Abstract
Analysis of the genetic changes in human tumors is often problematical because of the presence of normal stroma and the limited availability of pure tumor DNA. However, large amounts of highly reproducible “representations” of tumor and normal genomes can be made by PCR from nanogram amounts of restriction endonuclease cleaved DNA that has been ligated to oligonucleotide adaptors. We show here that representations are useful for many types of genetic analyses, including measuring relative gene copy number, loss of heterozygosity, and comparative genomic hybridization. Representations may be prepared even from sorted nuclei from fixed and archived tumor biopsies.
Analysis of the genetic changes in human tumors is often problematical because of the presence of normal stroma. Although either microdissection or flow cytometry can produce small samples highly enriched for tumor cells or nuclei, the extracted DNA is of insufficient quantity for most uses. Nevertheless, we have successfully performed a complex protocol, representational difference analysis (RDA), on such small samples. RDA is a subtractive DNA hybridization technique that discovers the differences between paired normal and tumor genomes (1). The first step of RDA is the preparation of “representations,” which are highly reproducible reformattings and amplifications of DNA populations. Typically, a representation is a set of restriction endonuclease fragments of a limited size range amplified by PCR. As much as 100 μg of DNA can be prepared from as little as 3 ng of DNA (≈1 × 103 cells).
In RDA, a representation with much lower complexity than the starting population is needed to enable a subtractive hybridization step to proceed effectively. Such low complexity representations (LCRs) do not “capture” enough (typically, ≤7%) of the genome to be useful for many of the more common types of analyses. However, we demonstrate here that high complexity representations (HCRs) can provide ample amounts of DNA in a sufficiently reproducible manner suitable for most conventional studies. We demonstrate one type of HCR that captures ≈70% of the genome and illustrate its use for determining gene copy number, deletion mapping, loss of heterozygosity (LOH), and comparative genomic hybridization (CGH). HCRs may be a generally useful means of “immortalizing” and archiving DNA for later analysis from nonrenewable sources.
MATERIALS AND METHODS
Materials.
Restriction endonucleases as well as T4 DNA ligase, T4 DNA polymerase, and T4 polynucleotide kinase were supplied by New England Biolabs. AmpliTaq was supplied by Perkin–Elmer. Oligonucleotide adaptors RBgl-12 and RBgl-24 [as used in Lisitsyn et al. (1)], oligonucleotides used for PCR of World-Wide Web probes, and tetranucleotide repeat D17S695 were synthesized by BioSynthesis (Lewisville, TX). Genescreen Plus was purchased from DuPont. Radioactive nucleotides, rediprime labeling kit, dNTPs, and hyperfilm were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden). Cell lines used were obtained through American Type Culture Collection, grown in culture, and DNA-prepared. Normal human placenta DNA was obtained from CLONTECH. Oligonucleotides and probes for use with the Applied Biosystems 7700 Sequence Detector were synthesized by Applied Biosystems. Nusieve agarose gels used for analysis of PCR products were purchased from FMC. World-Wide Web probe sequences were downloaded from the Massachusetts Institute of Technology human genome sequencing database (http://www-genome.wi.mit.edu/).
Production of Representations.
Genomic DNA (3–10 ng) was digested by the desired restriction endonucleases (DpnII and BglII) under conditions suggested by the supplier. The digest was purified by phenol extraction and then precipitated in the presence of 10 μg of tRNA. The digested DNA was resuspended by the addition of 444 pmol of each adaptor (RBgl24 and RBgl12), T4 DNA ligase buffer (diluted to 1×, provided with enzyme), and water to bring the volume to 30 μl. The reaction was placed in a 55°C heat block, and the temperature was decreased slowly to 15°C by placing the heat block at 4°C (for ≈1 hr). On reaching 15°C, the RBgl24 adaptor was ligated by the addition of 400 units of T4 DNA ligase and by incubation at 15°C for 12–18 hr. The ligated material was divided into two tubes, and the following was added: 80 μl of 5× PCR buffer [335 mM Tris⋅HCl, pH 8.8/20 mM MgCl2/80 mM (NH4)2SO4/50 mM β-mercaptoethanol/0.5 mg/ml of BSA], 2′-deoxynucleoside 5′-triphosphates to a final concentration of 0.32 mM, RBgl24 adaptor to a final concentration of 0.6 μM, and H2O to bring the volume to 400 μl. Each reaction was overlaid with 100 μl of mineral oil. The reaction was placed in a thermal cycler preheated at 72°C, and 15 units of AmpliTaq was added to the tubes. The thermal cycler was set to continue at 72°C for 5 min to allow for filling in the 3′ ends of the ligated molecules. This step was followed by 20 cycles lasting 1 min at 95°C and 3 min at 72°C, with an additional extension of 10 min at 72°C after the last cycle. The PCR was divided into two tubes (now a total of four tubes), and the following reagents were added: 40 μl 5× PCR buffer (as above), dNTP to a final concentration of 0.32 mM, RBgl24 adaptor to a final concentration of 0.6 μM, water to bring the volume to 400 μl, and 100 μl of mineral oil. The tubes then were amplified for an additional five cycles with extension according to the above conditions. The reactions were purified by phenol-chloroform and then precipitated by the addition of 1/10th the reaction volume of sodium acetate (3M pH 5.2) and by the addition of one reaction volume of isopropanol.
PTEN Tumor Suppressor Lori Genomic Sequencing.
Bacterial artificial chromosome DNA was purified by detergent lysis and polyethylene glycol precipitation. DNA (5 μg) was sheared and then repaired by using T4 DNA polymerase. Fragments in the range of 1.5–2 kb were isolated by gel fractionation and inserted into M13 by ligation. Sequencing reactions were done by using dye primer chemistry that uses energy transfer primers (2) and thermosequenase polymerase (3). Reactions were run on an Applied Biosystems 377 sequencer for 6- to 8-hr sequence runs for analysis. Base calling was carried out by using the program phred (P. Green, University of Washington). The collection of initial data was assembled into a contig by using the automated assembly program phrap (P. Green). The database generated was converted to xgap format and edited by using xgap (4, 5). After completion of ≈6-fold coverage, the finishing process was carried out by using the program finish (G. Marth, Genome Sequencing Center at Washington University) to fill gaps in the contig.
Southern Blotting.
Normal DNA from a tumor cell line or representations derived from sorted nuclei were digested with either DpnII or BglII. Digested genomic DNA (5.0 μg), digested HCR (5.0 μg) , or digested LCR (0.5 μg) was loaded on a 1.5% agarose gel. The gel was transferred to Genescreen Plus after electrophoresis was completed. After transfer, the blot was hybridized in GIBCO prehybridization solution at 67°C for 2 hr. The probe was produced by random priming with the rediprime kit in the presence of 32PαdCTP. After 12–18 hr of hybridization, the blots were washed with standard saline phosphate/EDTA buffer as described (6). The blots were used for producing either auto-radiograms and/or phosphorimages.
LOH Analysis.
LOH was assessed by using the tetranucleotide repeat marker D17S695 (listed as UT269 in the National Center for Biotechnology Information Web Site http://www.ncbi.nlm.nih.gov/Entrez/nucleotide.html), which maps to the 17p13 region of the human genome and has been used to determine the state of LOH at the p53 locus (7, 8). The reverse primer was labeled by phosphorylation in the presence of 32PγATP as described (7, 8). The labeled reverse primer in combination with both forward and reverse primers was used for PCR by using 50 ng of HCR as DNA template. PCRs were performed as described in the National Center for Biotechnology Information Web Site, and gel electrophoresis was performed as described (7, 8).
CGH.
CGH was performed according to standard procedure (9) by using HCR DNAs and genomic DNAs prepared from BT474 and MCF7 as target and normal female human lymphocyte as the reference. The genomic and HCR DNA samples were labeled by nick-translation with fluorescein isothiocyanate–12-dUTP and Texas Red 5-dUTP to produce DNA fragments ranging in size between 500 and 2,000 bp.
Quantitative PCR.
Primer and probe sequences were produced according to the Applied Biosystems software primer express. These primers and probes were used in a PCR that was placed in the Applied Biosystems 7700 Sequence Detector and amplified as described (10, 11). Samples used were HCRs produced from DNA derived from nuclei of sorted primary tumor biopsies. After the PCR, the data of fluorescence corresponding to cycle number were downloaded from the Applied Biosystems 7700 Sequence Detector to Microsoft excel and analyzed to give the graphs shown later.
RESULTS
The Basic Method.
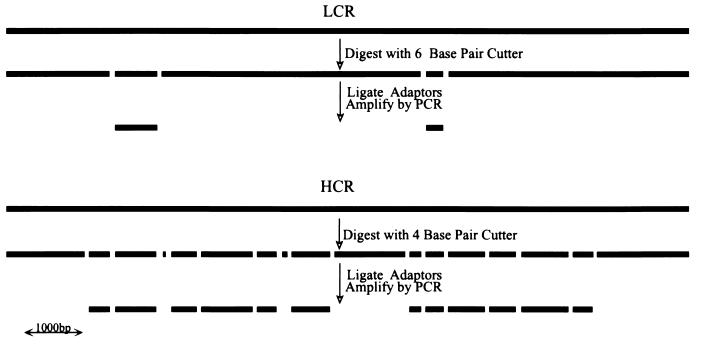
DNA from samples, usually paired tumor cells or nuclei and normal cells or nuclei, were processed in parallel to prepare representations. DNA was cleaved with a restriction endonuclease, such as DpnII or BglII, that is not blocked by 5-methylcytosine, and double-stranded, cohesive, adaptor oligonucleotides were ligated to the fragment ends. The adaptors were not phosphorylated and therefore could not be self-ligated to form interfering dimers. Because adaptors can be ligated only to the 5′ ends of the cleavage fragments, the ligated product then was treated with DNA polymerase to fill in the 3′ ends, forming the primer binding site for the subsequent PCR. Amplification was performed in stages, each stage using the adaptor oligonucleotides as primers and proceeding for no more than 20 rounds per stage. PCR does not amplify all fragments equally well, and high molecular weight fragments in particular are very poorly amplified. Thus, restriction endonucleases that cleave infrequently were used to prepare LCRs, and enzymes that cleave frequently were used to prepare HCRs (see Fig. 1). From 3 ng of starting material, we could obtain 40 μg of total HCR from a total of 25 rounds. We have not tested the usefulness of an HCR beyond a total of 35 rounds.
Figure 1.
Schematic comparison of LCR and HCR, illustrating the complexity reduction that occurs after cleavage with rare and frequent cutters, respectively.
Sampling the Complexity and Reproducibility of DpnII Representations.
We first sampled the reproducibility and complexity of DpnII HCRs. We analyzed 14 different HCRs, each made from 5 ng of DNA prepared from diploid nuclei separated from tumor biopsies by flow cytometry and each amplified by PCR for 25 rounds. In our first sampling, we designed pairs of PCR primers to detect Web Site sequence-tagged sites. We picked sequence-tagged sites that were not cleaved by DpnII and used primer pairs that amplified a single band from total genomic DNA controls. Of these, 18 of 25 pairs (72%) were able to amplify the same molecular weight fragment from each HCR, and 7 generally failed to amplify from any HCR. Our results suggest that DpnII HCRs reproducibly contain the same elements and ≈70% of the genome.
We performed a similar sampling with primer pairs derived from the locus encoding the PTEN tumor suppressor gene, for which we had the complete nucleotide sequence (unpublished data). In this way, we were able to use primers derived from DpnII fragments of known size. DpnII fragments were chosen at random, and PCR primer pairs were designed for each. Primer pairs (22 pairs) amplified single fragments by PCR from control genomic DNAs. These pairs were used with the same panel of 14 HCRs used above. Primer pairs (20 pairs) amplified the expected fragment from all HCRs, and 2 pairs failed to amplify from any. The fragments that were not in the HCRs were the largest, 3,916 bp, and one of the smallest, 97 bp. Totaling all fragment lengths, 16,039 bp were included in the HCRs, and 4,013 bp were excluded. Thus, assuming that our initial selection of DpnII fragments was random, the HCRs contained ≈75% of the PTEN region.
If DpnII cleavage is nearly complete during the preparation of an HCR, we expect that no PCR primer pairs should readily amplify from an HCR when the amplified sequence has an internal DpnII site. To test this, we chose four primer pairs from the PTEN locus that amplified a single fragment containing a single internal DpnII site. All four pairs amplified fragments from genomic DNA controls, and none amplified detectable fragments from the 14 HCRs. We conclude that HCRs prepared in parallel from samples processed in parallel are reasonably reproducible and represent ≈70% of the human genome.
Measuring Gene Copy Number in HCRs.
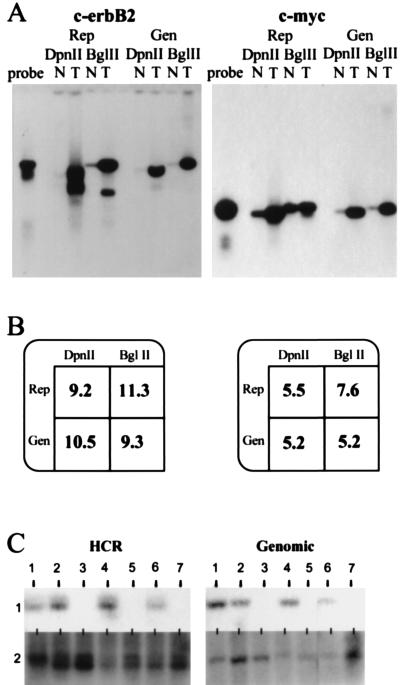
Tumor genomes often contain either extra copies of sequences caused by gene amplification or missing sequences caused by gene deletion. To explore the usefulness of representations for measuring gene copy number, we first compared Southern blots of genomic DNA to blots of HCRs and LCRs. For this purpose, we used human placental DNA as normal and prepared genomic DNA from tumor cell lines amplified at c-erbB2 (BT-474) (12) or c-myc (SK-BR-3) (13). HCRs and LCRs were made from cell line or placental DNAs by using DpnII or BglII, respectively. As probes we used small BglII fragments that we cloned from P1s containing inserts from the designated loci. The blots, shown in Fig. 2A, were quantitated by phosphorimaging. To normalize for loading differences, the blots were stripped and rehybridized with a single copy sequence probe. The normalized ratios of signal from tumor and normal are tabulated in Fig. 2B. The same relative copy number (tumor to normal) was determined from blots of representations as was determined from blots of genomic DNAs, indicating that there is a quantitatively reproducible amplification of these test sequences during the preparation of either HCRs or LCRs prepared in parallel from similar starting materials. Similar results were obtained for the cyclin D locus (data not shown).
Figure 2.
Analysis of copy number using representations (Rep) and genomic DNA (Gen) for c-erbB2 and c-myc. (A) Southern blot comparing tumor cell lines (T) to normal (N) cell lines. DpnII was used to prepare HCRs (DpnII), and BglII was used to prepare LCRs (BglII). As a reference, free probe also was run alongside the samples (probe). The hybridization probes were derived from small BglII fragments isolated from P1 clones specific for each locus respectively. (B) Quantitation of the Southern blot. Rep, representation; Gen, genomic DNA; DpnII, either DpnII HCR or DpnII digest of genomic DNA; BglII, either BglII HCR or BglII digest of genomic DNA. Blots in A were stripped and reprobed with a single copy probe to analyze loading differences (data not shown). Phosphorimage analysis of this probe was used to normalize the amplification differences. After this analysis, the resulting intensity of tumor was divided by the intensity of normal to give the fold amplification displayed. (C) Deletion mapping using HCRs and the deletion mapping of seven tumor cell lines comparing blots of HCRs (HCR) with blots of the corresponding genomic DNA (Genomic), each designated 1–7. The probe used for hybridization is from the human genomic region 20p11 (HCR). The Genomic panel shows the same DNAs blotted with an unrelated probe.
To explore the usefulness of HCRs for deletion mapping, we blotted both genomic and HCR DNAs from tumor cell lines for deletion at the 20p11 locus. This locus was discovered initially by using RDA and subsequently was found to be deleted frequently in gastrointestinal cancers (R.L., unpublished data). Fig. 2C illustrates that the probe hybridized to sequences in the HCRs when and only when it hybridized to sequences in the respective genomic DNA. An unrelated probe detects sequences present in all genomic samples and their representations.
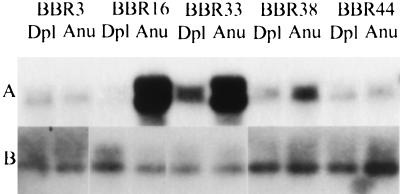
We tested the value of the HCRs made from limited amounts of DNA for quantitation of copy number. HCRs prepared from aneuploid and diploid nuclei sorted from several breast cancer biopsies were blotted for c-erbB2. Fig. 3A illustrates that c-erbB2 is amplified in the HCRs made from the aneuploid nuclei of some biopsy samples. Fig. 3B shows the blots with an unrelated probe. We obtained confirmation of the validity of the c-erbB2 amplifications by demonstrating that probes adjacent to but distinct from the c-erbB2 probe also were amplified in the same samples (M.N., unpublished results).
Figure 3.
Comparison of HCRs from primary tumor biopsies by Southern blotting. (A) HCRs from the designated tumor biopsies (denoted by an identification number preceded by BBR) were prepared from diploid (Dpl) and aneuploid (Anu) nuclei and compared by Southern blot analysis. The c-erbB2 probe was the same as that used in Fig. 2. (B) The same DNAs blotted with an unrelated probe.
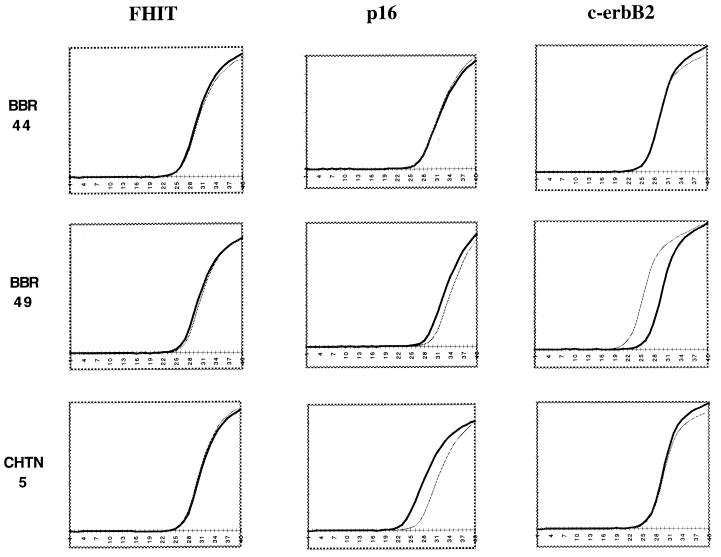
Finally, we tested some of the same samples by quantitative PCR. For this purpose, we used the dual-labeled fluorogenic hybridization probes and the Applied Biosystems 7700 Sequence Detector (10, 11) to compare HCR DNAs prepared from aneuploid and diploid nuclei pairs. The results, shown in Fig. 4, indicate that differences in copy number were detected by probes for the c-erbB2 oncogene and the p16 tumor suppressor. On the other hand, no differences in copy number were detected by probes from an uninvolved region on chromosome 3. The curve of amplification of the c-erbB2 fragment arises four cycles sooner in one aneuploid HCR than it does in the paired diploid HCR, indicating a higher copy number for c-erbB2 (≈16-fold higher in the aneuploid HCR sample). The curve for the aneuploid HCR deleted for p16 arises three cycles later than the paired diploid HCR (approximately one-eighth as much in the aneuploid HCR), probably reflecting ≈10% contamination of the aneuploid nuclei with diploid nuclei after sorting. These results were confirmed by Southern blotting (data not shown). One tumor/normal pair showed a shift of a single cycle for primer pairs detecting the p16 gene, which might reflect loss of a single allele in the tumor or experimental error.
Figure 4.
Quanitative PCR analysis of HCRs. Diploid (black) and aneuploid (gray) HCRs derived from sorted primary tumor biopsies were used as template for quanitative PCR analysis. Probes from several genomic loci (FHIT, p16, and c-erbB2) were used to determine copy number in three primary tumor pair HCRs (BBR44, BBR49, and CHTN5). The data from the Applied Biosystems 7700 Sequence Detector was analyzed with Microsoft excel to produce the graphs shown. (x axis, the cycle number during the reaction; y axis, the fluorescence detected.)
Detection of LOH in HCRs.
LOH is a common lesion found in cancer cells and may be indicative of genomic instability and/or the loss of function of a specific tumor suppressor gene (14–17). The detection of LOH often is obscured by the presence of normal stroma; hence, we tested whether HCRs prepared from minute amounts of samples highly enriched for tumor nuclei could be used for LOH analysis. PCR primers that amplify microsatellites and detect fragment length polymorphisms are used frequently for LOH mapping, and we chose to examine a primer pair that amplifies a highly polymorphic tetranucleotide repeat near the p53 locus (7, 8).
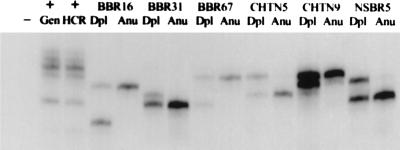
In preliminary experiments, we established that these PCR primers detected the same allele pattern in both genomic and HCR DNAs prepared from cell lines. Next, we examined 12 pairs of HCRs prepared from aneuploid and diploid nuclei. LOH at this locus was detected clearly in 9 of 10 informative pairs (see Fig. 5 for representative cases), which is greater than the reported proportion of LOH at this locus in breast cancer (50%) (14, 18, 19) but may reflect the selection of the highly aneuploid tumors that we sorted and/or the purity of our samples.
Figure 5.
Using HCRs for LOH analysis. LOH analysis was carried out on HCRs derived from sorted primary tumor biopsies. Dpl, diploid HCR; Anu, aneuploid HCR. The tumors are labeled as either BBR, CHTN, or NSBR followed by an identification number. The primers used in the reaction amplify a tetranucleotide repeat within the p53 locus. A mixed population of normal DNAs was used as a positive control. (+Gen, normal genomic DNA used as template; +HCR, HCR produced from this normal DNA used as template. −, reaction in which no template was added.)
Comparative Genome Hybridization with HCRs.
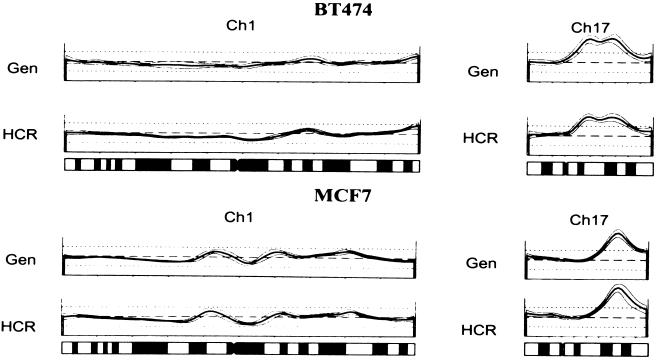
CGH is a powerful tool for analyzing the global genomic changes of tumors (20–25). It has been reported that chromosome painting could be performed with representations produced from each chromosome (26). We therefore tested whether CGH could be performed with HCRs. For this experiment, we chose to examine tumor cell lines so that we could directly compare CGH performed with genomic DNA with CGH performed with HCR. Little difference between HCR and genomic DNA could be discerned with either of the two cell lines examined, BT-474 and MCF7. Fig. 6 shows a sample of the chromosomal scanning profiles obtained with each DNA source.
Figure 6.
CGH analysis using HCRs. Shown are two representative chromosome spreads (Ch 1, and Ch17) comparing the genomic (Gen) with the HCR for two different cell lines, BT474, and MCF7. Deviation from the dashed line represents genomic change, a peak represents gain in copy number, and a trough represents loss of copy number. Underneath the profiles is an idiogram of each chromosome as a reference.
DISCUSSION
The idea of capturing essential features of the genome as a PCR product is not new. In 1989, Kinzler and Vogelstein (27) described “whole genome” PCRs to select for DNA sequences that were binding sites for DNA binding proteins. Their method was used in 1995 (28) for the same purpose. In the experiments from 1989, PCR adaptors were blunt-end ligated to the cleavage fragments of total genomic DNA. The usefulness of the resulting PCR products for genomic analysis was not explored; nor was its efficiency explored when starting with tiny amounts of material. An alternate approach to whole genome PCR is to use random priming with degenerate oligonucleotides (29–33). This method can produce large amounts of DNA starting from very minute amounts of sample, but the complexity of the DNA produced, the reproducibility of the representation, and the loss of natural restriction sites at the ends of the amplified material make the usefulness of this method somewhat limited. Restriction endonuclease-based representations have major advantages: Their complexity can be regulated by the choice of restriction enzyme; they can be readily reamplified; they can be analyzed by Southern blotting; and they are highly reproducible.
In this report, we have explored the uses of HCRs, including quantitative assessment of copy number, LOH, and CGH. All of these analytical methods require a high level of reproducibility in the representation. To achieve this level of reproducibility, we have prepared paired samples of HCRs from the same amount of starting material, used genomic DNAs extracted in the same manner, and performed PCR at the same time, under the same conditions, and in the same thermal cycler.
In principle, HCRs can be prepared from normal and tumor tissue stored as fixed, paraffin-embedded, archived biopsies, which would extend greatly the usefulness of such samples. We have made HCRs from DNAs extracted from pairs of aneuploid and diploid nuclei sorted from such sources (R.L., unpublished data). More rounds of PCR are required to obtain workable amounts of DNA, and HCRs from DNA extracted from fixed specimens have a markedly lower size distribution than HCRs prepared from fresh sources. Moreover, there is enormous variability between the HCRs from different specimens, reflected as varying size distributions. This diversity is probably caused by the variation in the quality of the DNA that can be extracted from specimens fixed and stored under different conditions (19, 34–36). Despite the variability between specimens, we found that the HCRs prepared from aneuploid and diploid nuclei from the same fixed specimen are similar to each other. Indeed, we have found that HCRs prepared from sorted nuclei of fixed specimens are useful for CGH (unpublished observations). These results suggest that normal and tumor HCRs will be useful even when prepared for analysis from microdissected specimens.
Both HCR and LCR render paired tumor and normal DNAs from minute specimens in a stable format that can be analyzed or further amplified at a later date. We have emphasized the usefulness of HCRs for gene copy, LOH, and global genomic analysis of tumor specimens. Most probes, chosen at random, will be present in an HCR but will not be present in the LCRs. However, precisely because LCRs are of lower complexity, hybridization-based assays that depend on completeness of hybridization are easier to perform. This ease is apparent in blotting analysis of LCRs. For the same reason, some global genomic analyses, such as microchip array analysis (37–40) or CGH (20–25), that depend on hybridization kinetics should be facilitated by the use of LCRs because reannealing times should be reduced and signal-to-noise ratios enhanced.
Acknowledgments
We thank Masaaki Hamaguchi, Eli Hatchwell, and Clifford Yen for useful discussions and probe sequences; Bert Vogelstein, Manuel Perucho, and Nikolai Lisitsyn for critical reading of the manuscript; Linda Rodgers and Mike Riggs for technical assistance; Jim Duffy for artwork, and Patricia Bird for secretarial assistance. This work was supported by grants to M.W. from the National Institutes of Health (OIG-CA39829 and 5P50-CA-68425); the United States Army (DAMD17-94-J-4247); 1 in 9: The Long Island Breast Cancer Action Coalition; and Tularik. M.W. is an American Cancer Society Research Professor.
ABBREVIATIONS
- HCR
high complexity representation
- LCR
low complexity representation
- RDA
representational difference analysis
- CGH
comparative genomic hybridization
- LOH
loss of heterozygosity
References
- 1.Lisitsyn N, Lisitsyn N, Wigler M. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 2.Ju J, Ruan C, Fuller C W, Galazer A N, Mathies R A. Proc Natl Acad Sci USA. 1995;92:4347–4351. doi: 10.1073/pnas.92.10.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1995;93:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dear S, Staden R. Nucleic Acids Res. 1991;19:3907–3911. doi: 10.1093/nar/19.14.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gleeson T J, Staden R. Comput Appl Biosci. 1991;7:398. doi: 10.1093/bioinformatics/7.3.398. [DOI] [PubMed] [Google Scholar]
- 6.Sambrook J, Fritsch E F, Maniatis T. In: Molecular Cloning: A Laboratory Manual. Ford N, Nolan C, Ferguson M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 9.38–9.56. [Google Scholar]
- 7.Gerken S C, Albertsen H, Elsner T, Ballard L, Holik P, Lawrence E, Moore M, Zhao X, White R. Am J Hum Genet. 1995;56:484–499. [PMC free article] [PubMed] [Google Scholar]
- 8.Stack M, Jones D, White G, Liscia D S, Venesio T, Casey G, Crichton D, Varley J, Mitchell E, Heighway J, et al. Hum Mol Genet. 1995;4:2047–2055. doi: 10.1093/hmg/4.11.2047. [DOI] [PubMed] [Google Scholar]
- 9.DeVries S, Gray J W, Pinkel D, Waldman F M, Sudar D. In: Current Protocols in Human Genetics. Boyle A L, editor. New York: Wiley; 1995. , Suppl. 6, Unit 4.6, pp. 1–18. [DOI] [PubMed] [Google Scholar]
- 10.Gibson U E, Heid C A, Williams P M. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 11.Isono K. Rinsho Byori. 1997;45:218–223. [PubMed] [Google Scholar]
- 12.Szollosi J, Balazs M, Feuerstein B G, Benz C C, Waldman F M. Cancer Res. 1995;55:5400–5407. [PubMed] [Google Scholar]
- 13.Feo S, Di L C, Jones T, Read M, Fried M. Oncogene. 1994;9:955–961. [PubMed] [Google Scholar]
- 14.Deng G, Chen L C, Schott D R, Thor A, Bhargava V, Ljung B M, Chew K, Smith H S. Cancer Res. 1994;54:499–505. [PubMed] [Google Scholar]
- 15.Patel U, Chen H C, Banerjee S. Cell Mol Biol Res. 1994;40:683–691. [PubMed] [Google Scholar]
- 16.Wada C, Shionoya S, Fujino Y, Tokuhiro H, Akahoshi T, Uchida T, Ohtani H. Blood. 1994;83:3449–3456. [PubMed] [Google Scholar]
- 17.Wieland I, Ammermuller T, Bohm M, Totzeck B, Rajewsky M F. Oncol Res. 1996;8:1–5. [PubMed] [Google Scholar]
- 18.Chen L C, Neubauer A, Kurisu W, Waldman F M, Ljung B M, Goodson W, III, Goldman E S, Moore D, II, Balazs M, Liu E, et al. Proc Natl Acad Sci USA. 1991;88:3847–3851. doi: 10.1073/pnas.88.9.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y H, Li C D, Yap E P, McGee J O. J Pathol. 1995;177:129–134. doi: 10.1002/path.1711770205. [DOI] [PubMed] [Google Scholar]
- 20.Bryndorf T, Kirchhoff M, Rose H, Maahr J, Gerdes T, Karhu R, Kallioniemi A, Christensen B, Lundsteen C, Philip J. Am J Hum Genet. 1995;57:1211–1220. [PMC free article] [PubMed] [Google Scholar]
- 21.Carter N P, Ferguson S M, Perryman M T, Telenius H, Pelmear A H, Leversha M A, Glancy M T, Wood S L, Cook K, Dyson H M, et al. J Med Genet. 1992;29:299–307. doi: 10.1136/jmg.29.5.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cher M L, MacGrogan D, Bookstein R, Brown J A, Jenkins R B, Jensen R H. Genes Chromosomes Cancer. 1994;11:153–162. doi: 10.1002/gcc.2870110304. [DOI] [PubMed] [Google Scholar]
- 23.Hermsen M A, Meijer G A, Baak J P, Joenje H, Walboomers J J. Hum Pathol. 1996;27:342–349. doi: 10.1016/s0046-8177(96)90106-9. [DOI] [PubMed] [Google Scholar]
- 24.Kallioniemi O P, Kallioniemi A, Piper J, Isola J, Waldman F M, Gray J W, Pinkel D. Genes Chromosomes Cancer. 1994;10:231–243. doi: 10.1002/gcc.2870100403. [DOI] [PubMed] [Google Scholar]
- 25.Storkel S, Simon R, Brinkschmidt C, Gronwald J, Bocker W. Pathologe. 1996;17:189–194. doi: 10.1007/s002920050154. [DOI] [PubMed] [Google Scholar]
- 26.Vooijs M, Yu Y–C, Johnson D, Tkachuk D, Pinkel D, Gray J. Am J Hum Genet. 1993;52:586–597. [PMC free article] [PubMed] [Google Scholar]
- 27.Kinzler K W, Vogelstein B. Nucleic Acids Res. 1989;17:3645–3653. doi: 10.1093/nar/17.10.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joulin V, Richard F H. Eur J Biochem. 1995;232:620–626. doi: 10.1111/j.1432-1033.1995.620zz.x. [DOI] [PubMed] [Google Scholar]
- 29.Telenius H, Carter N P, Bebb C E, Nordenskjold M, Ponder B A, Tunnacliffe A. Genomics. 1992;13:718–725. doi: 10.1016/0888-7543(92)90147-k. [DOI] [PubMed] [Google Scholar]
- 30.Xu K, Tang Y, Grifo J A, Rosenwaks Z, Cohen J. Hum Reprod. 1993;8:2206–2210. doi: 10.1093/oxfordjournals.humrep.a138004. [DOI] [PubMed] [Google Scholar]
- 31.Kristjansson K, Chong S S, Van den Veyver I B, Subramanian S, Snabes M C, Hughes M R. Nat Genet. 1994;6:19–23. doi: 10.1038/ng0194-19. [DOI] [PubMed] [Google Scholar]
- 32.Sun F, Arnheim N, Waterman M S. Nucleic Acids Res. 1995;23:3034–3040. doi: 10.1093/nar/23.15.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao Y, Slijepcevic P, Arkesteijn G, Darroudi F, Natarajan A T. Cytogenet Cell Genet. 1996;75:57–62. doi: 10.1159/000134457. [DOI] [PubMed] [Google Scholar]
- 34.Ben E J, Johnson D A, Rossi J, Cook N, Wu A. J Histochem Cytochem. 1991;39:351–354. doi: 10.1177/39.3.1704393. [DOI] [PubMed] [Google Scholar]
- 35.Greer C E, Lund J K, Manos M M. PCR Methods Appl. 1991;1:46–50. doi: 10.1101/gr.1.1.46. [DOI] [PubMed] [Google Scholar]
- 36.Greer C E, Peterson S L, Kiviat N B, Manos M M. Am J Clin Pathol. 1991;95:117–124. doi: 10.1093/ajcp/95.2.117. [DOI] [PubMed] [Google Scholar]
- 37.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 38.Schena M, Shalon D, Heller R, Chai A, Brown P O, Davis R W. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schena M. Bioessays. 1996;18:427–431. doi: 10.1002/bies.950180513. [DOI] [PubMed] [Google Scholar]
- 40.Shalon D, Smith S J, Brown P O. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]