Abstract
Considering the well established role of nonclassical HLA-G class I molecules in inhibiting natural killer (NK) cell function, the consequence of abnormal HLA-G expression in malignant cells should be the escape of tumors from immunosurveillance. To examine this hypothesis, we analyzed HLA-G expression and NK sensitivity in human malignant melanoma cells. Our analysis of three melanoma cell lines and ex vivo biopsy demonstrated that (i) IGR and M74 human melanoma cell lines exhibit a high level of HLA-G transcription with differential HLA-G isoform transcription and protein expression patterns, (ii) a higher level of HLA-G transcription ex vivo is detected in a skin melanoma metastasis biopsy compared with a healthy skin fragment from the same individual, and (iii) HLA-G protein isoforms other than membrane-bound HLA-G1 protect IGR from NK lysis. It thus appears of critical importance to consider the specific role of HLA-G expression in tumors in the design of future cancer immunotherapies.
The concept of immunosurveillance implies that transformed cells trigger an immune response intended to reject developing cancer lesions (1). It is well known that tumors frequently are infiltrated by T lymphocytes and natural killer (NK) cells (2). The effector functions of both of these cell types depend on the level of major histocompatibility complex (MHC) class I molecules expressed at the surfaces of tumor cells. Although cytotoxic T lymphocytes (CTL) recognize tumor-associated antigenic peptides presented by MHC class I molecules, NK cells are cytotoxic for tumor cells that have lost MHC class I expression. Expression and modulation of MHC class I molecules are thus important issues in tumor progression.
Treatment strategies for human tumors essentially have focused on triggering the immune response, and it is of critical importance to characterize the mechanisms that prevent such anti-tumor defense. Many tumors have developed selective mechanisms for down-regulating classical MHC class I expression (3, 4), allowing them to escape CTL immunosurveillance. Such class I-negative tumors consequently should become targets for NK lysis, leading to tumor regression (5). However, most tumors maintain their ability to grow in vivo. A recent study has demonstrated that classical HLA-Cw7 class I molecules expressed on a melanoma cell are able to mediate a tumor escape mechanism from immunosurveillance by interacting with killing inhibitory receptors (KIRs) present on CTL (6). Various KIRs were first identified on NK cells because of their ability to interact with specific classical HLA class I alleles (7–9). Several reports have shown that the nonclassical HLA-G class I molecule inhibits NK lytic activity upon interaction with KIRs that belong to both the C-type lectin and Ig superfamilies (10–14), and the existence of a different, still uncharacterized KIR able to interact specifically with HLA-G also was postulated (13, 15). Thus, expression of HLA-G by target cells would constitute a powerful mechanism by which tumors escape from NK immunosurveillance.
In contrast to classical HLA class I genes, the primary transcript of the HLA-G gene generates at least five mRNAs resulting from alternative splicing that potentially encode five protein isoforms: HLA-G1, HLA-G2, HLA-G3, and HLA-G4, which have the capacity to anchor to the cell membrane, and the soluble HLA-G5 isoform (16, 17). HLA-G is expressed physiologically on cytotrophoblasts at the feto-maternal interface, where it plays an important role in maternal tolerance of the semiallogenic fetus (18–20). We recently showed that, in addition to HLA-G1, the HLA-G2 isoform strongly inhibits NK cell lysis in vitro (15) and that HLA-G molecules protect cytotrophoblasts from the lytic activity of maternal uterine NK cells ex vivo (21, 22).
The fact that HLA-G is expressed selectively on trophoblast cells is of particular interest. Indeed, although the trophoblast is a normal tissue, it shares several common features with neoplastic cells, such as highly mitotic and invasive properties and the expression of growth factors, growth factor receptors, hormones, and proto-oncogenes, which have led to defining the trophoblast as a “pseudo-malignant” type (23, 24). Physiological expression of MAGE-3 and MAGE-4 genes, expressed in metastatic human melanoma, is restricted to placenta and testis (25). Melanoma cell adhesion molecules (Mel-CAM) are expressed in both melanoma and trophoblast cells (26). Taken together, these features suggest that genes preferentially expressed in trophoblast cells, such as HLA-G, also might be expressed preferentially in neoplastic cells.
The ectopic expression of HLA-G on tumor cells might be a mechanism by which malignant cells could escape from immunosurveillance. To address this question, we studied HLA-G isoform expression and function in human melanoma cell lines. To attest the biological relevance of HLA-G expression in tumors, we also carried out ex vivo experiments on melanoma metastasis biopsies. The results show that HLA-G is expressed in solid tumors and can protect them from NK cell lysis.
METHODS
Cell Lines.
The IGR (HLA-A2, -A3, B58/male), M74 (HLA-A1, -A2, -B8, -B14/female), M8 (HLA-A1, -A2, -B12, and B40/male) melanoma cell lines, kindly provided by F. Jotereau (INSERM U211, Nantes, France), the K562 human erythroleukemia cell line (American Type Culture Collection), and the nonadult T cell leukemia, NK-mediating YT2C2-PR subclone were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 1 μg/ml gentamicin, and fungizone (Sigma) and cultured in a 37°C, 5% CO2-humidified incubator. K562 transfectants were obtained as described (15). The human HLA-G-positive JEG-3 choriocarcinoma cell line (American Type Culture Collection) was cultured in DMEM (Sigma) supplemented with 10% heat-inactivated fetal calf serum, antibiotics, and 2 mM l-glutamine. SWEIG is an EBV-transformed B cell line (HLA-A29, -B61, and-Cw2). First trimester trophoblast tissues were obtained from elective termination of pregnancy, minced, and immediately used for RNA extraction. Human peripheral blood mononuclear cells (PBMC) were obtained from healthy male volunteer donors and isolated by Ficoll–Hypaque 1077 density gradient. These experiments were approved by the local ethics committee.
Tumor Samples.
Tissue samples from a patient, distinct from the ones from which the above cell lines were established, were obtained from the Institut Gustave Roussy (Villejuif, France) after obtaining the prior informed consent of the patient. Biopsy of a scalp melanoma metastasis was performed under aseptic conditions immediately after surgical excision. A biopsy was carried out on healthy skin obtained near the tumor site under the same conditions. Both samples were immediately frozen in liquid nitrogen. A diagnosis of melanoma metastasis was confirmed by cytological examination on formalin-fixed, hematoxylin/eosin-stained sections.
mAbs.
The following mAbs were used: W6/32, IgG2a anti-HLA class I heavy chains (HC) associated with β2-microglobulin (Sigma); HCA2, IgG anti-HLA-A and -G kindly provided by H. Ploegh (Harvard University, Cambridge, MA); and 4H84, IgG anti-HLA-G, kindly provided by S. Fisher and M. McMaster (University of California, San Francisco).
Reverse Transcriptase PCR (RT-PCR) Analysis of HLA Transcripts.
Total mRNA was extracted by using the RNA NOW reagent (Biogentex, Seabrook, TX) according to the manufacturer’s recommendations. The quality of RNA was checked by electrophoresis in a 1.5% agarose denaturing gel. Complementary DNAs were prepared from 10 μg of total RNA, by using oligo-(dT)12–18 primer and Moloney murine leukemia virus RT (GIBCO/BRL). HLA-G-specific RT-PCR amplifications were carried out as described by using G.257 (exon 2) and G3.U (3′-UT) (16, 17, 27) primers to detect all alternatively spliced HLA-G mRNA forms. Specific amplification of each HLA-G mRNA form was attempted with the following primer sets: G.526 (exon 3) and G3.U (3′-UT) for G1 (full length) and G4 (minus exon 4), G.526 (exon3), and G.i4b (intron 4) for G5 (full length with intron 4), G.-3 (overlapping exon 2 and 4) and G3U (3′-UT) for G2 (minus exon 3), and G6 (minus exon 3 with intron 4), G.-3–4 (overlapping exon 2 and 5), and G3.U (3′-UT) for G3. Classical HLA-class I cDNAs were amplified as described (28), by using a unique 5′ primer, HLA-5P2, and three distinct 3′ primers HLA-3pA, HLA-3pB, and HLA-3pC, which specifically amplify HLA-A, HLA-B, and HLA-C mRNA, respectively. The DRA-specific primers, 5′-GGCCATAAGTGGAGTCCC and 5′-CTATACTCCGATCACCAA for 3′ were used as described in Bull et al.(29). Coamplification of β-actin cDNA was carried out in each experiment with β-actin amplimer sets (CLONTECH) for 16 cycles to evaluate comparative amounts of RNA in samples. PCR products were analyzed by electrophoresis in 1% agarose gel and stained with ethidium bromide. The specificity of PCR products was confirmed by Southern blotting of the fragments onto nylon membranes (Hybond N+, Amersham). Hybridization was performed with several HLA-G-specific oligonucleotide probes: exon2-specific G.R (30), G.647 F (5′-CCACCACCCTGTCTTTGACT: exon 4-specific), G.I4 F (GAGGCATCATGTCTGTTAGG: intron 4-specific), and G.927 F (5′-ATCATGGGTATCGTTGCTGG: exon 5-specific (27) an HLA-A-specific probe (5′-GGAGGACCAGACCCAGGACACG), an HLA-B-specific probe (5′-AGCTCCGATGACCACAACTGC), an HLA-C-specific probe (5′-TGTCCTAGCTGCCTAGGAG), and an HLA-DRA specific probe (TGTGATCATCCAGGCCGAG). The same membranes then were probed with an actin-specific probe (5′-ATCATGTTTGAGACCTTCAACACCCCAGCC). The filters were exposed to Kodak (Biomax) films with amplifying screens for 4–16 h at −80°C.
Metabolic Labeling, Immunoprecipitation, and Gel Electrophoresis.
Cells were labeled metabolically with 0.5 mCi [35S] methionine plus [35S] cysteine (Amersham)/2 × 107 cells for 30 min or 1 h in methionine- and cysteine-free medium containing 5% fetal calf serum. Cells were washed twice with cold PBS and lysed in 1% Triton X-100/PBS for 30 min at 4°C. After preclearing with protein A–Sepharose CL-4B (Pharmacia Biotech) and normal serum, the lysates were incubated with specific mAbs (W6/32, 4H84, and HCA2). Immunoprecipitated products were washed five times in 0.1% Triton X-100, 20 nM Tris (pH 7.5), 150 mM NaCl, and 0.05% NaN3 and once in 0.1% Triton X-100/0.1% SDS. The bound proteins were eluted from the beads by boiling for 5 min in 2% SDS sample buffer. The samples then were analyzed on SDS 12 or 10%/PAGE. The gels were dried and exposed to Kodak XAR-5 film at −70°C.
Immunoprecipitation of Cell Surface Biotinylated Proteins and Western Blotting.
Cell surface proteins were labeled with biotin. After washes in PBS, 1.5 × 107 cells were incubated in 1 ml of cold PBS containing 5 mg of N-hydroxysuccinimide/standard saline/biotin (Pierce) for 15 min at 4°C. Residual active groups were quenched in 50 mM NH4Cl for 10 min at 4°C. Cells were lysed in 1% Triton X-100/PBS. W6/32 immunoprecipitates were separated on SDS 12%/PAGE, transferred onto nitrocellulose membranes (Hybond enhanced chemiluminescence, Amersham), and probed with peroxidase-conjuqated streptavidin.
Cytotoxicity Assays.
The cytolytic activity of the YT2C2-PR subclone effector cells against melanoma cell lines and K562 transfectants was assessed in 4-h 51Cr-release assays in which effector cells were mixed with 5 × 103 51Cr-labeled targets [100 μCi of 51Cr sodium chromate (Amersham)] in U-bottomed microtiter plates. For inhibition assays, excess inhibitor cells were incubated previously with effector cells and 51Cr-labeled targets subsequently were added. After 4 h at 37°C in a humidified 5% CO2 incubator, 50 μl of the supernatant was collected for liquid scintillation counting (Wallac 1410, Pharmacia). The percentage of specific lysis was calculated as follows: percentage specific lysis = [(cpm experimental release − cpm spontaneous release)/(cpm maximum release − cpm spontaneous release)] × 100. Spontaneous release was determined by incubation of labeled target cells with medium. Maximum release was determined by solubilizing target cells in 0.1 M HCl. Results are presented as a mean of five experiments.
RESULTS
Identification of Alternatively Spliced HLA-G mRNA Transcripts in Human Melanoma Cell Lines and Biopsies.
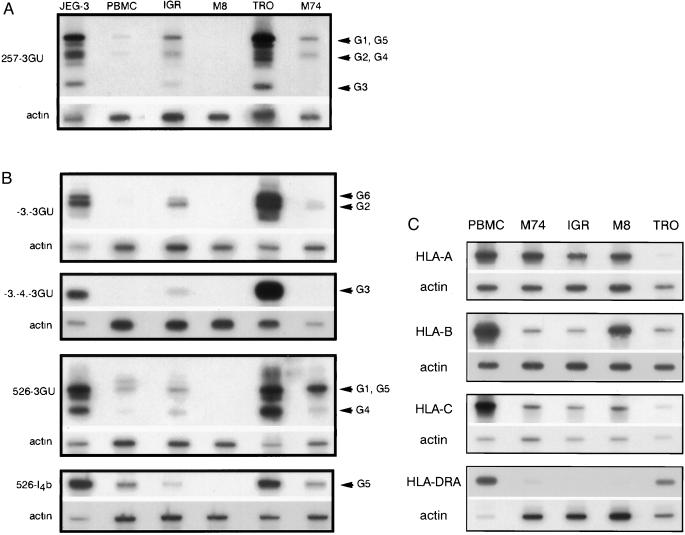
HLA-G cDNA from three human melanoma cell lines (IGR, M8, and M74) were amplified, by using previously described pan-HLA-G PCR primers derived from HLA-G-specific exon 2 and 3′ untranslated sequences (16, 17). The JEG-3 choriocarcinoma cell line and trophoblast cells, both of which exhibit high levels of the various HLA-G transcripts, were used as positive controls, and peripheral blood mononuclear cells (PBMC) from healthy volunteer donors were used as a control for a low level of HLA-G transcripts. Hybridization of PCR products led to the identification of high levels of HLA-G mRNAs in two melanoma cells, IGR and M74, whereas no detectable signal could be detected in the M8 melanoma cell line. As expected, in trophoblast and JEG-3 cells, all HLA-G isoforms (HLA-G1, G2, G3, G4, and G5) were detected (Fig. 1A). In IGR melanoma cells, all HLA-G hybridizing bands also were detected by pan-HLA-G primers (Fig. 1A). However, pan-HLA-G primers do not permit discrimination between HLA-G1 and HLA-G5 signals, both of which are found in a 1,000-bp band and between HLA-G2 and HLA-G4, which comigrate as 600-bp fragments (16). We therefore conducted further reverse transcriptase PCR (RT-PCR) identification of isolated isoforms by using specific primers (27). IGR cells expressed all HLA-G isoforms, except for a relatively lower level of the HLA-G4 and G5 isoforms (Fig. 1B). In the M74 melanoma cell line, pan-HLA-G primers detected bands corresponding to HLA-G1 and HLA-G5 (1,000 bp) at high intensity, a signal for HLA-G2 and G4 (600 bp), and no signal for HLA-G3 (300 bp) (Fig. 1A). Primers for specific isoforms revealed that, in M74 melanoma cells, the G1 and G4 isoforms were more abundant than in PBMC whereas the level of the soluble G5 transcript was comparable to that observed in PBMC. Only weak levels of HLA-G2 and HLA-G6 mRNA, which correspond to the soluble HLA-G2 isoform, were detected in M74, whereas specific amplification of the HLA-G3 transcript confirmed the absence of HLA-G3 observed with pan-HLA-G primers in these cells (Fig. 1 A and B). No detectable HLA-G hybridizing signal was observed in M8 (Fig. 1 A and B).
Figure 1.
(A) RT-PCR analysis of HLA-G mRNA isoforms in melanoma cells. Pan-HLA-G primers 257 (exon 2) and 3GU (3′ untranslated) were used for PCR amplification of HLA-G transcripts corresponding to all known HLA-G isoforms. cDNA from choriocarcinoma JEG-3, first trimester trophoblast (TRO), and PBMC were used as controls for high and basal HLA-G transcription. IgR, M8, and M74 correspond to cDNA amplification of melanoma cell lines. HLA-G-specific bands were revealed by hybridization with the GR-specific probe located in exon 2. Bands corresponding to HLA-G1, G2, G3, G4, and G5 transcripts are indicated by arrows. PCR products co-amplified in the same reaction by β-actin primers were detected on the same membrane by a β-actin probe. (B) Specific RT-PCR detection of alternative HLA-G transcripts in melanoma cells. Primer –3. is specific for HLA-G2 and soluble G2 (G6) isoforms that lack exon 3. Primers -3.-4. discriminate HLA-G4 mRNA transcripts. Primers 526 and I4b specifically amplify the HLA-G5 transcript, which corresponds to the soluble isoform. PCR products co-amplified in the same reaction by β-actin primers were detected on the same membrane by a β-actin probe. (C) RT-PCR analysis of classical HLA class I mRNA. cDNA from PBMC, trophoblast cells, and melanoma cell lines were amplified by RT-PCR by using locus-specific HLA-A, HLA-B, HLA-C, or HLA-DRA locus-specific primers. Locus-specific bands were revealed by using HLA-A, -B, -C, and DRA-specific probes. PCR products co-amplified in the same reaction by β-actin primers were detected on the same membrane by a β-actin probe used as a quantitative control for RNA levels.
PCR analysis of classical MHC transcripts using HLA-A, HLA-B, HLA-C (28), and HLA-DR (29) specific primers revealed high levels of classical HLA class I transcripts in all melanoma cell lines, whereas no or extremely low levels of HLA-DRA transcripts were detected in these cells (Fig. 1C). These results were confirmed by flow cytometry analysis, by using mAbs for HLA class I and II antigens, which showed that HLA-A, -B, and -C molecules were expressed on the surfaces of melanoma cell lines, where no HLA class II molecules could be detected (data not shown).
Analysis of HLA-G Transcription in ex Vivo Melanoma Biopsies.
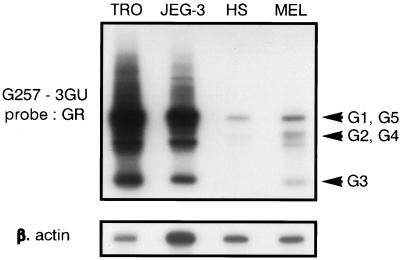
To address the in vivo relevance of HLA-G transcription in melanoma compared with normal skin, we analyzed HLA-G transcription of mRNAs levels in an ex vivo skin melanoma metastasis and compared it with a healthy skin biopsy from the same patient. All of the HLA-G transcripts were detected at high levels in the melanoma biopsy, whereas only the 1,000-bp band was present in the healthy skin (Fig. 2). These results were confirmed on other melanoma biopsies (data not shown) and suggest that the high levels of transcription observed in melanoma cells were not the result of higher HLA-G transcription in the skin but were specific to the tumor site.
Figure 2.
RT-PCR analysis of HLA-G mRNA isoforms in an ex vivo skin melanoma metastasis biopsy. The pan-HLA-G primers 257 and 3GU were used for RT-PCR amplification of HLA-G transcripts from an ex vivo skin metastasis (MEL) and from healthy skin biopsy (HS) obtained from the same patient. JEG-3 and first trimester trophoblast (TRO) were used as controls of high HLA-G transcription. HLA-G-specific bands were revealed by hybridization with the GR-specific probe located in exon 2. Bands corresponding to HLA-G1, G2, G3, G4, and G5 transcripts are indicated by arrows. PCR products co-amplified in the same reaction by β-actin primers were detected on the same membrane by a β-actin probe.
Analysis of HLA-G Protein in Melanoma Cells.
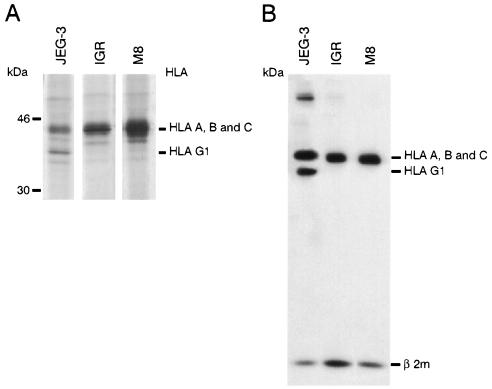
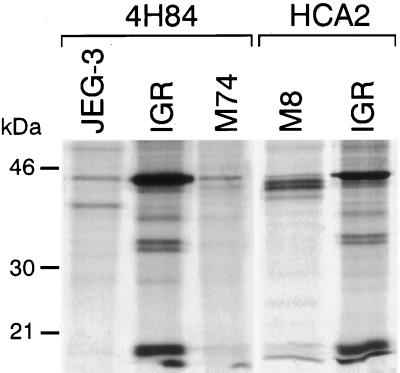
To determine whether the HLA-G mRNA transcripts detected in melanoma are translated as HLA-G proteins, we carried out immunoprecipitation studies with different anti-HLA class I mAbs by using lysates from [35S] methionine/cysteine metabolically labeled cells. Comparison was made with the positive control cell line JEG-3 and with negative M8 control melanoma cells that do not transcribe any HLA-G mRNA. Immunoprecipitation results of W6/32 mAb reacting with HLA-A, -B, -C, and -G1 β2-microglobulin-associated HC are shown in Fig. 3. As expected, in JEG-3, W6/32 immunoprecipitated two proteins of 45 kDa (HLA-C molecule) and 39 kDa, characterized as the membrane-bound HLA-G1 isoform (31, 32) (Fig. 3A). In IGR and M8 melanoma cells, only a 45-kDa HLA-A,-B, and -C HC was detected. Similar results were obtained by immunoprecipitation of biotinylated cell surface proteins (Fig. 3B). These data indicate that the HLA-G1 protein was not expressed in IGR cells, even though these cells express the corresponding mRNA. However, the absence of HLA-G1 protein in IGR does not exclude the expression of the three other HLA-G protein isoforms (HLA-G2, -G3, and -G4). These protein isoforms could not be revealed by the W6/32 mAb because of their incapacity to associate with β2-microglobulin (16). Therefore, we carried immunoprecipitation of [35S] methionine-labeled proteins, by using mAbs that recognize the free HLA-G HC, HCA2 mAb that recognizes the denatured HLA-G and HLA-A HC (33, 34), and the 4H84 mAb, which recognizes an epitope located in the α1 domain shared by all of the HLA-G protein isoforms (S. Fisher and M. McMaster personal communication). The 4H84 mAb revealed the presence of the 39-kDa HLA-G1 protein in JEG-3 cells that was not detected in IGR (Fig. 4). Additional specific bands migrating at 32–34 kDa and at 18 kDa, which most likely corresponded to the size of the HLA-G2 and/or HLA-G4 and HLA-G3 isoforms, were detected in IGR by both the 4H84 and HCA2 anti-HLA-G mAbs (Fig. 4). These additional HLA-G-specific bands could not be observed in M74 and M8 cells, which do not present the corresponding HLA-G isoform transcripts (Fig. 4).
Figure 3.
Detection of HLA-G1 proteins in JEG-3 cells but not in IGR and M8 melanoma cells by W6/32 mAb reacting against β2-microglobulin-associated HLA-A,-B, -C, and -G HC. (A) Immunoprecipitation of metabolically labeled proteins. Cells were metabolically labeled with 0.5 mCi of [35S] methionine plus [35S] cysteine for 1 h, and cell lysates were precipitated with W6/32, analyzed by SDS 12%/PAGE and autoradiographed. (B) Biotinylated cell surface proteins of melanoma and the JEG-3 cell lines were immunoprecipitated with W6/32 mAb, and immunoprecipitates then were separated by SDS 12%/PAGE and transferred to nitrocellulose membranes. Surface class I molecules were detected by using peroxidase-conjugated streptavidin.
Figure 4.
Immunoprecipitation of the alternative HLA-G isoforms in IGR melanoma cells by mAb reacting against the free HLA-G HC, 4H84, and HCA2 mAbs. Cells were labeled for 30 min and immunoprecipitated with specific mAbs, and the eluted immunoprecipitates were analyzed in SDS 10%/PAGE.
Protection of the HLA-G-Positive IGR Melanoma Cell Line from NK Cytolysis.
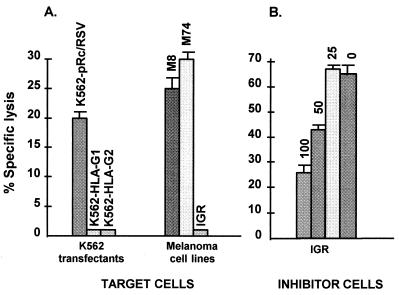
We then investigated whether such HLA-G isoform protein expression in melanoma cell lines could be associated with decreased susceptibility to NK lysis. For this purpose, the NK-like YT2C2-PR subclone was used as a model of the NK effector cell, which does not bear any of the KIR known to interact with classical HLA class I molecules, including CD94, and in which we identified a new killing inhibitory receptor involved in HLA-G-specific recognition (15). The abolition of YT2C2-PR NK lysis previously was found to be mediated by expression of either the HLA-G1 or HLA-G2 membrane-bound isoforms in transfected K562 target cells, whereas the K562 pRc/RSV control cell line remained sensitive to such lysis (15) (Fig. 5A). In this study, we show that only the IGR melanoma target cell line, which can express the HLA-G2 and/or G4 and HLA-G3 protein isoforms, abolished YT2C2-PR NK lysis. The M74 melanoma cell line, which expresses classical HLA class I but demonstrates a selective defect in HLA-G2 and HLA-G3 isoform transcription and protein expression, was lysed by YT2C2-PR. Lysis also was observed with the M8 cell line, which expresses classical MHC class I molecules but does not transcribe any HLA-G mRNA (Fig. 5A). To further exclude the involvement of classical HLA class I molecules present on IGR melanoma cells in the inhibition of YT2C2-PR lysis, we used several HLA-G-negative B-EBV cell lines, each sharing at least one classical HLA-A, -B, or -C allele with IGR, as target cells. All of these B-EBV cell lines were lysed by the YT2C2-PR subclone, indicating that HLA-A, -B, and -C molecules were not involved in the protection of IGR melanoma from YT2C2-PR lysis (data not shown).
Figure 5.
(A) Effect of HLA-G expression by IGR melanoma on sensitivity to lysis by the YT2C2-PR subclone. K562 transfected with either the vector alone (K562-pRc/RSV), HLA-G1 (K562-HLA-G1), or HLA-G2 (K562-HLA-G2) and the M8, M74, and IGR melanoma cell lines were used as targets. The YT2C2-PR subclone was used as an effector cell at a 50:1 effector/target (E/T) ratio. Results are expressed as the percentage of specific lysis recorded in a 4-h 51Cr-release assay. Spontaneous release never exceeded 10% of the maximum release. This experiment was repeated at least five times, yielding the same pattern of protection. (B) Inhibition of YT2C2-PR lysis is caused by an “off signal” transmitted by IGR melanoma cells. The M8 melanoma cell line was used for Cr-labeled target cells. The YT2C2-PR subclone was used for effector cells at a 50:1 E/T ratio. The IGR melanoma cell line was added as inhibitor cells at 100, 50, and 25:1 inhibitor/target ratios. Zero indicated that no IGR cells were added to the test.
To demonstrate that the inhibition of YT2C2-PR lysis by IGR was caused by an “off signal” transmitted by this melanoma cell line, IGR cells were used as inhibitors in a cytotoxicity assay of Cr-labeled M8 target cells and YT2C2-PR effector cells. As shown in Fig. 5B, IGR efficiently inhibited M8 lysis by the YT2C2-PR subclone, and the inhibition of this lysis was proportional to the number of IGR cells used in the competition assay.
DISCUSSION
Several reports agree that HLA class I molecule down-regulation in melanoma cells correlates with the degree of de-differentiation of the tumor and with biological signs of increased malignancy (3). The defective classical MHC class I expression could favor escape from cytotoxic T cell lysis, rendering such cells more susceptible to elimination by NK cells. To our knowledge, this is the first evidence of the ectopic expression of the nonclassical HLA-G gene in solid tumor cells.
The ability of the IGR HLA-G-positive melanoma cell line to inhibit lysis by the HLA-G-specific YT2C2-PR NK clone suggests that HLA-G plays a functional role in protecting tumor cells from NK destruction. Although an HLA-G1 mRNA transcript is detected in IGR, the corresponding 39-kDa HLA-G1 protein was not observed in this melanoma cell line. This result is in accordance with the absence of HLA-G protein recognition at the surface of IGR cells by the 87G anti-HLA-G antibody (data not shown), which specifically detects the HLA-G1 isoform on JEG-3- and HLA-G-transfected cells (16). However, low molecular weight proteins were detected in the cytoplasm of IGR cells by using mAbs directed against denatured HLA-G HC, suggesting that HLA-G2/G4 and G3 protein isoforms are indeed translated in this melanoma cell line, although their surface expression cannot be verified because of the lack of antibodies that detect such membrane-bound HLA-G isoforms. In contrast to IGR, the M74 melanoma cell line was efficiently lysed by the NK clone. Transcription of HLA-G2 and -G3 mRNAs was not detected in M74, suggesting that HLA-G protein expression and function is impaired in this melanoma cell line. These results show that, although the HLA-G1 protein isoform is not expressed in IGR cells, other protein isoforms, namely HLA-G2 and/or -G4 or HLA-G3, can function as inhibitory signals for NK cells. This observation is in accordance with work showing that the membrane-bound HLA-G2 isoform, which lacks the α2 domain of the molecule, can, as HLA-G1, inhibit NK lysis (15).
Our findings strongly support the idea that, in addition to its immunomodulatory role in materno-fetal tolerance, the HLA-G class I molecule can participate in the escape of tumors from immunosurveillance. The regulatory mechanisms that allow ectopic expression of HLA-G in melanoma cells remain to be elucidated. Abnormal activation of genes in tumor cells, including the MAGE-1 tumor antigen, has been correlated to genome-wide demethylation mechanisms that affect various promoters and could lead to deregulation of HLA-G transcription in melanoma cells (35). HLA-G expression usually is restricted to pregnancy, in which hormone impregnation may be a determinant factor. Accordingly, many controversial observations suggest the possible dependence of human melanoma on endocrine influences (36). The presence of estrogen receptors on melanoma cells and the use of the anti-estrogen tamoxifen in the treatment of melanoma have been reported, although only a small percentage of melanomas respond to hormone therapy (37). Furthermore, pregnancy has long been suspected as a poor prognostic factor in the clinical course of melanoma, although recent reviews minimize its impact on patient survival, retaining only increased incidence of nodal metastasis, which appears more rapidly in pregnant women (38). Considered together, these observations suggest that changes in hormone status may enhance the ability of malignant melanoma cells to metastasize. Consequently, HLA-G expression could be induced in tumor cells by hormones, favoring the formation of malignant metastatic clones that can escape immune surveillance.
HLA-G expression in tumor cells amounts to a new appreciation of the role of HLA class I molecules in tumor invasiveness because HLA-G, in contrast to classical class I molecules: (i) exhibits low polymorphism (39) and interacts as a public ligand with KIRs that belong to both C-type lectin-like and Ig superfamilies, allowing functional inhibition of HLA-G-positive tumors from a wider repertoire of NK cells; (ii) can be expressed as several membrane-bound and soluble isoforms that play a functional role in inhibiting NK function, thereby enhancing the ability of tumors to resist NK lysis; and (iii) also may interact with KIRs recently described on the surface of CTL (6, 9), thereby further enlarging the capacity of HLA-G-expressing tumor cells to evade immmunosurveillance by both CTL and NK cells.
It thus appears important to consider the specific role of HLA-G expression in tumors when considering future therapeutic choices of immunomodulation strategies intended to complement conventional cancer treatments. In this regard, melanoma vaccines currently used to generate tumor-specific cytotoxic T lymphocytes would not be effective against tumors that have lost classical class I MHC antigens, a frequent phenomenon in metastatic tumor cells. Adoptive NK immunotherapy could be an important alternative to consider when designing new cancer therapies for class I-deficient cells. Negative modulation of HLA-G expression in tumors also could constitute a new therapeutic approach to limiting tumor progression.
Acknowledgments
We thank Drs. Daniel Geraghty, Hidde Ploegh, and Susan Fisher and Michael McMaster for generously providing us with HLA-G antibodies. We are grateful to Francisco Adrian Cabestre and Elisabeth Nelson for excellent scientific assistance and to Drs. Alvaro Margulis (Service de Chirurgie Générale, Institut Gustave Roussy, Villejuif, France), Sylvaine Mercier, and Pierre Duvillard (Service d’Anatomo-pathologie, Institut Gustave Roussy, Villejuif, France) for providing us with histopathology reports and tumor sample biopsies. We thank Dr. Noah Hardy for rereading and correcting the manuscript. This study was supported by grants from the French Commissariat à l’Energie Atomique and the Association pour la Recherche sur le Cancer.
ABBREVIATIONS
- CTL
cytotoxic T lymphocyte
- MHC
major histocompatibility complex
- NK
natural killer
- PBMC
peripheral blood mononuclear cell
- KIR
killing inhibitory receptor
- RT-PCR
reverse transcriptase PCR
- HC
heavy chain
References
- 1.Burnet F M. Prog Exp Tumor Res. 1970;13:1–27. doi: 10.1159/000386035. [DOI] [PubMed] [Google Scholar]
- 2.Tefany F J, Barnetson R S, Halliday G M, McCarthy S W, McCarthy W H. J Invest Dermatol. 1991;97:197–202. doi: 10.1111/1523-1747.ep12479662. [DOI] [PubMed] [Google Scholar]
- 3.Garrido F, Ruiz-Cabello F, Cabrera T, Peres-Villar J J, Lopez-Botet M, Duggan-Keen M, Stern P L. Immunol Today. 1997;18:89–95. doi: 10.1016/s0167-5699(96)10075-x. [DOI] [PubMed] [Google Scholar]
- 4.Bodmer W F, Browning M J, Rowan A, Bicknell D C, Bodmer J G. Ann N Y Acad Sci. 1993;12:42–49. doi: 10.1111/j.1749-6632.1993.tb43994.x. [DOI] [PubMed] [Google Scholar]
- 5.Porgador A, Mandelboim O, Restifo N, Strominger J L. Proc Natl Acad Sci USA. 1997;94:13140–13145. doi: 10.1073/pnas.94.24.13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikeda H, Lethe B, Lehmann F, van Baren N, Baurain J F, de Smet C, Chambost H, Vitale M, Moretta A, Boon T, et al. Immunity. 1997;6:199–208. doi: 10.1016/s1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 7.Lanier L L. Immunity. 1997;6:371–378. doi: 10.1016/s1074-7613(00)80280-0. [DOI] [PubMed] [Google Scholar]
- 8.Reyburn H, Mandelboim O, Vales-Gomez M, Sheu E G, Pazmany L, Davis D M, Strominger J L. Immunol Rev. 1997;155:119–125. doi: 10.1111/j.1600-065x.1997.tb00944.x. [DOI] [PubMed] [Google Scholar]
- 9.Moretta A, Biassoni R, Bottino C, Pende D, Vitale M, Poggi A, Mingari M C, Moretta L. Immunol Rev. 1997;155:105–117. doi: 10.1111/j.1600-065x.1997.tb00943.x. [DOI] [PubMed] [Google Scholar]
- 10.Pazmany L, Mandelboim O, Vales-Gomez M, Davis D M, Reyburn H T, Strominger J L. Science. 1996;274:792–795. doi: 10.1126/science.274.5288.792. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Villar J J, Melero I, Navarro F, Carretero M, Bellon T, Llano M, Colonna M, Geraghty D E, Lopez-Botet M. J Immunol. 1997;158:5736–5743. [PubMed] [Google Scholar]
- 12.Munz C, Holmes N, King A, Loke Y W, Colonna M, Schild H, Rammensee H-G. J Exp Med. 1997;185:385–391. doi: 10.1084/jem.185.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pende D, Sivori S, Accame L, Pareti L, Falco M, Geraghty D, Le Bouteiller P, Moretta L, Moretta A. Eur J Immunol. 1997;27:1875–1880. doi: 10.1002/eji.1830270809. [DOI] [PubMed] [Google Scholar]
- 14.Soderstrom K, Corliss B, Lanier L, Phillips J. J Immunol. 1997;159:1072–1075. [PubMed] [Google Scholar]
- 15.Rouas-Freiss N, Marchal R E, Kirszenbaum M, Dausset J, Carosella E D. Proc Natl Acad Sci USA. 1997;94:5249–5254. doi: 10.1073/pnas.94.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishitani A, Geraghty D E. Proc Natl Acad Sci USA. 1992;89:3947–3951. doi: 10.1073/pnas.89.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirszenbaum M, Moreau P, Gluckman E, Dausset J, Carosella E. Proc Natl Acad Sci USA. 1994;91:4209–4213. doi: 10.1073/pnas.91.10.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carosella E D, Dausset J, Kirszenbaum M. Immunol Today. 1996;17:407–409. doi: 10.1016/0167-5699(96)30055-8. [DOI] [PubMed] [Google Scholar]
- 19.Kovats S, Main E K, Librach C, Stubblebine M, Fisher S J, DeMars R. Science. 1990;248:220–223. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 20.King A, Loke Y W, Chaouat G. Immunol Today. 1997;18:64–66. doi: 10.1016/s0167-5699(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 21.Rouas-Freiss N, Marchal-Bras Goncalves R, Menier C, Dausset J, Carosella E D. Proc Natl Acad Sci USA. 1997;94:11520–11525. doi: 10.1073/pnas.94.21.11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King A, Hiby S E, Verma S, Burrows T, Gardner L, Loke Y W. Am J Reprod Immunol. 1997;37:459–462. doi: 10.1111/j.1600-0897.1997.tb00260.x. [DOI] [PubMed] [Google Scholar]
- 23.Ohlsson R. Cell Differ Dev. 1989;28:1–15. doi: 10.1016/0922-3371(89)90018-x. [DOI] [PubMed] [Google Scholar]
- 24.Strickland S, Richards W G. Cell. 1992;71:355–357. doi: 10.1016/0092-8674(92)90503-5. [DOI] [PubMed] [Google Scholar]
- 25.De Plaen E, Naerhuyzen B, De Smet C, Szikora J P, Boon T. Genomics. 1997;40:305–313. doi: 10.1006/geno.1996.4566. [DOI] [PubMed] [Google Scholar]
- 26.Shih I M, Kurman R J. Lab Invest. 1996;75:377–388. [PubMed] [Google Scholar]
- 27.Moreau P, Carosella E, Gluckman E, Gourand L, Prost S, Dausset J, Kirszenbaum M. C R Acad Sci. 1995;318:837–842. [PubMed] [Google Scholar]
- 28.King A, Boocock C, Sharkey A M, Gardner L, Beretta A, Siccardi A G, Loke Y W. J Immunol. 1996;156:2068–2076. [PubMed] [Google Scholar]
- 29.Bull M, Van Hoef A, Gorski J. Mol Cell Biol. 1990;10:3792–3796. doi: 10.1128/mcb.10.7.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houlihan J M, Biro P A, Fergar-Payne A, Simpson K L, Holmes C H. J Immunol. 1992;149:668–675. [PubMed] [Google Scholar]
- 31.Lee N, Malacko A R, Ishitani A, Chen M C, Bajorath J, Marquardt H, Geraghty D E. Immunity. 1995;3:591–600. doi: 10.1016/1074-7613(95)90130-2. [DOI] [PubMed] [Google Scholar]
- 32.Schust D J, Hill A B, Ploegh H L. J Immunol. 1996;157:3375–3380. [PubMed] [Google Scholar]
- 33.Stam N J, Vroom T M, Peters P J, Pastors E B, Ploegh H L. Int Immunol. 1990;2:113–125. doi: 10.1093/intimm/2.2.113. [DOI] [PubMed] [Google Scholar]
- 34.Grandea A G, Androlewicz M J, Athwal R S, Geraghty D E, Spies T. Science. 1995;270:105–108. doi: 10.1126/science.270.5233.105. [DOI] [PubMed] [Google Scholar]
- 35.De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. Proc Natl Acad Sci USA. 1996;93:7149–7153. doi: 10.1073/pnas.93.14.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neifeld J P. Semin Surg Oncol. 1996;12:402–406. doi: 10.1002/(SICI)1098-2388(199611/12)12:6<402::AID-SSU5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 37.Jones J A, Albright K D, Christen R D, Howell S B, McClay E F. Can Res. 1997;57:2657–2660. [PubMed] [Google Scholar]
- 38.Driscoll M, Grin-Jorgensen C M, Grant-Kels J M. J Am Acad Dermatol. 1993;29:619–630. doi: 10.1016/0190-9622(93)70229-m. [DOI] [PubMed] [Google Scholar]
- 39.Kirszenbaum M, Djoulah S, Hors J, LeGall I, de Oliveira E B, Prost S, Dausset J, Carosella E D. Hum Immunol. 1997;53:140–147. doi: 10.1016/S0198-8859(97)00038-4. [DOI] [PubMed] [Google Scholar]