Abstract
Interaction of the αβ T cell receptor (TCR) with major histocompatibility (MHC) molecules occupied with any of a large collection of peptides derived from self proteins is a critical step in driving T cell “positive” selection in the thymus. Interaction with this same pool of self-peptide/MHC ligands deletes T cells with potential self-reactivity. To examine how T cells survive both of these processes to form a self-tolerant mature repertoire, mice were constructed whose entire class II MHC IEk specific repertoire was positively selected on a single peptide covalently attached to the IEk molecule. In these mice T cells were identified that could respond to a variant of the positively selecting peptide bound to IEk. The affinities of the TCRs from these T cells for the positively selecting ligand were extremely low and at least 10-fold less than those for the activating ligand. These results support the theory that positive selection is driven by TCR affinities lower than those involved in T cell deletion or activation and that, if present at high concentration, even very low affinity ligands can positively select.
Thymocytes are positively selected to become mature T cells by interaction of their αβ T cell receptors (TCRs) with combinations of peptides derived from self proteins and major histocompatibility complex (MHC) proteins expressed in the thymus (1–5). Paradoxically, interactions with this same set of ligands is required to remove self-reactive T cells from the selected repertoire (negative selection) (6, 7). Some difference between these two processes allows a cadre of positively selected T cells to escape negative selection to form the mature T cell peripheral repertoire, enriched in T cells whose TCRs are specific for the same MHC alleles occupied by peptides derived from foreign proteins.
The favored hypothesis to explain these processes proposes that positive selection is driven by weaker TCR/ligand interactions than is negative selection or mature T cell activation (3–5, 8). Therefore, a window is created allowing survival of T cells whose reactivity with self peptide/MHC is strong enough to drive positive selection, but not to trigger negative selection or activation. Replacement of the self peptide with the appropriate foreign peptide can increase the strength of interaction to a level required for T cell activation.
There have been two ideas about which quality of the TCR/MHC interaction determines the distinction between positive and negative selection, each based on a different set of experimental data. The first idea is that the kinetics of TCR interaction with the selecting ligand determines the outcome and that the half-life of the complex is particularly important (3, 9). Complexes with a relatively short half-life trigger positive selection through incomplete signaling, while those with longer half-lives trigger negative selection via a more complete signal. Because TCR signaling outcome is determined by TCR dissociation rate, the process is predicted to be relatively insensitive to ligand concentration. The second idea is that total receptor occupancy is the key to signaling outcome, i.e., the same qualitative signal can be achieved from a low affinity or high affinity TCR engagement with ligand by an adjustment in ligand concentration (4, 5). Low total occupancy results in positive selection, while high occupancy results in negative selection.
Even though the techniques for measuring TCR affinities and binding kinetics accurately have improved markedly (10–12), it has been difficult to test these hypotheses because the positively selecting combinations of MHC and peptide are known for only a few T cells. To rectify this problem we created a collection of transgenic mice in which the peptide/MHC combination involved in positive selection was known because the grooves of a particular MHC molecule were totally occupied with a single, covalently bound peptide (13, 14). We have studied several T cells that developed in such a mouse and compared the interaction of their TCRs with either a positively selecting or activating class II MHC/peptide ligand.
MATERIALS AND METHODS
Peptides.
Peptide 88–103 from moth cytochrome c (MCC), peptide 64–76 from the d allele of mouse hemoglobin β chain (Hb), and the MCC peptide with position 99 changed from lysine to alanine (MCC99A) were produced and purified in the Biological Resource Center at the National Jewish Medical Research Center.
T Cell Hybridomas and Production of Interleukin (IL)-2.
The T cell hybridomas used in this study were produced by standard techniques (15). KMAC-19, KMAC-92, and KMAC-126, specific for IEk + MCC, were described previously (14). KC99A and KH-2 were produced from B10.BR mice immunized with MCC99A or Hb, respectively. T cell hybridomas were stimulated with peptide presented by the IEk-bearing B cell lymphoma CH12 (16), or by immobilized soluble IEk-peptide in microtiter wells. The IL-2 produced was assayed at 24 hr (15, 17).
Production of Soluble MHC-Peptide and TCR Molecules.
Soluble IEk covalently bound to MCC, MCC99A, or Hb was produced and purified as described (18, 19).
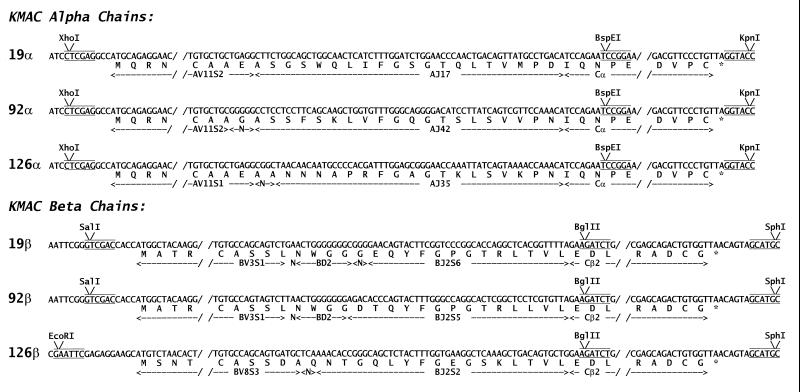
The genes for the Vα and Vβ portions of the TCRs of three T cell hybridomas (KMAC-19, KMAC-92, and KMAC-126) were synthesized by PCR using, as template, cDNA prepared from RNA isolated from the hybridomas. As shown in Fig. 1, the 5′-oligonucleotides used in the PCRs introduced restriction enzyme sites (XhoI, EcoRI or SalI) just 5′ to the V region start ATGs. The 3′-oligonucleotides introduced restriction enzyme sites described (18–21) at the 5′ end of either Cα (BspEI) or Cβ (BglII). The genes were cloned in a baculovirus transfer vector (18–21) in frame with sequences encoding Cα and Cβ. These C region sequences were truncated just past the codons for the cysteines forming the interchain disulfide, thus removing sequence encoding the transmembrane and cytoplasmic tail regions. Using this two promoter vector both the α chain and β chain of each TCR was then introduced into a single baculovirus. Using high titered stocks of these viruses the KMAC soluble TCRs were produced by infection of High Five insect cells (Invitrogen) in TMN-FH culture media. The receptors were purified from culture supernatants by immunoaffinity chromatography with an anti-Cβ mAb, H597 (22). The eluted TCRs were further purified by size exclusion chromatography. A control soluble TCR (20, 21) from the T cell hybridoma DO-11.10, specific for an ovalbumin peptide plus IAd, was also prepared.
Figure 1.
Sequences of the cloned KMAC TCRs. The figure shows part of the sequences of the KMAC α and β chains as they were cloned into a baculovirus transfer vector. The restriction enzyme sites introduced by the 3′ and 5′ oligonucleotides used to amplify the Vα and Vβ segments are shown. The Cα and Cβ gene segments were truncated as shown just past the codons for cysteines forming the interchain disulfide. In addition the codons for asparagines at position 5 and 118 of Cβ and 80 of Cα were changed to those for aspartic acid to eliminate three N-linked glycosylation sites (not shown).
Surface Plasmon Resonance Studies.
Interactions between soluble KMAC TCRs and IEk-peptide complexes were evaluated by surface plasmon resonance by using the BIAcore system (Piscataway, NJ). TCRs purified as described above were immobilized in flow cells of a CM-5 BIAcore biosensor chip by capture with a hamster anti-mouse Cα mAb, ADO-304, which had previously been covalently attached to the dextran matrix of the flow cell. Because the original immunoaffinity purifications of the TCRs were performed with an anti-Cβ mAb only complete αβTCRs were captured in the flow cell and the small amount of β chain dimers contaminating the preparations were not retained. Because of the very slow dissociation rate of TCR from this antibody (kd ≈ 10−4/s), binding of subsequently injected soluble IEk-peptide to the immobilized TCR could be followed. The running buffer was phosphate buffered saline (pH 7.2) supplemented with 5 mM NaN3 and 0.005% P20 surfacant. All measurements were performed at 25°C. Binding kinetics were analyzed with the standard BIAcore biaevaluation software. To correct for differences in bulk refractive index, all IEk-peptide preparations were also injected through a flow cell with an irrelevant immobilized TCR (DO-11.10). These control data were subtracted from the experimental data.
RESULTS
Properties of the 99ATg Transgenic Mouse and IEk-MCC Reactive Hybridomas Derived from Its T Cells.
We have previously described a mouse (99ATg) that carried a transgenic IEk molecule attached covalently to its β chain N terminus (14). In this peptide the lysine at position 99 of MCC, a key residue in T cell recognition, was changed to an alanine. These mice also had the normal H-2f MHC encoding a class II IAf molecule and had the invariant chain locus knocked out. The 99ATg mice were characterized extensively to assure that the occupancy of IEk by MCC99A was virtually 100%. For example, antigen presenting cells from these mice are 10,000–100,000 times worse than those from wild-type IEk expressing mice in presenting exogenously added IEk binding peptide antigens to T cells.
To introduce functional IEk bearing antigen presenting cells into the 99ATg mice, they were irradiated and repopulated with fetal liver from mice bearing wild-type IEk transgenes. Thus, in these chimeric mice the IEk restricted repertoire was selected on IEk–MCC99A expressed in the radioresistant thymus, but the peripheral antigen promoting cells were capable of presenting other IEk binding peptides. These mice were immunized with the wild-type MCC peptide to produce a set of MCC-specific T cell hybridomas (14). Extensive control experiments established the importance of MCC99A in the thymus for the development of this MCC reactive repertoire.
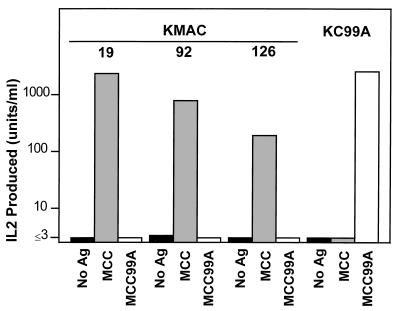
In the present study we have selected three representative T cell hybridomas from this series to examine in detail, KMAC-19, KMAC-72, and KMAC-126. Because the IEk + MCC reactive T cells that gave rise to these hybridomas developed by positive selection on IEk-MCC99A, they should be reactive to IEk plus MCC, but tolerant to IEk plus MCC99A. These points are established in Fig. 2. The three hybridomas produced IL-2 in response to IEk bearing cells in the presence of MCC. They failed to respond to even very high concentrations of MCC99A, although this peptide stimulated very well a T cell hybridoma, KC99A, produced from conventional IEk mice immunized with MCC99A. Therefore, T cells bearing these TCRs were fully tolerant to IEk bound to the positively selecting MCC99A peptide.
Figure 2.
The KMAC T cell hybridomas respond to IEk/MCC, but not IEk/MCC99A. Shown is the IL-2 produced by the three IEk+MCC specific KMAC T cell hybridomas and the T cell hybridoma, KC99A, specific for IEk+MCC99A in response to IEk-bearing CH12 B lymphoma cells in the presence of no antigen (▪), 4 μg/ml of MCC peptide (░⃞), or 100 μg/ml MCC99A peptide (□).
Binding of IEk-MCC and IEk-MCC99A to Soluble αβTCRs.
As described in Materials and Methods, the genes for the Vα and Vβ portions of the TCRs of the three KMAC hybridomas were amplified by PCR and cloned into baculovirus. The virus was used to produce soluble versions of each of the TCRs. Surface plasmon resonance was then used to assess the kinetics of binding of various IEk-peptides to the soluble αβTCRs from the KMAC hybridomas. The soluble TCR was immobilized in the flow cell of a biosensor chip via an anti-Cα mAb and a bolus of various concentrations of soluble IEk with covalently attached MCC, MCC99A or, as a negative control, Hb, were passed through the flow cell and the binding kinetics recorded.
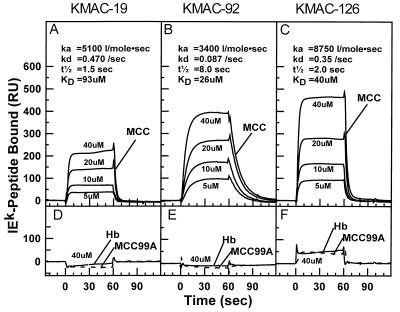
Soluble IEk with covalently bound MCC bound to each of the immobilized KMAC TCRs in a dose dependent manner (Fig. 3 A–C). Each receptor showed characteristic binding kinetics. The average association rates were all quite slow at 3,400 l/mol⋅s for the KMAC-92 TCR, 5,100 l/mol⋅s for KMAC-19 TCR, and 8,800 l/mol⋅s for the KMAC-126 TCR. The complexes with the KMAC-19 and KMAC-126 TCRs had extremely rapid dissociation rates of 0.47/s and 0.35/s, respectively. Thus the half lives of these complexes were only 1.5 s and 2.0 s. These are among the shortest dissociation rates yet measured for T cell activating ligands (9–12, 21, 23–27). The dissociation rate for the complex with KMAC-92 was somewhat slower at 0.087/s for a complex half life of 8.0 s. These kinetic data led to calculated dissociation constants of 93, 26, and 40 μM, respectively for the KMAC-19, KMAC-92, and KMAC 126 TCRs. These values are within the range reported for other TCR interactions with activating MHC/peptide ligands (9–12, 21, 23–27).
Figure 3.
Kinetics of the interaction of soluble IEk-peptide with immobilized KMAC TCRs. Various concentrations (5–40 μM) of soluble IEk-MCC (solid line) were injected at 10 μl/min for 60 s through biosensor flow cells in which (A) KMAC-19, (B) KMAC-92, or (C) KMAC-126 TCR had been immobilized and the binding kinetics recorded. As a control for bulk fluid phase refractive index the IEk-MCC preparations were also injected through a fourth flow cell with an immobilized irrelevant TCR from the IAd/ovalbumin specific T cell hybridoma, DO-11.10. The differences between these two curves are shown for each IEk-MCC concentration. Average association and dissociation rates were derived from the data by using standard BIAcore biaevaluation software, and these were used to calculate the half-life (t½) and dissociation constant (Kd) of the TCR/MHC complex. (D–F) A single concentration of 40 μM IEk-MCC99A (dashed line) or IEk-Hb (dotted line) was injected through the same flow cells containing immobilized the KMAC TCRs. The data were corrected for bulk fluid phase refractive index as above.
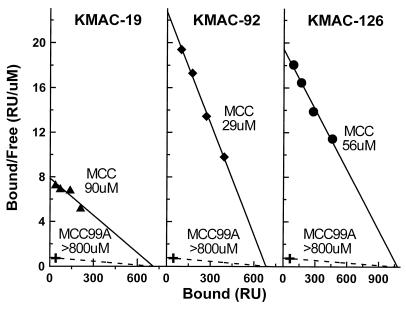
Because of the fast dissociation of IEk-MCC from the KMAC TCRs the binding reactions all reached equilibrium rapidly. Therefore, the plateau binding values were used to construct a Scatchard plot (Fig. 4) as a check of the dissociation constants calculated from the kinetic data. The dissociation constants calculated from these plots were 90, 29, and 56 μM for KMAC-19, KMAC-92, and KMAC-126, respectively, which were in good agreement with those calculated from the kinetic data.
Figure 4.
Scatchard analysis of the interaction of soluble IEk-peptide with immobilized KMAC TCRs. The equilibrium resonance units values in Fig. 3 (A–C) were used to construct Scatchard plots. The Kd of the KMAC-126 TCR interaction with IEk-MCC was estimated from −1/m, where m was the slope of the least squares regression line (solid line) fit to the data. The maximum binding capacity of the flow cells was estimated from the intercept of this line with the x axis. Assuming this potential binding capacity and a minimum significant binding of 30 resonance units at a 40 μM injection (X) the minimal Kd for IEk-MCC99A was estimated at >800 μM (dashed line).
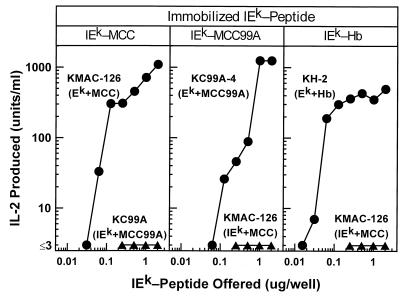
We did not detect any significant binding to the KMAC TCRs of IEk occupied by the positively selecting peptide, MCC99A, when compared with an irrelevant Hb peptide, at any dose tested up to 40 μM (Fig. 3 D–F). If we assumed the same number of potential binding sites in the KMAC TCR flow cells, and a conservative minimal significant binding of 30 resonance units, the dissociation constants of these interactions could be estimated at >800 μM (Fig. 4 A–C). In control experiments we demonstrated that all the soluble IEk-peptide proteins were functional, because they could all, when immobilized, specifically activate T cell hybridomas bearing the TCRs of the appropriate specificity (Fig. 5).
Figure 5.
Stimulation of T cell hybridomas by immobilized IEk bearing appropriate covalently bound peptides. Various amounts of IEk-MCC, IEk-MCC99A, or IEk-Hb were immobilized by absorption to the surface of tissue culture microtiter wells. A constant number of T cell hybridoma cells of the appropriate specificity (•) was added and the stimulation of IL-2 secretion assessed 24 hr later. As a negative control, similar wells containing a T cell hybridoma of inappropriate specificity (▴) were assayed. Each panel is labeled with the immobilized IEk molecule at the top. The names of the T cell hybridomas are shown with their known MHC peptide specificity given in parentheses.
DISCUSSION
The idea that the extent of TCR occupancy determines signaling outcome derives from two sets of experiments. In the first, cultures of fetal thymuses taken from TAP knockout mice were studied (4, 5). Because of the defect in peptide transport, these thymuses lacked MHC class I expression and failed to support mature CD8 T cell development. Positive selection of T cells bearing a transgenic TCR taken from a mature T cell clone of known peptide/MHC class I specificity was restored by the rescue of MHC class I expression by introduction into the cultures of very low doses of the activating peptide. Higher doses of the peptide resulted in negative selection of the developing T cells. An in vivo study with CD4 T cells (28) was consistent with these results. In this case the IEk molecule in invariant chain knockout mice was shown to lack the appropriate self-peptides for the positive selection of mature CD4 T cells capable of responding to IEk/MCC. Introduction of apparently very low levels of the MCC peptide itself into the thymus via a viral vector restored positive selection of the IEk/MCC reactive cells.
In these examples the level of the restored positively selecting MHC/peptide complex in the thymus was below the level of detection. The conclusion from these experiments was that ligands with high affinities can positively select T cells provided that their concentrations are low enough to keep TCR occupancy low. Therefore, a particular type of receptor binding kinetics was not required for positive selection. Along the same lines, other experiments have shown that the levels of CD8 (29, 30) or CD4 (31) in the thymus can determine whether a T cell clone will be positively or negatively selected in a thymus bearing particular MHC alleles. These experiments again suggest that some integration of total TCR complex signal determines the fate of the developing T cell, rather than a specific type of TCR/ligand kinetics.
Our results are consistent with this view. The level of expression of IEk-MCC99A in the thymuses of our transgenic mice was very high (14) and comparable to those of IEk in normal mice. This placed the concentration of the MCC99A peptide on the cell surface of 99ATg mice at a much higher level than that of even the most abundant peptides in wild-type class II MHC molecules (32–34). Therefore, if low receptor occupancy is required for positive, rather than negative selection, we could predict that only T cells bearing TCRs with very low affinities for IEk-MCC99A should be selected in these mice. This perhaps explains why the KMAC TCRs had affinities for IEk-MCC99A below the limits of detection (Kd > 800 μM) and yet still were positively selected on this ligand.
The alternate view is that TCR signaling outcome is controlled by the kinetics of TCR interaction with its MHC + peptide ligands, with different kinetics leading to qualitative, not just quantitative, differences in signaling (3). This idea has its roots in experiments with mature T cells. For some CD4 or CD8 T cell clones that are specifically activated by a particular peptide/MHC pair, variant peptides have been identified that can only partially activate the T cell even when used at very high concentration (35–37). Furthermore, exposure to these variant peptides often renders the T cell clone refractory to subsequent or simultaneous activation by the original peptide, a phenomenon often referred to as antagonism.
Several studies have shown that the TCRs from these clones have lower affinities for these variant MHC/peptide ligands than for the fully activating MHC/peptide complex. One study with a TCR from a CD4 T cell specific for IEk/MCC suggested that the difference between activation and antagonism is related to the dissociation rate of the TCR from the MHC/peptide complex (9). Antagonism was associated with half-lives of TCRs bound to the MHC + peptide ligands shorter than ≈10 s. It was concluded that 10s may be the minimum time required for receptor multimerization and assembly of all of the secondary molecules required for a complete intracellular signal and that shorter half lives for TCR-ligand interactions led to partial, inhibitory signals to the mature T cell. Our results do not support this view, because for two of the three IEk/MCC specific TCRs we examined the fully activating agonist ligand dissociated from the receptor with half-life of <2 s.
This idea of receptor kinetics controlling signaling outcome was extended in one set of experiments studying positive selection in cultures of fetal thymuses taken from β2-microglobulin knockout mice that lack MHC class I expression (3). In these thymuses positive selection of T cells bearing a transgenic TCR taken from a mature T cell clone specific for a peptide from ovalbumin bound to the MHC class I molecule, Kb, was studied after restoration of Kb expression by introduction into the cultures of β2-microglobulin and peptides related to the activating peptide. There was a direct relation between the activity of the peptide on mature T cells bearing the TCR and the fate of the T cell in the thymic cultures. Those with the ability to activate fully the mature T cell led to negative selection of the T cell in the thymus while those that acted as antagonists with the mature T cell led to positive selection of the T cell in the thymus. Subsequent experiments showed that affinity of the TCR of this T cell for these peptide/Kb complexes showed a similar hierarchy, i.e., highest affinities were seen with activating/deleting peptides and while lower, but clearly measurable affinities, were seen with antagonizing/positively selecting peptides (12).
These results suggested the appealing idea that positive selection may require a precise intermediate TCR affinity so that only a partial signal is sent to the developing T cell. Higher affinities may then lead to negative selection as a more complete signaling complex assembles, while lower affinities may not produce the minimum signal required for positive selection. Our results do not formally disprove this hypothesis. However, taken together with the data from other laboratories described above they support the view that positive selection can be driven by TCR ligands of widely varying affinities. There appears to be an inverse relationship between this affinity and the ligand concentration that allows positive selection without negative selection.
Acknowledgments
We thank Drs. Christopher Benoist, Diane Mathis, Elizabeth Bikoff, and Ronald Germain for their generous gifts of plasmids and/or mice. We also thank Dean Becker for production of the transgenic mice. This work was supported in part by National Institutes of Health Grants AI-18785, AI-17134, and AI-22295.
ABBREVIATIONS
- MHC
major histocompatibility complex
- TCR
αβ T cell receptor
- MCC
moth cytochrome c
- IL
interleukin
- Hb
hemoglobin β chain
References
- 1.Bevan M J. Nature (London) 1977;269:417–418. [Google Scholar]
- 2.Zinkernagel R M, Callahan G N, Althage A, Cooper S, Klein P A, Klein J. J Exp Med. 1978;147:882–896. doi: 10.1084/jem.147.3.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogquist K A, Jameson C S, Heath W R, Howard J L, Bevan M J, Carbone F R. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 4.Ashton-Rickardt P G, Bandiera A, Delaney J R, van Kaer L, Pircher H P, Zinkernagel R M, Tonegawa S. Cell. 1994;76:651–663. doi: 10.1016/0092-8674(94)90505-3. [DOI] [PubMed] [Google Scholar]
- 5.Sebzda E, Wallace V A, Mayer J, Yeung R S M, Mak T W, Ohashi P S. Science. 1994;263:1615–1618. doi: 10.1126/science.8128249. [DOI] [PubMed] [Google Scholar]
- 6.Kappler J W, Roehm N, Marrack P. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- 7.Kisielow P, Bluthmann H, Staerz U D, Steinmetz M, von Boehmer H. Nature (London) 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- 8.Lo D, Ron Y, Sprent J. Immunol Res. 1986;5:221–232. doi: 10.1007/BF02919203. [DOI] [PubMed] [Google Scholar]
- 9.Lyons D S, Lieberman S A, Hampl J, Boniface J J, Chien Y, Berg L J, Davis M M. Immunity. 1996;5:53–61. doi: 10.1016/s1074-7613(00)80309-x. [DOI] [PubMed] [Google Scholar]
- 10.Matsui K, Boniface J J, Steffner P, Reay P A, Davis M M. Proc Natl Acad Sci USA. 1994;91:12862–12866. doi: 10.1073/pnas.91.26.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corr M, Slanetz A E, Boyd L F, Jelonek M T, Khilko S, al-Ramadi B K, Kim Y S, Maher S E, Bothwell A L, Margulies D H. Science. 1994;265:946–949. doi: 10.1126/science.8052850. [DOI] [PubMed] [Google Scholar]
- 12.Alam S M, Travers P J, Wung J L, Nasholds W, Redpath S, Jameson S C, Gascoigne N R. Nature (London) 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- 13.Ignatowicz L, Kappler J, Marrack P. Cell. 1996;84:521–529. doi: 10.1016/s0092-8674(00)81028-4. [DOI] [PubMed] [Google Scholar]
- 14.Liu C-P, Parker D, Kappler J, Marrack P. J Exp Med. 1997;186:1441–1450. doi: 10.1084/jem.186.9.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White J, Blackman M, Bill J, Kappler J, Marrack P, Gold D P, Born W. J Immunol. 1989;143:1822–1825. [PubMed] [Google Scholar]
- 16.Arnold L, LoCascio N, Lutz P, Pennell C, Klapper D, Haughton G. J Immunol. 1983;131:2064–2068. [PubMed] [Google Scholar]
- 17.Mosmann T. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 18.Kozono H, White J, Clements J, Marrack P, Kappler J W. Nature (London) 1994;369:151–154. doi: 10.1038/369151a0. [DOI] [PubMed] [Google Scholar]
- 19.Kozono H, Parker D, White J, Marrack P, Kappler J. Immunity. 1995;3:187–196. doi: 10.1016/1074-7613(95)90088-8. [DOI] [PubMed] [Google Scholar]
- 20.Kappler J, White J, Kozono H, Clements J, Marrack P. Proc Natl Acad Sci USA. 1994;91:8462–8466. doi: 10.1073/pnas.91.18.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seibel J L, Wilson N, Kozono H, Marrack P, Kappler J. J Exp Med. 1997;185:1919–1927. doi: 10.1084/jem.185.11.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubo R T, Born W, Kappler J W, Marrack P, Pigeon M. J Immunol. 1989;142:2736–2742. [PubMed] [Google Scholar]
- 23.Weber S, Traunecker A, Oliveri F, Gerhard W, Karjalainen K. Nature (London) 1992;356:793–796. doi: 10.1038/356793a0. [DOI] [PubMed] [Google Scholar]
- 24.Sykulev Y, Brunmark A, Jackson M, Cohen R J, Peterson P A, Eisen H N. Immunity. 1994;1:15–22. doi: 10.1016/1074-7613(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 25.Seth A, Stern L J, Ottenhoff T H M, Engel I, Owen M J, Lamb J R, Klausner R D, Wiley D C. Nature (London) 1994;369:324–327. doi: 10.1038/369324a0. [DOI] [PubMed] [Google Scholar]
- 26.Garcia K C, Scott C A, Brunmark A, Carbone F, Peterson P A, Wilson I A, Teyton L. Nature (London) 1996;384:577–581. doi: 10.1038/384577a0. [DOI] [PubMed] [Google Scholar]
- 27.Garcia K, Tallquist M, Pease L, Brunmark A, Scott C, Degano M, Stura E, Peterson P, Wilson I, Teyton L. Proc Natl Acad Sci USA. 1997;94:13838–13843. doi: 10.1073/pnas.94.25.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano N, Rooke R, Benoist C, Mathis D. Science. 1997;275:678–683. doi: 10.1126/science.275.5300.678. [DOI] [PubMed] [Google Scholar]
- 29.Robey E A, Ramsdell F, Kioussis D, Sha W, Loh D, Axel R, Fowlkes B J. Cell. 1992;69:1089–1096. doi: 10.1016/0092-8674(92)90631-l. [DOI] [PubMed] [Google Scholar]
- 30.Fung-Leung W P, Wallace V A, Gray D, Sha W C, Pircher H, The H S, Loh D Y, Mak T W. Eur J Immunol. 1993;23:212–216. doi: 10.1002/eji.1830230133. [DOI] [PubMed] [Google Scholar]
- 31.Rahemtulla A, Fung-Leung W P, Schilham M W, Kundig T M, Sambhara S R, Narenran A, Arabian A, Wakeham A, Paige C J, Zinkernagel R M, et al. Nature (London) 1991;353:180–184. doi: 10.1038/353180a0. [DOI] [PubMed] [Google Scholar]
- 32.Rudensky A, Preston-Hurlburt S, Hong S, Barlow A, Janeway C., Jr Nature (London) 1991;353:622–625. doi: 10.1038/353622a0. [DOI] [PubMed] [Google Scholar]
- 33.Hunt D H, Michel H, Dickinson T A, Shabanowitz J, Cox A L, Sakaguchi K, Appella E, Grey H M, Sette A. Science. 1992;256:1817–1821. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 34.Marrack P, Ignatowicz L, Kappler J W, Boymel J, Freed J H. J Exp Med. 1993;178:2173–2183. doi: 10.1084/jem.178.6.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evavold B D, Allen P M. Science. 1991;252:1308–1310. [PubMed] [Google Scholar]
- 36.De Magistris M T, Alexander J, Coggeshall M, Altman A, Gaeta F C A, Grey H M, Sette A. Cell. 1992;68:625–634. doi: 10.1016/0092-8674(92)90139-4. [DOI] [PubMed] [Google Scholar]
- 37.Racioppi L, Rochese F, Matis L A, Germain R N. J Exp Med. 1993;177:1541–1550. doi: 10.1084/jem.177.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]