02% diaminobenzidine with 0.02% hydrogen peroxide. For photo documentation, Adobe photoshop was used.
Flow Cytometry.
Thymi from embryonic day (E) 18.5 mice were removed and single cell suspensions (1.5 × 106 cells per sample) were incubated for 10 min on ice with the appropriate mAb or streptavidin-Red 670 (GIBCO/BRL) and washed with PBS containing 0.45% glucose, 3% fetal calf serum, and 0.01% NaN3. The following mAbs were used: H129.19 (anti-mouse CD4, GIBCO/BRL), 53–6.7 (anti-mouse CD8, GIBCO/BRL), 29B (anti-mouse CD3, GIBCO/BRL), H57–597 [anti-mouse T cell receptor αβ (TCRαβ), PharMingen], GL3 (anti-mouse TCRγδ, PharMingen), F23.1 (anti-mouse Vβ8.1,2,3, PharMingen), IM7 (anti-mouse CD44, PharMingen), and 7D4 (anti-mouse CD25, PharMingen). Analysis was performed on a FACScan cytometer (Becton Dickinson). A minimum of 5,000 cells per sample were analyzed.
RESULTS
Targeting Strategy.
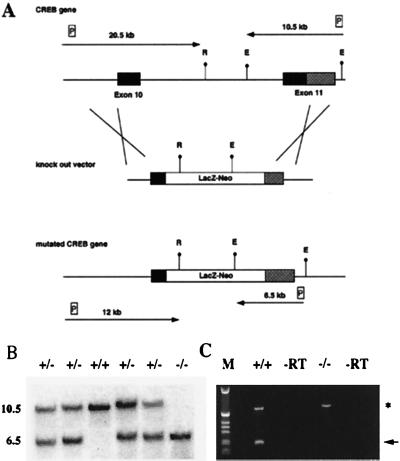
The targeting strategy used for the generation of CREB null mice is shown in Fig. 1A. We deleted part of the DNA binding domain and all of the leucine zipper of the CREB protein, thereby creating a protein that can no longer dimerize or bind to DNA. RT–PCR analysis (Fig. 1C), immunocytochemical studies, and Western blot analyses with antibodies 3′ and 5′ of the deletion (data not shown) indicate that we have generated a null mutation.
Figure 1.
Targeting strategy for the generation of a CREB null allele. (A) (Top) Gene structure of the 3′ region of the CREB locus. Exons are shown as boxes. Solid boxes, coding regions; shaded boxes, nontranslated regions. (Middle) Targeting vector. (Bottom) Gene structure of the targeted allele. E, EcoRI; R, EcoRV; LacZ-Neo, fusion gene encoding β-galactosidase and neomycin phosphotransferase; P, probes used for detection of homologous recombination events. Arrows show the expected size of the restriction fragments detected with these probes. (B) Southern blot analysis of EcoRI-digested genomic DNA from E18.5 newborns. The 3′ probe was used, the size of the restriction fragments is indicated. (C) RT–PCR analysis using a cDNA derived from total brain RNA of a CREB −/− mouse and wild-type littermate, respectively, demonstrating the absence of the 3′ part of the CREB transcript in mutant mice. Primers for the CREB gene were derived from the 3′ end of the gene; primers for Hprt (hypoxanthine guanine phosphoribosyltransferase) were used as control for RNA integrity. The CREB PCR product is indicated by an arrow, and the Hprt PCR product is indicated by an asterisk. −RT, −reverse transcriptase.
No surviving CREB −/− mice were found at 4 weeks of age, and CREB heterozygous mice were viable and reproduced normally. However, CREB −/− mice were identified at birth, although they were present at a reduced Mendelian frequency (15%). This suggests that CREB −/− mice have a developmental disadvantage and some of them were lost before birth. E18.5 CREB −/− mice had a reduced birth weight (70%) compared with their littermates. All CREB −/− mice were cyanotic and died within 15 min after birth from respiratory distress. Histological analyses of lungs (Fig. 2 A and B) from E18.5 animals showed a severe atelectasis of the lung in CREB −/− mice.
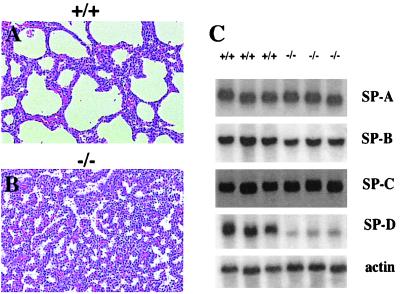
Figure 2.
Severe atelectasis of the lung and reduced expression of SP-D in CREB null mice. Histological analysis of lungs from a CREB −/− animal (B) and a control wild-type littermate (A), stained with hematoxylin/eosin and showing a severe atelectasis of the lung in CREB mutant animals. (C) Northern blot analyses of total lung RNA (5 μg) using probes specific for SP-A, SP-B, SP-C, and SP-D. Filters were rehybridized with β-actin as a loading control.
Because respiratory distress is very often related to an insufficient production of surfactant, we analyzed the expression of surfactant-associated proteins SP-A, SP-B, SP-C, and SP-D by Northern blot analyses (Fig. 2C). CREB-deficient mice showed a reduction in the expression of surfactant-associated protein SP-D alone (20% of the wild-type level), suggesting an impairment of surfactant function in CREB −/− mice.
The cyanosis and early respiratory failure of CREB −/− animals prompted us to study the brain stem region known to be of importance in the regulation of vegetative functions such as respiration. Histological (Nissl and hematoxylin/eosin staining) and immunocytochemical studies with antiserum specific for SCIP/Oct-6, a marker for neuronal populations of the brain stem and spinal cord involved in respiratory control (18), and calcitonin gene-related peptide (CGRP), a marker for motoneurons and fiber systems of brain stem and spinal cord (19), were performed on serial sections through the whole brain stem and the cervical spinal cord. However, SCIP/Oct-6 and CGRP expression was not altered in CREB −/− mice (data not shown), suggesting no structural defects in these respective neuron populations.
Nissl- and hematoxylin/eosin-stained serial sections on brains of E18.5 CREB −/− mice demonstrated that all major neuronal populations of the CNS were present. However, two commissural structures were clearly reduced: the corpus callosum and the anterior commissure (Fig. 3 A and B), a defect not observed in adult CREBαΔ mutant mice. In contrast, other commissures, such as the hippocampal commissure, exhibited a reduction in proportion to the other brain areas of CREB null mice (data not shown). Comparison of brain sections from E18.5 −/− mice with those of E17.5 wild-type animals, which are similar in size to CREB −/− brains showed that both structures were clearly more developed in E17.5 wild-type mice than in E18.5 CREB mutant mice (data not shown), suggesting that the reduction observed in CREB −/− mice does not represent a general developmental retardation.
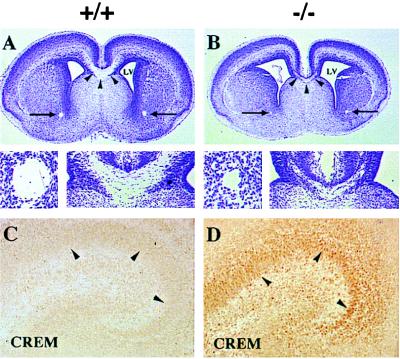
Figure 3.
Histological analysis of the CNS of CREB −/− mice on E18.5. (A and B) A severe reduction in the corpus callosum (arrowheads and lower right of micrographs) and the anterior commissures (arrows and lower left of micrographs) was observed in CREB −/− mice. LV, lateral ventricle. (C and D) A marked up-regulation of CREM was observed in the developing hippocampus as well as other forebrain areas (data not shown) of CREB −/− animals. Wild-type littermates only showed faint CREM immunoreactivity. Arrowheads indicate immunostained nuclei of the pyramidal neurons of the hippocampus.
As in CREBαΔ mice, we also observed a marked up-regulation of the transcription factor CREM in many neuronal populations, e.g., hippocampus, cortex, amygdala, striatum, and thalamus (Fig. 3 C and D and data not shown). Histologically other organs in CREB mutant mice appeared to be normal.
T Cell Development in CREB Null Mice.
Macroscopic analysis of thymi from CREB −/− mice on E18.5 showed a reduction in size, which was more pronounced than the overall reduced body size of the mutant animals. Thymocyte yield from CREB −/− thymi was reduced 5-fold compared with littermate controls (Table 1). Interestingly, thymi from E18.5 CREBαΔ mutant mice were of normal size and did not show a decrease in thymocyte yield (Table 1).
Table 1.
Thymocyte yield (×106) from embryos of gestational age E18.5
| Thymocytes, no. (×106)
|
|||
|---|---|---|---|
| −/− | +/− | +/+ | |
| CREBαΔ | 8.64 ± 2.27 | 8.85 ± 2.62 | 10.18 ± 4.09 |
| (n = 9) | (n = 26) | (n = 5) | |
| CREB | 1.55 ± 0.87 | 6.84 ± 3.90 | 8.14 ± 3.80 |
| (n = 15) | (n = 13) | (n = 8) | |
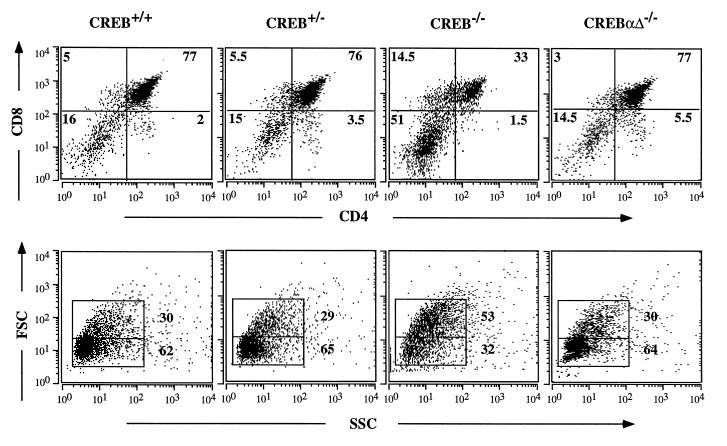
To investigate thymic development in CREB −/−, +/−, and +/+ mice, surface expression of CD4 and CD8 was examined by microfluorimetry. CREB null mice showed a 3- to 4-fold increase in the percentage of double-negative (DN) cells, and the percentage of double-positive (DP) cells was reduced 2- to 3-fold (Fig. 4 Upper). DN thymocytes contain a high number of proliferating blasts. Accordingly, CREB −/− thymi contained a larger percentage of cells with a higher forward side scatter, a measure of cell size (Fig. 4 Lower). In comparison, thymi from CREBαΔ mutant mice did not differ from littermate controls (Fig. 4).
Figure 4.
Fetal T cell development in CREB null mice and CREBαΔ mice on E18.5. (Upper) Thymocytes from E18.5 CREB null mice, wild-type and heterozygous littermates, and CREBαΔ mice were analyzed by flow cytometry using antibodies against CD4 and CD8. The thymocyte subsets from one representative animal of each genotype are shown. (Lower) Forward side scatter (FSC) versus side scatter (SSC) plots are shown for thymocytes from the indicated genotypes. CREB null mice had a larger percentage of thymocytes with a higher FSC. CREBαΔ mice did not differ from the control animals.
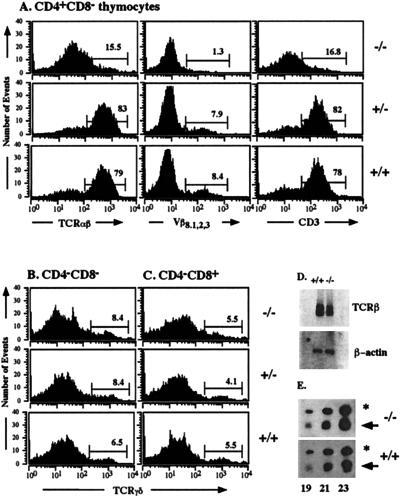
Expression of genes for the TCRα and β chain is controlled by T cell-specific transcriptional enhancers that bind a variety of ubiquitous and lymphoid-specific transcription factors, which include members of the CREB/ATF family (for review, see ref. 20). CRE elements have been identified in the enhancer elements of the TCRα and β genes but not in the enhancer elements of the TCRγ and δ genes. We therefore analyzed TCR expression. CD4+ CD8− single-positive (SP) thymocytes were stained for surface expression with antibodies against the TCRαβ surface complex, the TCRVβ8.1,2,3 chain, and CD3ɛ (Fig. 5A). CREB null mice showed a severe reduction in mature CD3high TCRαβhigh CD4+ CD8− SP cells. On E18.5, mature CD3high TCRαβhigh CD8+ CD4− SP cells are not yet present. They appear shortly thereafter, however (21). Some γδ T cells express the accessory molecule CD8 on the surface but the majority of these T cells expresses neither CD4 nor CD8. Therefore, we examined the CD4− CD8− and CD4− CD8+ subpopulation of thymocytes in CREB mutant mice for surface expression of the TCR γδ complex (Fig. 5 B and C). In contrast to the impairment in the development of mature αβ thymocytes, differentiation of γδ thymocytes was not affected in CREB −/− mice.
Figure 5.
Normal fetal γδ T cell differentiation but severely reduced numbers of mature CD3high TCRαβhigh CD4+ CD8− SP thymocytes in CREB null mice on E18.5. (A) CD4+ CD8− thymocytes from CREB null mice and control littermates were stained with antibodies against the TCRαβ complex, the TCRVβ8.1,2,3 chain, and CD3ɛ as indicated and were analyzed by flow cytometry. CREB null mice had a severely reduced percentage of mature CD4+ SP cells characterized by high TCRαβ, TCRVβ8.1,2,3, and CD3ɛ surface expression. CD4− CD8− (B) and CD4−CD8+ (C) thymocytes from CREB null mice and control littermates were stained with an antibody against TCRγδ and analyzed by flow cytometry. No difference in γδ T cell differentiation was observed between CREB −/− mice and their control littermates. (D) Northern blot analysis of RNA (500 ng) from sorted DN thymocytes derived from five animals of each genotype. A TCRβ-chain-specific probe containing exon cβ2 was used (Upper). The filter was rehybridized with β-actin as a loading control (Lower). (E) Quantitative RT–PCR using a cDNA derived from sorted DP cells of mutant or wild-type animals, respectively. One representative experiment of five is shown. Gene-specific primers were used for the TCRα chain. Primers for β-actin were used as a standard. Aliquots of the PCR products were taken after 19, 21, and 23 cycles as indicated, separated on an agarose gel, and blotted. Filters were hybridized with internal oligonucleotides to detect the two PCR products: the TCRα product is indicated by an arrow and the β-actin product is indicated by an asterisk.
Because most CD4+ SP cells in CREB null mice lacked surface staining for the TCRαβ complex, we analyzed the mRNA expression of the TCRβ and α chains in CREB mutant mice. A slight decrease in the expression of the TCRβ chain was observed in CREB null mice by Northern blot analyses (Fig. 5D). TCRα chain expression, determined by quantitative RT–PCR, was not affected in CREB mutant mice (Fig. 5E). We also analyzed mRNA expression of other components of the TCR surface complex such as CD3ɛ and CD3ζ, which are essential for TCR surface expression and T cell development (22–24), and of CD3δ and pTα, which are required for the transition from DP cells to SP cells or the transition from DN cells to DP cells, respectively (25, 26). No difference in the expression of these genes was observed between control littermates and CREB mutant mice (data not shown).
Because we observed a reduction in the percentage of DP cells but an increase in the percentage of DN cells in CREB deficient mice on E18.5, we suspected a developmental defect in the differentiation of DN cells. CD25 (interleukin 2 receptor α chain) and CD44 (phagocyte glycoprotein 1) are useful markers to distinguish developmentally relevant subsets within the DN population of thymocytes. However, analyzing the DN subset of thymocytes from CREB mutant mice for surface expression of CD44 and CD25 did not reveal a particular block in the differentiation of DN cells (data not shown).
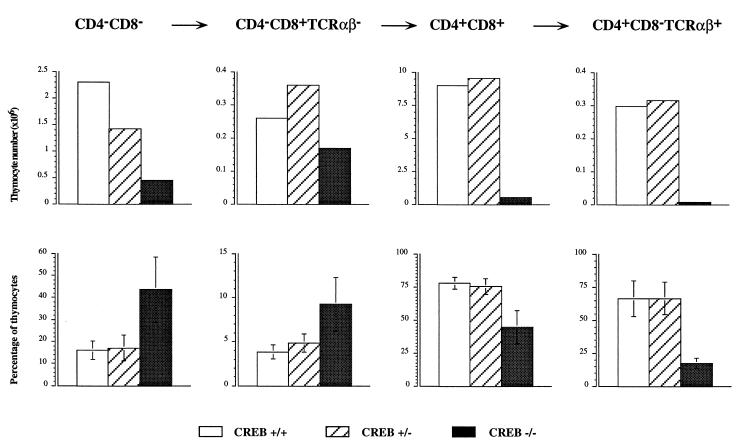
In CREB −/− mice, the absolute number of each individual thymocyte subset was severely reduced (Fig. 6 Upper). However, it should be noticed that the different thymocyte subsets did not contribute equally to the overall reduction of thymocyte number. In particular, the most immature subpopulations (CD4− CD8− and CD4− CD8+ TCRαβ−) were relatively overrepresented in the thymi of CREB −/− mice, whereas the more mature ones (CD4+ CD8+ and CD4+ CD8− TCRαβ+) were concordantly reduced in both relative percentage and absolute number (Fig. 6). These data suggest a gradual and progressive hindrance in the development of the TCRαβ lineage in CREB mutant mice.
Figure 6.
Altered thymocyte subsets defined by CD4/CD8 surface staining in CREB null mice on E18.5. (Upper) The absolute thymocyte numbers (×106) of each of the thymocyte populations indicated above for one representative animal of each genotype. Open bars, +/+ animal; hatched bars, +/− animal; solid bars, the CREB null mouse. The absolute number of the thymocytes in each of the subsets was decreased in CREB null mice. (Lower) Relative percentage of the thymocyte subsets given as the mean from n = 8 +/+ (open bars), n = 13 +/− (hatched bars), and n = 15 CREB −/− mice (solid bars). The standard deviation is indicated. The relative percentage of CD4− CD8− cells (P < 0.00005, Wilcoxon rank sum test) and immature CD4− CD8+ TCRαβ− SP cells (P < 0.00005, Wilcoxon rank sum test) was increased in CREB null mice, whereas the relative percentage of the more mature CD4+ CD8+ (P < 0.00005, Wilcoxon rank sum test) and the CD4+ CD8− TCRαβ+ SP cells (P < 0.0159, Wilcoxon rank sum test, n = 5) was reduced compared with the littermate controls. We did not observe statistically significant differences between wild-type and heterozygous littermates.
DISCUSSION
Phenotypic Differences Between CREBαΔ and CREB Null Mice Are Due to the Presence of the CREBβ Isoform in CREBαΔ Mice.
Herein we report the generation of CREB null mice, which carry a mutation that removes all functional isoforms of the CREB gene. These mice show a much more dramatic phenotype compared with the CREBαΔ mice, which only have the isoforms CREBα and Δ deleted but still contain the previously uncharacterized CREBβ isoform. Both mutations lead to an up-regulation of CREM, which with CREB and ATF1 constitutes a subfamily of the CREB/ATF family. Because both mouse strains have a similar genetic background (129/C57BL/6), we conclude that the phenotypic differences are due to the presence or absence of the CREBβ isoform. CREBβ provides residual CREB activity, which ensures viability of CREBαΔ mice after birth and is responsible for unimpaired growth and the lack of a phenotype in fetal T cell differentiation of CREBαΔ mice.
Comparison of CREB Dominant Negative Transgenic Mice with CREB Null Mice.
Transgenic mice expressing a dominant negative CREB protein under the control of a T cell-specific CD2 promoter/enhancer (13) show normal T cell development. This discrepancy can be attributed to the different genetic approach. By using homologous recombination, CREB is inactivated in early precursors entering the thymus, whereas expression of a dominant negative transgene under the control of the CD2 promoter/enhancer occurs later during T cell maturation.
Target Gene Analysis Suggests Compensation Within the CREB/ATF Family.
The strong effect of the CREB null mutation on T cell development suggests a critical role of CREB in expression of genes involved in T cell development. However, mRNA expression of components of the TCR surface complex such as TCRα, TCRβ, CD3ɛ, CD3ζ, CD3δ, pTα, or signaling molecules downstream of the TCR complex such as the Src family kinase p56lck or calcineurin B were not affected in CREB null mice (data not shown), suggesting that expression of genes involved in the assembly or surface expression of the TCR complex could be affected in CREB −/− mice.
Recombinant CREB activates the TCRα enhancer on nucleosome-assembled DNA in vitro. Furthermore, a Gal4/CREB protein can activate a TCRα enhancer in which the CRE was replaced by a Gal4 site and confer protein kinase A responsiveness to the enhancer in T cell lines (27). However, TCRα chain expression is not altered in CREB mutant mice. TCRβ chain expression is only mildly affected even though functional CRE consensus sequences have been reported in the TCRVβ promoter (11) and the enhancer 3′ of the locus (12). Although reports on binding activities in fetal thymic extracts are still missing, Barton et al. (13) showed that several members of the CREB/ATF family are present in adult thymus, which could fulfill distinct roles. Even though CREB is essential for TCRα chain expression in vitro, in vivo other CRE-binding proteins are capable of regulating T cell receptor gene expression.
Preliminary analysis of mice lacking both CREB and ATF1 clearly demonstrates that there is compensation within the gene family; although ATF1 −/− mice are viable (J. B. Blendy and G.S., unpublished data) and CREB null mice die at birth, ATF1/CREB double mutant mice die early during embryogenesis (D.R. and S. B. Bleckmann, unpublished data). Therefore, the presence of ATF1 during embryogenesis guarantees survival of CREB null mice until birth. Even though CREB is widely expressed, CREB null mice show a phenotype that is restricted to organs such as lung, brain, and thymus, suggesting that a lack of phenotype in other organs is compensated by other members of the family.
CREM is up-regulated in CREB null mice and in CREBαΔ mice (7), which is thought to occur posttranscriptionally (10). The physiological significance of the CREM up-regulation remains to be determined. We are currently generating CREB/CREM double knock out mice to assess the issue.
Analysis of the Lung Phenotype.
CREB null mice die due to respiratory distress. Even though CREB is highly expressed in the brain, we did not observe any structural abnormalities of the brain stem region, which is implicated in respiratory control. These data suggest that the breathing defect is intrinsic to the lung. Surfactant deficiency in prematurely born infants is a major factor in the respiratory distress syndrome. CREB null mice show a decreased mRNA expression of one of the surfactant-associated proteins, SP-D, implicating an impaired surfactant function. However, because pulmonary surfactant is a complex mixture of various components, we cannot conclude that this reduction is sufficient to explain respiratory distress in CREB null mice.
In conclusion, the generation of CREB null mice has given insights into the developmental und physiological role of CREB up to birth. In the future, generation of tissue-specific CREB knock out mice using the Cre-loxP system should allow a detailed analysis of CREB function in adult animals.
Acknowledgments
We are grateful to Drs. P. A. Monaghan and T. Mantamadiotis for critically reading the manuscript. We thank S. Ridder, A. Klewe-Nebenius, K. Anlag, E. Schmid, H. Glaser, A. Klevenz, J. Weik, and K. Hexel for expert technical assistance; Dr. W. Rittgen for statistical analysis; and W. Fleischer for oligonucleotide synthesis. A probe for TCR cβ2 was provided by A. Wilson (Ludwig Institute for Cancer Research, Epalinges, Switzerland). This work was supported by the Deutsche Forschungsgemeinschaft through SFB 405, by the Fonds der Chemischen Industrie, the Bundesministerium für Bildung and Forschung through the Human Genome Project Grant 01 KW 9606/7, by the Volkswagen-Stiftung, and by Boehringer Ingelheim.
ABBREVIATIONS
- DN
double negative
- DP
double positive
- SP
single positive
- E
embryonic day(s)
- RT–PCR
reverse transcriptase-coupled PCR
- CNS
central nervous system
- TCR
T cell receptor
- CREM
cAMP reponse element modulator
References
- 1.Montminy M. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 2.Sun P, Enslen H, Myung P S, Maurer R A. Genes Dev. 1994;8:2527–2539. doi: 10.1101/gad.8.21.2527. [DOI] [PubMed] [Google Scholar]
- 3.Xing J, Ginty D D, Greenberg M E. Science. 1996;273:959–962. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez G A, Montminy M R. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 5.Sheng M, Thompson M A, Greenberg M E. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- 6.Ginty D D, Bonni A, Greenberg M E. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 7.Hummler E, Cole T J, Blendy J A, Ganss R, Aguzzi A, Schmid W, Beermann F, Schütz G. Proc Natl Acad Sci USA. 1994;91:5647–5651. doi: 10.1073/pnas.91.12.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schütz G, Silva A. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 9.Maldonado R, Blendy J A, Tzavara E, Gass P, Roques B P, Hanoune J, Schütz G. Science. 1996;273:657–659. doi: 10.1126/science.273.5275.657. [DOI] [PubMed] [Google Scholar]
- 10.Blendy J A, Kaestner K H, Schmid W, Gass P, Schütz G. EMBO J. 1996;15:1098–1106. [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson S J, Miyake S, Loh D Y. Mol Cell Biol. 1989;9:4835–4845. doi: 10.1128/mcb.9.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottschalk L R, Leiden J M. Mol Cell Biol. 1990;10:5486–5495. doi: 10.1128/mcb.10.10.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barton K, Muthusamy N, Chanyangam M, Fischer C, Clendenin C, Leiden J M. Nature (London) 1996;379:81–85. doi: 10.1038/379081a0. [DOI] [PubMed] [Google Scholar]
- 14.Kaestner K H, Montoliu L, Kern H, Thulke M, Schütz G. Gene. 1994;148:67–70. doi: 10.1016/0378-1119(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 15.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:152–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Kaestner K H, Silberg D G, Traber P G, Schütz G. Genes Dev. 1997;11:1583–1595. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]
- 17.Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, Grosveld F, Meijer D. Science. 1996;273:507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- 18.Bermingham J R J, Scherer S S, O’Connell S, Arroyo E, Kalla K A, Powell F L, Rosenfeld M G. Genes Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- 19.Hares K A, Foster G A. J Chem Neuroanat. 1991;4:187–203. doi: 10.1016/0891-0618(91)90002-t. [DOI] [PubMed] [Google Scholar]
- 20.Leiden J M. Annu Review Immunol. 1993;11:539–570. doi: 10.1146/annurev.iy.11.040193.002543. [DOI] [PubMed] [Google Scholar]
- 21.Fowlkes B J, Pardoll D M. Adv Immunol. 1989;44:207–264. doi: 10.1016/s0065-2776(08)60643-4. [DOI] [PubMed] [Google Scholar]
- 22.Love P E, Shores E W, Johnson M D, Tremblay M L, Lee E J, Grinberg A, Huang S P, Singer A, Westphal H. Science. 1993;261:918–921. doi: 10.1126/science.7688481. [DOI] [PubMed] [Google Scholar]
- 23.Liu C-P, Ueda R, She J, Sancho J, Wang B, Weddell G, Loring J, Kurahara C, Dudley E C, Hayday A, et al. EMBO J. 1993;12:4863–4875. doi: 10.1002/j.1460-2075.1993.tb06176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malissen M, Gillet A, Ardouin L, Bouvier G, Trucy J, Ferrier P, Vivier E, Malissen B. EMBO J. 1995;14:4641–4653. doi: 10.1002/j.1460-2075.1995.tb00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dave V P, Cao Z, Browne C, Alarcon B, Fernandez-Miguel G, Lafaille J, de la Hera A, Tonegawa S, Kappes D J. EMBO J. 1997;16:1360–1370. doi: 10.1093/emboj/16.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fehling H J, Kotkava A, Saint-Ruf C, Boehmer H. Nature (London) 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 27.Mayall T P, Sheridan P L, Montminy M R, Jones K A. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]