Abstract
The ST6Gal sialyltransferase controls production of the Siaα2-6Galβ1-4GlcNAc (Sia6LacNAc) trisaccharide, which is the ligand for the lectin CD22. Binding of CD22 to Sia6LacNAc is implicated in regulating lymphocyte adhesion and activation. We have investigated mice that lack ST6Gal and report that they are viable, yet exhibit hallmarks of severe immunosuppression unlike CD22-deficient mice. Notably, Sia6LacNAc-deficient mice display reduced serum IgM levels, impaired B cell proliferation in response to IgM and CD40 crosslinking, and attenuated antibody production to T-independent and T-dependent antigens. Deficiency of ST6Gal was further found to alter phosphotyrosine accumulation during signal transduction from the B lymphocyte antigen receptor. These studies reveal that the ST6Gal sialyltransferase and corresponding production of the Sia6LacNAc oligosaccharide are essential in promoting B lymphocyte activation and immune function.
Sialyltransferases are a family of glycosyltransferase enzymes that add sialic acid residues during oligosaccharide diversification (reviewed in ref. 1). Sialic acid addition occurs in the Golgi apparatus and generally terminates further oligosaccharide chain elongation. The outer position of sialic acid linkages places these residues in a location to provide key structural determinants in ligand formation for endogenous and pathogenic lectins. Three sialic acid linkage types commonly exist among vertebrates and the corresponding sialyltransferase genes have been previously isolated. The most abundant sialic acid linkage found among mammalian cell surface oligosaccharides is of the α2-3 variety and can be produced independently by four sialyltransferases that each, nonetheless, bear unique substrate preferences among glycolipids, asparagine (N)-linked glycans, and serine/threonine (O)-linked glycans (2). Sialyltransferases have also been found to be developmentally regulated and differentially expressed among various cell types (1–6). For example, expression of α2-8 linked sialic acids is much less common than α2-3 linkages and appears restricted to a small subset of glycoproteins (7–10).
α2-6-linked sialic acids are also less abundant than α2-3-linked forms and are generated by at least four distinct gene products. However, the ST6Gal sialyltransferase appears solely responsible for producing the Siaα2-6Galβ1-4GlcNAc (Sia6LacNAc) terminus on various N glycans, and perhaps on some O glycans (1, 11). High levels of ST6Gal RNA have been found to preferentially accumulate in hematopoietic cells, as well as in the liver (3–5). Moreover, ST6Gal gene transcription is regulated by multiple promoters and altered by glucocorticoids and cytokines (12–14). Although the physiologic role of the ST6Gal sialyltransferase has not been defined previously by available genetic approaches, it has been shown to be unique in producing the ligand for the CD22 lectin molecule expressed on B lymphocytes.
CD22 is a transmembrane glycoprotein lectin found exclusively on B lymphocytes and is known to play a role in the immunologic activation of these cells (15–17). CD22 has been found associated with the antigen receptor and is a target for tyrosine kinase phosphorylation on the cytoplasmic domain, which thereby recruits various signal transduction molecules (18, 19). The extracellular domain of CD22 specifically binds the Sia6LacNAc trisaccharide (20–22). This trisaccharide ligand exists on several lymphoid molecules. Lymphocyte interactions involving CD22 binding to CD45 have been reported (23). As CD22 itself carries Sia6LacNAc, homotypic binding interactions have been shown to occur and may play a regulatory role in immune function (24, 25). These results suggest that CD22 and Sia6LacNAc are a lectin–ligand pair with the potential to control immune cell surface interactions. However, a relatively simple model for CD22 function has not developed from analyses of CD22 null mice by several laboratories (26–29). Results obtained have inferred both positive and negative roles for CD22 in B lymphocyte immune function, suggesting that CD22 may modulate threshold signaling responses from the antigen-receptor complex.
To investigate ST6Gal-dependent physiology we have chosen a complementary approach involving the generation of mice deficient in the carbohydrate ligand for CD22 by inactivating the ST6Gal sialyltransferase gene implicated in it’s synthesis. We report that such mice develop normally but harbor an immunodeficient phenotype that is distinct from CD22 null mice. These studies describe an essential role for the ST6Gal sialyltransferase in B lymphocyte immune responses.
MATERIALS AND METHODS
ST6Gal Gene Targeting.
The ST6Gal targeting vector was assembled from a 129/Sv genomic clone by inserting the 1.9-kb AccI–BamHI fragment containing exon 2 of ST6Gal encoding the first 200 amino acids into the BamHI site of the pflox vector as described (30). Adjacent 129/Sv ST6Gal genomic sequences were added by subcloning the 1.8-kb NheI–AccI fragment into the SalI site and the BamHI–BamHI 12-kb fragment into the HindIII site of pflox, respectively. Ten micrograms of NotI-linearized targeting vector were electroporated into R1 embryonic stem (ES) cells (31) and G418 resistant transfectants (120 μg/ml), positive by PCR for homologous recombination and retaining all three loxP sites were transfected with pCreHygro expression vector. Following 4 days of gancyclovir (2 μM) selection, subclones were isolated and those bearing either the ST6GalF allele (B3) or the ST6GalΔ allele (B9) were resolved by Southern blotting with loxP and HindIII–HindIII genomic probes. B3 and B9 ES cells were used to generate chimeric mice in C57BL/6 host embryos. Offspring were genotyped by Southern blotting with BglII-digested tail DNA hybridized to the HindIII–HindIII 500-bp genomic probe. Heterozygous offspring were mated to C57BL/6 mates and the mutations were propagated in this strain for at least two generations before crosses to produce homozygotes for experimentation. The phenotypes described were found linked to mice bearing the homozygous ST6GalΔ mutant genotype in subsequent studies involving more than four generations. Mice homozygous for the ST6GalF genotype did not display any phenotypic consequences and as expected contained normal levels of Sia6LacNAc (data not shown and D.C. and J.D.M., unpublished work).
Fluorescence-Activated Cell Sorter (FACS) Analysis.
Single cell suspensions from the spleen, thymus, lymph node, and bone marrow were subjected to lysis of red blood cells by ammonium chloride. Cells were counted with a hemocytometer and 500,000 cells were labeled in a final volume of 100 μl with either SNA–fluorescein isothiocyanate (FITC) lectin (5 μg/ml; Vector Laboratories) or MAL II–FITC lectin (5 μg/ml; Vector Laboratories), and/or 1 μl of mAb or CD22–Ig chimera for 10 min. All incubations and washes were performed on ice in FACS buffer [2% fetal calf serum (FCS) in PBS]. Cells were analyzed using a FACScan flow cytometer and cellquest software (Becton Dickinson). Serum isotype-specific antibody titers were determined by ELISA using plates coated with anti-mouse isotype-specific antibodies. A standard curve was generated using purified mouse IgM, IgA (Sigma), and IgG1 (PharMingen) to convert OD values to micrograms per milliliter. Antibodies and lectins used in the course of these and other studies included here were from PharMingen unless otherwise noted): anti-CD3 (2C11), anti-CD4 (RM4-5), anti-CD8 (53-6.7), anti-CD19 (1D3), anti-CD21/35 (7G6), anti-CD23 (B3B4), anti-CD22 (Cy34.1), anti-CD43 (S7), anti-B220 (RA3-6B2), anti-mouse IgM (II/41), anti-mouse IgD (11-26c.2a), anti-CD24 (HSA; heat-stable antigen) (M1/69 and J11d), anti-CD40 (HM40-3), anti-CD44 (IM7), anti B7.2 (GL1), anti-major histocompatibility complex class II I-Ab (AF6[hyphen120.1), anti-erythroid (Ter-119), anti-Ly-6G (Gr-1) (RB6-8C5), and SNA–FITC (Vector Laboratories). In FACS analyses, biotinylated antibodies were detected using Streptavidin-Tri-Color (Caltag, South San Francisco, CA). The CD22–hIg fusion chimera (23) was provided by A. Varki (University of California, San Diego).
ST6Gal Sialyltransferase Activity Measurements.
Tissue samples were homogenized in 3 ml of 500 mM sucrose, 1 mM MgCl2, 1% dextran, 5 mM 2-mercaptoethanol. After removal of cell debris at 10,000 × g, membranes were isolated by centrifugation at 40,000 rpm in a Beckman SW50.1 rotor for 1 hr. Membranes were solubilized in 25 mM cacodylate buffer (pH 6.8)/2% Triton X-100 for 15 min on ice. The membrane and cytosolic fractions (12.5 μl) were analyzed for sialyltransferase activity in 25-μl assays with 25 mM cacodylate (pH 6.8), 0.4 mM CMP–sialic acid (Sigma), 105 cpm of CMP–[14C]sialic acid (170 pmol) (Amersham), and 1 mM LacNAc–octyl (provided by R. Ohrlein, Novartis, Switzerland). Reactions were incubated at 37°C for 1 hr. Products were separated from unincorporated sialic acid by chromatography on Sep-Pak C18 cartridges (Waters), dried by centrifugal evaporation, redissolved in 50 μl of 50 mM sodium citrate buffer (pH 6.0), and incubated for 1 hr at 37°C after the addition of 2 units Salmonella typhimurium LT2 sialidase (New England Biolabs). The digestion products were applied on Sep-Pak C18 cartridges, washed with 15 ml of H2O, and eluted with 5 ml of methanol. The amount of [14C]sialic acid in the methanol eluates was measured in a liquid scintillation counter (RackBeta, Pharmacia). Separately, a recombinant soluble form of the human ST6Gal enzyme produced in Pichia pastoris (provided M. Malissard, University of Zurich, Switzerland) was used in sialic acid transfer to the LacNAc–octyl acceptor and followed by the sialidase treatment described above. No significant cleavage was observed by loss of radiolabeling, thereby confirming the specificity of the LT2 sialidase for the α2-3 linkage when used in the above conditions. ST6Gal activity measurements undertaken with cytosolic fractions did not reveal any significant activity in either wild-type (wt) or mutant extracts (data not shown).
B Cell Proliferation Assays.
B lymphocyte isolation was accomplished by subjecting splenocytes or lymph node cell suspensions to complement mediated lysis with anti-Thy1.2 (Becton Dickinson), anti-Ly-6G (Gr-1), and rabbit complement (Accurate Chemicals), followed by a cushion centrifugation onto NycoPrep (Life Technologies, Gaithersburg, MD) to remove debris and dead cells. Viable cells were identified by FACS as >90% B lymphocytes using anti-B220. Equivalent numbers of B cells of each genotype (1 × 105) were cultured in complete RPMI 1640 medium containing 2-mercaptoethanol (0.1 mM), 10% FCS, and l-glutamine with the indicated concentrations of goat F(ab′)2 anti-mouse IgM antiserum (Jackson, West Grove, PA), anti-CD40 with or without IL-4 (Genzyme), or lipopolysaccharide (LPS) (Sigma). Proliferation was measured by [3H]thymidine incorporation (2.5 μCi per well) during the last 16 hr of a 64-hr assay period.
Calcium Measurements.
Single cell suspensions of splenocytes were isolated and subjected to ammonium chloride red cell lysis. Cells were then counted using a hemocytometer and 107 cells were incubated with 10 μM Indo-1 AM ester (Molecular Probes) at 37°C for 30 min in 1 ml of complete RPMI 1640 medium. Cells were washed in FACS buffer and stained with FITC-conjugated anti-B220 (PharMingen) for 10 min at room temperature, washed again and resuspended in 1 ml of complete RPMI 1640 medium containing 10 mM Hepes (pH 7.4). Two hundred microliters of this suspension was added to 750 μl of RPMI–Hepes medium and B220+ cells were analyzed by FACS using a Coulter Elite (Coulter) with multitime software (Phoenix Flow Systems, San Diego). Baseline fluorescence ratios (525/405 nm) were collected for 1 min before the addition of antibodies. The final concentrations of goat F(ab′)2 anti-mouse IgM used were 10 μg/ml and 30 μg/ml.
Protein Phosphotyrosine Analyses.
Splenic B cells were isolated by complement-mediated lysis (see above) and 5 × 105 isolated B cells were resuspended in 50 μl of RPMI 1640 medium with 0.5% FCS. Cells were warmed to 37°C before stimulation with 10 μl of goat F(ab′)2 anti-mouse IgM (120 μg/ml). At the indicated times, 15 μl of lysis buffer was added to give a final concentration of 1% Triton X-100, 50 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 10 mM NaF, 1 mM Na3VO4, 1 mM Na2MoO4, 2 μg/ml leupeptin, 2 μg/ml pepstatin, 2 μg/ml aprotinin, 5 μg/ml soybean trypsin inhibitor, and 40 μg/ml phenylmethylsulfonyl fluoride. Whole cell extracts representing 1 × 105 splenic B cells were run on a SDS/10% PAGE gel and transferred to nitrocellulose. Proteins phosphorylated on tyrosine were detected with monoclonal anti-phosphotyrosine antibody 4G10 (Upstate Biotechnology, Lake Placid, NY) by immunoblotting.
Immunizations and Serum Antibody Assays.
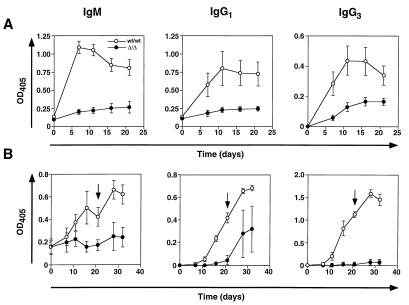
Mice were prebled to obtain preimmune sera and subsequently immunized by i.p. injection of either 10 μg or 100 μg of dinitrophenyl (DNP)–keyhole limpet hemocyanin (KLH) (Calbiochem) in Freund’s complete adjuvant, or 0.1 μg or 10 μg of DNP–Ficoll (Biosearch) in PBS. Serum was collected at the indicated times and anti-DNP titers were determined by ELISA using plates coated with 20 μg of DNP–BSA and blocked with 10% FCS in PBS. Mice receiving the DNP–KLH antigen were boosted at the indicated times with the same amount of antigen in Freund’s incomplete adjuvant. Sera were diluted to various concentrations and analyzed using anti-mouse isotype-specific antibodies conjugated to alkaline phosphatase (for IgM, Sigma; for IgG1, Caltag; for IgG3, Southern Biotechnology Associates). OD405 values were obtained using a microplate reader (Molecular Devices). Results shown in Fig. 4 comprise the indicated sera dilution in the linear range for OD405 values obtained.
Figure 4.
Deficient antibody production following immunization of Sia6LacNAc-deficient mice. (A) T-independent antigen immunization (10 μg of DNP–Ficoll) was followed by analysis of anti-DNP antibody levels in sera at indicated times. (B) T-dependent antigen DNP–KLH immunization (10 μg and 100 μg) was accomplished. Anti-DNP antibody levels were measured before and subsequent to a second boost immunization (arrow). Results obtained with 10 μg of DNP–KLH antigen were essentially identical to those depicted with 100 μg (data not shown). Multiple dilutions of sera were assayed to find the linear range of response by OD405 measurements (not shown). Sera dilution factors for results depicted are as follows: IgM (1/200), IgG1 against T-independent antigen (1/200), IgG3 against T-independent antigen (1/800), IgG1 against T-dependent antigen (1/1,000), and IgG3 against T-dependent antigen (1/200). Data are presented as the mean ± SEM from three mice of the indicated genotypes. Use of 0.1 μg of DNP–Ficoll in immunizations resulted in the absence of anti-DNP antibody production specifically in ST6Gal deficient mice, whereas low levels of anti-DNP antibodies were observed in wt mice (data not shown).
RESULTS
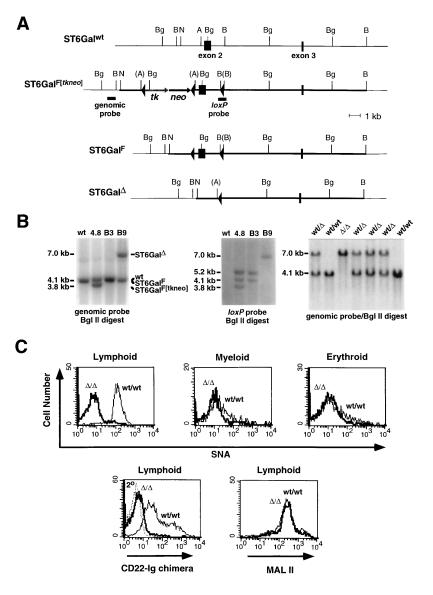
ST6Gal alleles were mutagenized in embryonic stem cells by homologous recombination and Cre recombinase action (Fig. 1 A and B). To achieve a systemic mutation, exon 2 was chosen for deletion. This exon contains the N-terminal 200 amino acids and over 50% of the ST6Gal coding sequence. Deletion of exon 2 results in loss of cytoplasmic, transmembrane, and considerable catalytic domain sequences, such that any resulting stably translated, truncated, and active enzyme would be incapable of entry into the lumen of the endoplasmic reticulum and Golgi apparatus. Mice homozygous for the deleted ST6GalΔ allele were born at normal frequency, were fertile, and did not exhibit abnormalities in weight or overt behavior (Fig. 1B and data not shown). Histologic studies of various tissues including liver, brain, kidney, spleen, and thymus were unremarkable. Additionally, a normal hematologic profile involving leukocytes, erythrocytes, and platelets was observed among peripheral blood samples (data not shown). Enzymatic studies of liver and splenocyte extracts using the Galβ1-4GlcNAc–octyl acceptor (see Materials and Methods) revealed that mice homozygous for the ST6GalΔ allele were deficient in ST6Gal activity (Table 1), indicating that the exon 2 deletion produced a null mutation.
Figure 1.
ST6Gal gene mutagenesis and function. (A) A partial ST6Gal genomic structure (ST6Galwt) was cloned and used with pflox in constructing a targeting vector (thick line) shown recombined at the ST6Gal locus (ST6GalF[tkneo] locus in ES cell clone 4.8). Following Cre recombinase expression and gancyclovir selection, ES cell subclones B3 and B9, bearing the ST6GalF allele or the ST6GalΔ allele, respectively, were isolated and used in generating chimeric mice. (B) Genomic Southern blots of ST6Gal alleleic structure in wt ES cells, targeted ES cell clone 4.8 (ST6GalF[tkneo]), 4.8 ES cell subclone B3 (ST6GalF), and 4.8 ES cell subclone B9 (ST6GalΔ) using either a genomic probe outside the targeting vector (Left) or a loxP probe (Center). (Right) A genomic Southern blot analysis of offspring derived from matings of mice heterozygous for the ST6GalΔ allele. Genotypes include the presence of mice homozygous for the exon 2 deletion (ST6GalΔ allele). (C) Using SNA and CD22-Ig lectins in FACS analyses, splenic CD3+ and B220+ lymphocytes normally express high levels of Sia6LacNAc in comparison with Gr-1+ myeloid and Ter-119+ erythroid cells. Lymphocytes from mice homozygous for the B9-derived ST6GalΔ allele were deficient in Sia6LacNAc (n = 7). α2-3-linked sialic acids were detected using the MAL II lectin and were found to be expressed at low levels normally and unaltered among cells from mice homozygous for the ST6GalΔ allele (n = 7). Mice homozygous for the B3-derived ST6GalF allele displayed wt profiles in these lectin-based analyses (data not shown). Similar results were obtained with mesenteric lymph node-derived lymphocytes (data not shown). Fluorescence signal intensity using an anti-human IgG–FITC conjugate is shown (2° and dotted line, Lower Left).
Table 1.
ST6Gal activity levels in membranes derived from wild-type (wt/wt) and homozygous-mutant (Δ/Δ) ST6Gal genotypes
| Genotype | ST6Gal activity, pmol/ min per mg of protein
|
|
|---|---|---|
| Liver | Splenocytes | |
| wt/wt | 13.4 ± 1.0 | 33.1 ± 9.5 |
| Δ/Δ | 0.3 ± 0.1 | 0.3 ± 0.2 |
Results are mean ± SD determined using three distinct mice of the indicated genotypes. ST6Gal activity is defined as detailed in Materials and Methods. Activity observed in Δ/Δ samples is not significantly distinct from background and is at the level of assay sensitivity.
Expression of the ST6Gal product was assessed using the Sambucus nigra-derived lectin SNA and a recombinant soluble CD22-Ig lectin chimera, which both bind the Sia6LacNAc trisaccharide with high specificity (20–22). In contrast to erythroid and myeloid cells analyzed, lymphocytes were found to highly express cell surface Sia6LacNAc (Fig. 1C). Lymphocytes from mice homozygous for the ST6GalΔ mutation were deficient in binding to SNA and CD22-Ig, whereas mice homozygous for the loxP-flanked ST6GalF allele expressed normal levels of Sia6LacNAc on their cell surfaces (Fig. 1C and data not shown). Levels of α2-3-linked sialic acids were visualized using the Maackia amurensis-derived lectin MAL II. No significant changes in α2-3-linked sialic acid levels were thereby observed among lymphoid cells isolated from mice homozygous for the ST6GalΔ allele (Fig. 1C). Residual or low-level SNA and CD22-Ig binding may reflect the presence of Siaα2-6GalNAc on O glycans (25), although it also remains possible that additional enzymes exist that produce low levels of Sia6LacNAc. Nevertheless, the deletion generated in the ST6Gal locus results in loss of ST6Gal activity and a deficiency in Sia6LacNAc production at the cell surface, consistent with the described biochemical role of ST6Gal and its’ inactivation in mice homozygous for the ST6GalΔ allele.
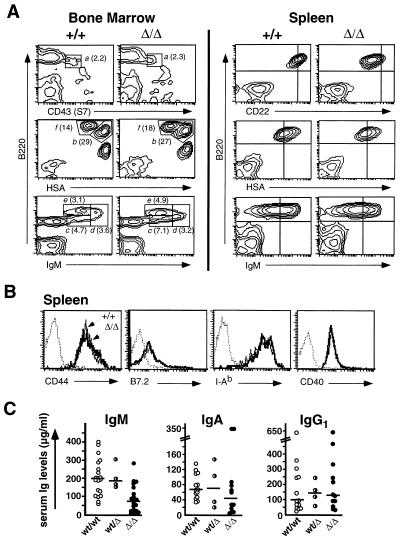
Sia6LacNAc-deficient mice contained normal numbers of Gr-1+ myeloid, Ter-119+ erythroid, B220+ B lymphoid, and T lymphoid (CD3+, CD4+, CD8+) cells in the thymus, spleen, lymph nodes, and bone marrow (data not shown). B lymphocyte development in the bone marrow of ST6Gal-deficient mice was unaltered. The percentage of pro-B cells in the ST6Gal-deficient bone marrow (B220lo CD43+, ref. 32) was normal (Fig. 2A, gate a). Furthermore, the frequency of immature B cells (B220lo IgMint, ref. 33) was not affected by ST6Gal inactivation (Fig. 2A, gates b and c). Similarly, no statistically significant changes were found in the abundance of mature B cells in the marrow (B220hi IgMint and B220hi HSAlo, ref. 33; Fig. 2A, gates e and f) or in the previously defined transitional B cell population undergoing maturation and negative selection (B220lo-hi IgMhi, ref. 33; Fig. 2A, gate d).
Figure 2.
B lymphocyte characterization and serum immunogloblin analyses. (A Left) Bone marrow lymphocytes were analyzed for cell surface expression of IgM, CD24 (HSA), CD43 (S7), and B220 by flow cytometry. No deviations were observed in ST6Gal-deficient mice among the percentages (denoted in parentheses) of pro-B cells (B220lo CD43(S7)+, gate a; ref. 32), pre-B cells (B220lo HSAhi, gate b; ref. 33), immature B cells (B220lo IgMint, gates b and c; ref. 33), transitional B cells (B220lo-hi IgMhi, gate d; ref. 33), and mature B cells (B220hi IgMint, gate e, and B220 hi HSAlo, gate f; ref. 33). Percentages shown are the mean of four different analyses with calculated Student’s t test, P > 4 for all genotypic comparisons indicating no significant variations. (A Right) ST6Gal-deficient splenic B cells exhibited reductions in cell surface CD22 and IgM, but not in HSA. Reductions in cell surface IgM levels were found to be at 65 ± 20% of controls (Student’s t test; P < 0.001) and CD22 levels at 38 ± 9% of controls (P < 0.001) as determined by comparisons of peak fluorescence (n = 8). (B) Sia6LacNAc-deficient splenic B cells expressed normal levels of activation markers (CD44, B7.2, and I-Ab) and CD40. Dotted lines represents fluorescence of cells stained using an isotype control antibody (n = 8). (C) Serum Ig levels were measured in 4–6-month-old unimmunized mice of indicated genotypes. The median Ig levels are depicted as horizontal bars. Results revealed a 63% reduction (P < 0.001) in circulating IgM and no statistically significant reduction in IgA or IgG. Genotypes of 4–6-month-old mice are provided on the x-axis. Points represent measurements from individual animals.
Although B lymphocyte abundance and development appeared unaltered in Sia6LacNAc-deficient mice, their B cells invariably exhibited reductions in cell surface IgM and CD22. Although HSA levels were unaffected, ST6Gal deficiency resulted in peak IgM levels at 65% of controls and CD22 levels 38% of normal (Fig, 2A, Right). However, no evidence for B lymphocyte activation was found in analyses of cell surface activation markers CD44, B7.2, and major histocompatibility complex class II (I-Ab) (Fig. 2B). Additionally, CD40 expression was unaltered with ST6Gal deficiency (Fig. 2B). Additional analyses of splenic B cells from Sia6LacNAc-deficient mice revealed normal expression of cell surface CD19, CD21, CD23, and CD45RA+ (B220) (data not shown).
Reductions observed in cell surface IgM and CD22 prompted analyses of serum Ig levels to initially assess B cell function in unimmunized mice. Sia6LacNAc-deficient mice exhibited significant decreases in IgM levels to a median value of 37% of normal while statistically unaltered levels of IgA and IgG were observed (Fig. 2C). Although Sia6LacNAc deficiency does not impede lymphocyte development or trafficking, dysfunctional B lymphocytes might reside in mice homozygous for the ST6GalΔ allele.
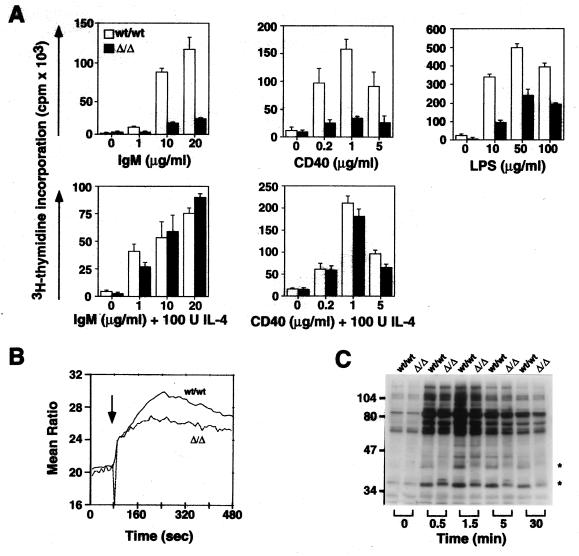
B lymphocyte immune activation proceeds by increased tyrosine phosphorylation and Ca2+ mobilization leading to gene activation and proliferation as necessary for effective humoral immunity (34–37). B lymphocytes from control and ST6Gal-deficient mice were isolated from the spleen or lymph nodes and activated by crosslinking cell surface IgM, CD40, or by addition of LPS. Proliferation of ST6Gal-deficient B lymphocytes in response to these stimuli was found to be significantly reduced (Fig. 3A). Interestingly, normal responsiveness was observed when interleukin-4 (IL-4) was present during the stimulation at various and suboptimal concentrations of stimulatory anti-IgM or anti-CD40 antibodies. IL-4 promotes Ig class switching from IgM to IgG and IgE (38, 39) and can synergize with suboptimal antigen-receptor activation likely through coactivation of “downstream” cytosolic and nuclear signal transduction events. Results observed with IL-4 reveal the intrinsic ability of ST6Gal-deficient B lymphocytes to respond normally in some conditions; however, activation of B lymphocytes through the antigen-receptor complex and following CD40 ligation is adversely affected.
Figure 3.
Attenuated B cell activation responses in mice deficient in ST6Gal function. (A) B lymphocytes were isolated and stimulated by LPS or by crosslinking IgM or CD40 in the absence and presence of IL-4. Reduced proliferation of Sia6LacNAc-deficient B cells was observed in all experiments in the absence of IL-4. The addition of IL-4 as indicated rescued the defective proliferation response to IgM and CD40 ligation. Proliferation is shown as triplicate measurements of [3H]thymidine incorporation indicating the mean and SD. Results shown are representative of three different experiments (P < 0.005 for IgM crosslinking, P < 0.025 for CD40 crosslinking, and P < 0.001 for LPS stimulation). (B) B lymphocytes were analyzed for Ca2+ mobilization in response to IgM crosslinking (arrow). Sia6LacNAc-deficient B cells exhibited a reduction in amount of cytosolic Ca2+ mobilization (63 ± 4% of control value, P < 0.01, n = 4) (C) Phosphotyrosine accumulation following IgM crosslinking is altered in Sia6LacNAc-deficient B lymphocytes. Splenic B lymphocytes were isolated and stimulated with anti-IgM antibody. At indicated times, cells were lysed and extracts were subsequently analyzed by SDS/PAGE and immunoblotting with anti-phosphotyrosine antibody (n = 6). Positions of proteins exhibiting continuously altered levels of phosphotyrosine are denoted by asterisks.
The efficacy of early signal transduction events was assessed by measuring Ca2+ mobilization and phosphotyrosine accumulation immediately following anti-IgM stimulation. Cytosolic mobilization of Ca2+ from intracellular compartments occurs rapidly upon IgM crosslinking by hydrolysis of inositol phospholipids (36, 37). B lymphocytes from ST6Gal null mice failed to mobilize Ca2+ as efficiently as controls with a decrease in both the rate of Ca2+ mobilization and the amount of Ca2+ mobilized (Fig. 3B). ST6Gal-deficient B lymphocytes were next analyzed for their ability to accumulate phosphotyrosine on cellular proteins in response to anti-IgM stimulation. CD22 is among those proteins phosphorylated on tyrosine following B cell antigen-receptor activation, and this phosphorylation is reported to recruit the SHP tyrosine phosphatase and other effector molecules to the antigen-receptor complex by SH2 binding interactions (40, 41). Phosphotyrosine accumulation on CD22 was observed following anti-IgM stimulation of ST6Gal-deficient B cells (data not shown); however, reduced levels of cell surface CD22 (Fig. 2) necessitate additional quantitative studies on CD22 localization and relative phosphorylation potential. Nevertheless, among total cellular proteins analyzed following anti-IgM stimulation, a reduction in phosphotyrosine accumulation was noted on a protein migrating at approximately 42 kDa, with varied alterations on proteins of approximately 37 kDa (Fig. 3C).
The potential physiologic relevance of these alterations was addressed by analyzing the ability of ST6Gal-deficient mice to mount an immune response to T-independent and T-dependent antigens as judged by antibody production. Control and ST6Gal-deficient mice were immunized with either DNP–Ficoll (T-independent antigen) or DNP conjugated to (KLH) (T-dependent antigen). Anti-DNP antibody titers were measured at various times after immunization. Mice deficient in Sia6LacNAc consistently failed to generate high titers of anti-DNP antibody following immunization with the T-independent antigen DNP–Ficoll (Fig. 4A). Control mice produced high anti-DNP IgM titers within 7–10 days of immunization, whereas mice lacking a functional ST6Gal allele were deficient in anti-DNP IgM and IgG1 antibodies, and a reduced response was evident involving anti-DNP IgG3 antibody production. In response to immunization with the T-dependent antigen DNP–KLH, ST6Gal-deficient mice were again severely impaired in their ability to generate an anti-DNP IgM antibody (Fig. 4B). Following a second “boost” immunization with DNP–KLH, Sia6LacNAc-deficient B cells still yielded reduced anti-DNP IgG1 antibody levels and did not generate significant anti-DNP IgG3 antibody. From these studies it is clear that mice deficient in the Sia6LacNAc trisaccharide are impaired in their ability to mount immune responses.
DISCUSSION
The effector role of the ST6Gal sialyltransferase in B lymphocyte activation discloses a mechanism of biologic control operating at the immune cell surface through oligosaccharide variation. Production of the Sia6LacNAc trisaccharide does not appear to be necessary for B lymphocyte development but is required for efficacious immune responses. Although B cells from ST6Gal-deficient mice display a significant reduction in CD22 cell surface levels, this is unlikely to account for the immune deficiencies observed because reduced CD22 expression in mice bearing a heterozygous CD22 null allele did not affect B cell immune function (26–29). Moreover, ST6Gal-deficient mice present a severe and widespread immunodeficiency much unlike that reported for CD22-deficient mice. Our results indicate that the function of ST6Gal encompasses distinct and unique activities in regulating immune responsiveness that may only partially overlap with the function of CD22.
B cell activation and acquisition of an anergic state reduces cell surface IgM levels, and activation is also reported to down-regulate CD22 (42, 43). Sia6LacNAc-deficient B cells exhibit characteristics that may reflect prior aberrant antigen-receptor signal transduction leading to an anergic phenotype. Reduced IgM levels on ST6Gal-deficient B lymphocytes is similar to the anergic phenotype that develops following naive B cell stimulation by endogenous transgenic antigen production (44). Nevertheless, Sia6LacNAc-deficient B cells do not exhibit an activated phenotype. Hypotheses as to how Sia6LacNAc may normally function in mature B lymphocytes requires knowledge of Sia6LacNAc expression.
A subset of secreted or cell surface lymphoid glycoproteins carries the Sia6LacNAc ligand of CD22. Some of these have been identified and include IgM, CD45, and CD22 itself (23–25, 45–47). Although selective high-affinity binding of CD22 to soluble IgM has been reported (25), the stoichiometry of cell surface CD22–IgM interaction is reported to be low both before and after B cell stimulation (19, 48, 49). Because conditions employed in the latter type of experiment can dissociate lectin binding, homotypic and heterotypic cell interactions involving antigen receptors of B and T lymphocytes may normally occur by CD22–CD22 or CD22–CD45 Sia6LacNAc-dependent binding. Within B cells, loss of Sia6LacNAc on IgM and CD22 may thereby lead to aberrant antigen-receptor complex assembly at the cell surface. Perhaps Sia6LacNAc plays a role in the structural stabilization of one or more B lymphocyte membrane molecules that participate in antigen-receptor signal transduction. The observed reductions in serum IgM and cell surface CD22 levels could reflect a destabilizing effect of Sia6LacNAc deficiency, including increased clearance of serum glycoproteins.
Altered protein phosphotyrosine accumulation following ST6Gal-deficient B cell activation nevertheless implies the dysfunction of signal transduction events emanating from the B cell antigen-receptor complex. The identification of altered phosphoproteins would further define the mechanisms by which ST6Gal acts in B cell activation. Such uncertainties are common and presently exist regarding the mechanism of CD22 function (50). It is of importance to understand how molecules that collaborate in signal transduction are able to associate with the B cell antigen-receptor complex in a normal membrane milieu without covalent binding interactions.
The position of oligosaccharides such as Sia6LacNAc at the extracellular side of the plasma membrane indicates that such molecules have evolved to function in cell–cell and receptor subunit interactions (51, 52). Additionally, the functions of glycosyltransferases and glycosidases have been found in some cases to be focused to a particular cell lineage (53). In these studies, we have observed the preferential accumulation of Sia6LacNAc on lymphoid cell surfaces among hematopoietic cell types. We have further observed what appears to be a cell-type specific role for ST6Gal and Sia6LacNAc in regulating B lymphocyte immune function. No evidence for T cell dysfunction using in vitro anti-CD3 activation methods has been found thus far (data not shown). Further research in understanding the B lymphoid role of Sia6LacNAc should illuminate the reason for the more severe and widespread immune deficiency in ST6Gal-deficient mice than is noted in studies of CD22 null mice. It is possible that the presence of CD22 in the absence of its ligand may be especially deleterious to B lymphocyte function by inappropriate localization and sequestration of intracellular signaling molecules. Alternatively, a redundancy in CD22 function may be invoked with the presence of yet unidentified Sia6LacNAc-binding lectins operating at the B lymphocyte cell surface. This latter hypothesis reflects the biologic paradigm exemplified by the sialyl-Lewis X oligosaccharide and the E-, L-, and P-selectins in the inflammatory response involving leukocyte intravasation (54, 55). While these possibilities can be tested, the immunodeficient phenotype of mice lacking Sia6LacNAc provides strong rationale for investigating ST6Gal-dependent signal transduction in B cell activation and Sia6LacNAc expression in syndromes of immune dysfunction.
Acknowledgments
We thank Ajit Varki for helpful comments on the manuscript and the generous gift of CD22–Ig chimera and Mary-Ann Campbell for the anti-CD22 antibody. This research was supported by the Howard Hughes Medical Institute (J.D.M.). T.H. was supported by scholarships from the Swiss National Science Foundation and the Swiss Foundation for Medical and Biological Fellowships.
ABBREVIATIONS
- Sia6LacNAc
Siaα2-6Galβ1-4GlcNAc
- FACS
fluorescence-activated cell sorting
- ES
embryonic stem
- wt
wild type
- KLH
keyhole limpet hemocyanin
- DNP
dinitrophenyl
- LPS
lipopolysaccharide
- IL-4
interleukin 4
References
- 1.Harduin-Lepers A, Recchi M-A, Delannoy P. Glycobiology. 1995;5:741–758. doi: 10.1093/glycob/5.8.741. [DOI] [PubMed] [Google Scholar]
- 2.Kono M, Ohyama Y, Lee Y-C, Hamamoto T, Kojima N, Tsuji S. Glycobiology. 1997;7:469–479. doi: 10.1093/glycob/7.4.469. [DOI] [PubMed] [Google Scholar]
- 3.O’Hanlon T P, Lau K M, Wang X-C, Lau J Y. J Biol Chem. 1989;264:17389–17394. [PubMed] [Google Scholar]
- 4.Wen D X, Svensson E C, Paulson J C. J Biol Chem. 1992;267:2512–2518. [PubMed] [Google Scholar]
- 5.Kitagawa H, Paulson J C. J Biol Chem. 1994;269:17872–17878. [PubMed] [Google Scholar]
- 6.Sjoberg E R, Kitagawa H, Glushka J, van Halbeek H, Paulson J C. J Biol Chem. 1996;271:7450–7459. doi: 10.1074/jbc.271.13.7450. [DOI] [PubMed] [Google Scholar]
- 7.Finne J. J Biol Chem. 1982;257:11966–11970. [PubMed] [Google Scholar]
- 8.Hoffman S, Sorkin B C, White P C, Brackenbury R, Mailhammer R, Edelman G M. J Biol Chem. 1982;257:7720–7729. [PubMed] [Google Scholar]
- 9.Zuber C, Lackie P M, Catterall W A, Roth J. J Biol Chem. 1992;267:9965–9971. [PubMed] [Google Scholar]
- 10.Qu B, Ziak M, Zuber C, Roth J. Proc Natl Acad Sci USA. 1996;93:8995–8998. doi: 10.1073/pnas.93.17.8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinstein J, Lee E U, McEntee K, Lai P-H, Paulson J C. J Biol Chem. 1987;262:17735–17743. [PubMed] [Google Scholar]
- 12.Wang X C, Smith T J, Lau J T Y. J Biol Chem. 1990;265:17849–17853. [PubMed] [Google Scholar]
- 13.Hanasaki K, Varki A, Stamenkovic I, Bevilacqua M P. J Biol Chem. 1994;269:10637–10643. [PubMed] [Google Scholar]
- 14.Lo N-W, Lau J T Y. Glycobiology. 1996;6:271–279. doi: 10.1093/glycob/6.3.271. [DOI] [PubMed] [Google Scholar]
- 15.Campana D, Janossy G, Bofill M, Trejdosiewicz L K, Ma D, Hoffbrand A V, Mason D Y, Lebacq A M, Forster H K. J Immunol. 1985;134:1524–1530. [PubMed] [Google Scholar]
- 16.Stamenkovic I, Seed B. Nature (London) 1990;345:74–77. doi: 10.1038/345074a0. [DOI] [PubMed] [Google Scholar]
- 17.Dorken B, Moller A, Schwartz-Albiez R, Moldenhauer G. In: Leucocyte Typing IV: White Cell Differentiation Antigens. Knapp W, Dorken B, Gilks W R, Rieber E P, Schmidt R E, Stein H, von dem Borne A, editors. Oxford: Oxford Univ. Press; 1989. pp. 63–64. [Google Scholar]
- 18.Schulte R J, Campbell M-A, Fischer W H, Sefton B M. Science. 1992;258:1001–1004. doi: 10.1126/science.1279802. [DOI] [PubMed] [Google Scholar]
- 19.Leprince C, Draves K E, Geahlen R L, Ledbetter J A, Clark E A. Proc Natl Acad Sci USA. 1993;90:3236–3240. doi: 10.1073/pnas.90.8.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell L D, Sgroi D, Sjoberg E R, Stamenkovic I, Varki A. J Biol Chem. 1993;268:7019–7027. [PubMed] [Google Scholar]
- 21.Sgroi D, Varki A, Braesch-Andersen S, Stamenkovic I. J Biol Chem. 1993;268:7011–7018. [PubMed] [Google Scholar]
- 22.Powell L D, Varki A. J Biol Chem. 1994;269:10628–10636. [PubMed] [Google Scholar]
- 23.Stamenkovic I, Sgroi D, Aruffo A, Sy M S, Anderson T. Cell. 1991;66:1133–1144. doi: 10.1016/0092-8674(91)90036-x. [DOI] [PubMed] [Google Scholar]
- 24.Braesch-Andersen S, Stamenkovic I. J Biol Chem. 1994;269:11783–11786. [PubMed] [Google Scholar]
- 25.Hanasaki K, Varki A, Powell L D. J Biol Chem. 1995;270:7533–7542. doi: 10.1074/jbc.270.13.7533. [DOI] [PubMed] [Google Scholar]
- 26.O’Keefe T L, Williams G T, Davies S L, Neuberger M S. Science. 1996;274:798–801. doi: 10.1126/science.274.5288.798. [DOI] [PubMed] [Google Scholar]
- 27.Otipoby K L, Andersson K B, Draves K E, Klaus S J, Farr A G, Kerner J D, Perlmutter R M, Law C-L, Clark E A. Nature (London) 1996;384:634–637. doi: 10.1038/384634a0. [DOI] [PubMed] [Google Scholar]
- 28.Sato S, Miller A S, Inaoki M, Bock C B, Jansen P J, Tang M L K, Tedder T F. Immunity. 1996;5:551–562. doi: 10.1016/s1074-7613(00)80270-8. [DOI] [PubMed] [Google Scholar]
- 29.Nitschke L, Carsetti R, Ocker B, Kohler G, Lamers M C. Curr Biol. 1997;7:133–143. doi: 10.1016/s0960-9822(06)00057-1. [DOI] [PubMed] [Google Scholar]
- 30.Priatel J J, Sarkar M, Schachter H, Marth J D. Glycobiology. 1997;7:45–56. doi: 10.1093/glycob/7.1.45. [DOI] [PubMed] [Google Scholar]
- 31.Nagy A, Rossant J, Nagy R, Abramov-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardy R R, Carmack C E, Shinton S A, Kemp J D, Hayakawa K. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carsetti R, Kohler G, Lamers M C. J Exp Med. 1995;181:2129–2140. doi: 10.1084/jem.181.6.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell M-A, Sefton B M. EMBO J. 1990;9:2125–2131. doi: 10.1002/j.1460-2075.1990.tb07381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gold M R, Law D A, DeFranco A L. Nature (London) 1990;345:810–813. doi: 10.1038/345810a0. [DOI] [PubMed] [Google Scholar]
- 36.Pleiman C M, D’Ambrosio D, Cambier J C. Immunol Today. 1994;15:393–399. doi: 10.1016/0167-5699(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 37.Cooke M P, Heath A W, Shokat K M, Zeng Y, Finkelman F D, Linsley P S, Howard M, Goodnow C C. J Exp Med. 1994;179:425–438. doi: 10.1084/jem.179.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul W E. Blood. 1991;77:1859–1870. [PubMed] [Google Scholar]
- 39.Coffman R L, Lebman D A, Rothman P. Adv Immunol. 1993;54:229–270. doi: 10.1016/s0065-2776(08)60536-2. [DOI] [PubMed] [Google Scholar]
- 40.Campbell M-A, Klinman N R. Eur J Biochem. 1995;25:1573–1579. doi: 10.1002/eji.1830250616. [DOI] [PubMed] [Google Scholar]
- 41.Law C-L, Sidorenko S P, Chandran K A, Zhao Z, Shen S-H, Fischer E H, Clark E A. J Exp Med. 1996;183:547–560. doi: 10.1084/jem.183.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pezzutto A, Rabinovitch P S, Dorken B, Moldenhauer G, Clark E A. J Immunol. 1988;140:1791–1795. [PubMed] [Google Scholar]
- 43.Dorken B, Moldenhauer G, Pezzutto A, Schwartz R, Kiesel S, Hunstein W. J Immunol. 1988;136:4470–4479. [PubMed] [Google Scholar]
- 44.Goodnow C C. Proc Natl Acad Sci USA. 1996;93:2264–2271. doi: 10.1073/pnas.93.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hickman S, Kornfeld R, Osterland C K, Kornfeld S. J Biol Chem. 1972;247:2156–2163. [PubMed] [Google Scholar]
- 46.Anderson D R, Atkinson P H, Grimes W J. Arch Biochem Biophys. 1985;243:605–618. doi: 10.1016/0003-9861(85)90538-7. [DOI] [PubMed] [Google Scholar]
- 47.Sato T, Furukawa K, Autero M, Gahmberg C G, Kobata A. Biochemistry. 1993;32:12694–12704. doi: 10.1021/bi00210a019. [DOI] [PubMed] [Google Scholar]
- 48.Hanasaki K, Powell L D, Varki A. J Biol Chem. 1995;270:7543–7550. doi: 10.1074/jbc.270.13.7543. [DOI] [PubMed] [Google Scholar]
- 49.Peaker C J G, Neuberger M S. Eur J Biochem. 1993;23:1358–1363. doi: 10.1002/eji.1830230626. [DOI] [PubMed] [Google Scholar]
- 50.Cyster J G, Goodnow C C. Immunity. 1997;6:509–517. doi: 10.1016/s1074-7613(00)80339-8. [DOI] [PubMed] [Google Scholar]
- 51.Marth J D. Glycoconjugate J. 1994;11:3–8. doi: 10.1007/BF00732424. [DOI] [PubMed] [Google Scholar]
- 52.Varki A, Marth J D. Semin Dev Biol. 1995;6:127–138. [Google Scholar]
- 53.Chui D, Oh-Eda M, Liao Y-F, Panneerselvam K, Lal A, Marek K, Freeze H H, Moremen K W, Fukuda M N, Marth J D. Cell. 1997;90:157–167. doi: 10.1016/s0092-8674(00)80322-0. [DOI] [PubMed] [Google Scholar]
- 54.Maly P, Thall A, Petryniak B, Rogers C E, Smith P L, Marks R M, Kelly R J, Gersten K M, Cheng G, Saunders T L, et al. Cell. 1996;86:643–653. doi: 10.1016/s0092-8674(00)80137-3. [DOI] [PubMed] [Google Scholar]
- 55.Frenette P S, Mayadas T N, Rayburn H, Hynes R O, Wagner D D. Cell. 1996;84:563–574. doi: 10.1016/s0092-8674(00)81032-6. [DOI] [PubMed] [Google Scholar]